A New Genus, Luciargentis gen. nov. Revealed by Morphological and Phylogenetic Evidence in the Family Lecithoceridae from Tibet, China
Simple Summary
Abstract
1. Introduction
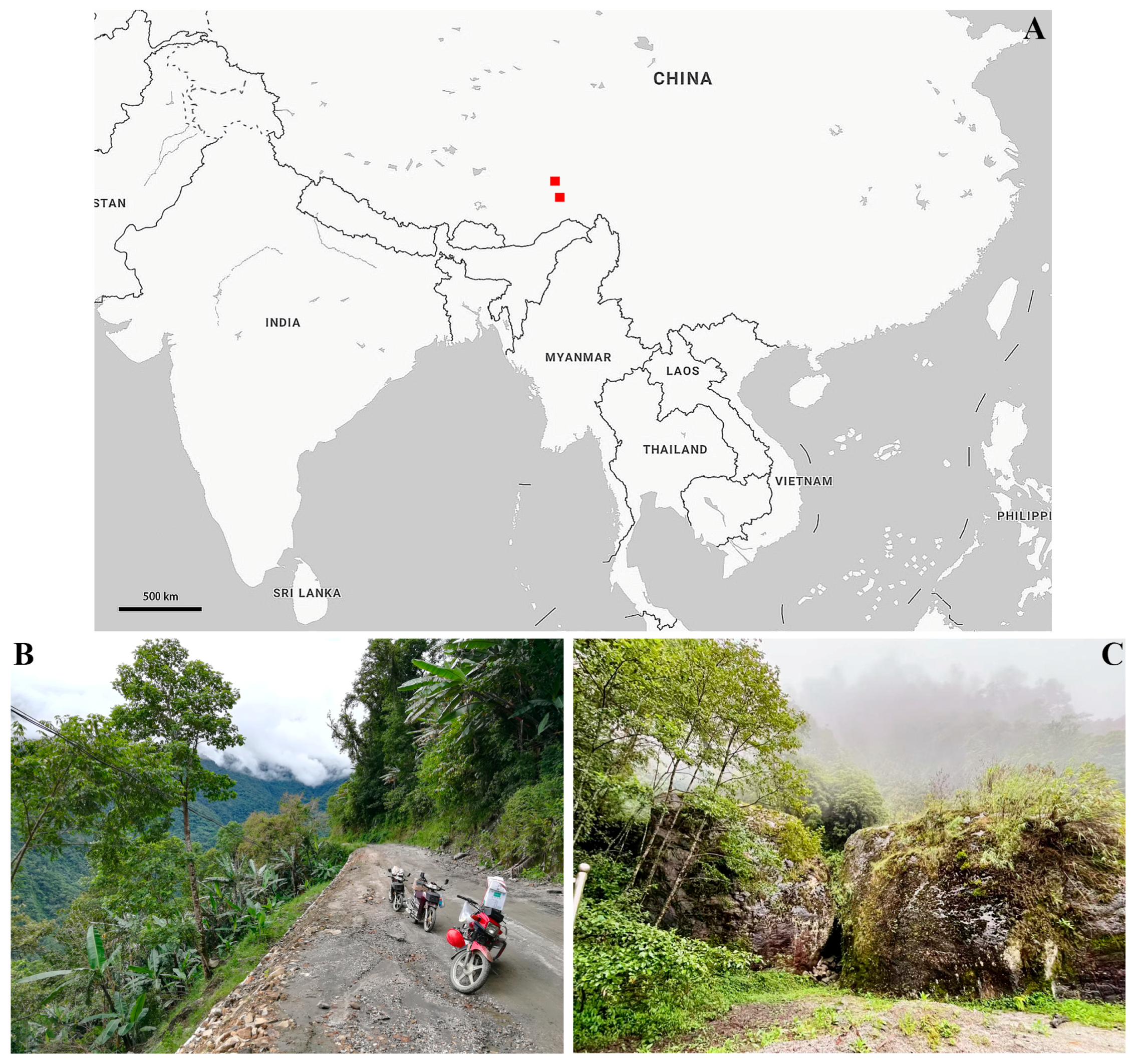
2. Materials and Methods
3. Results
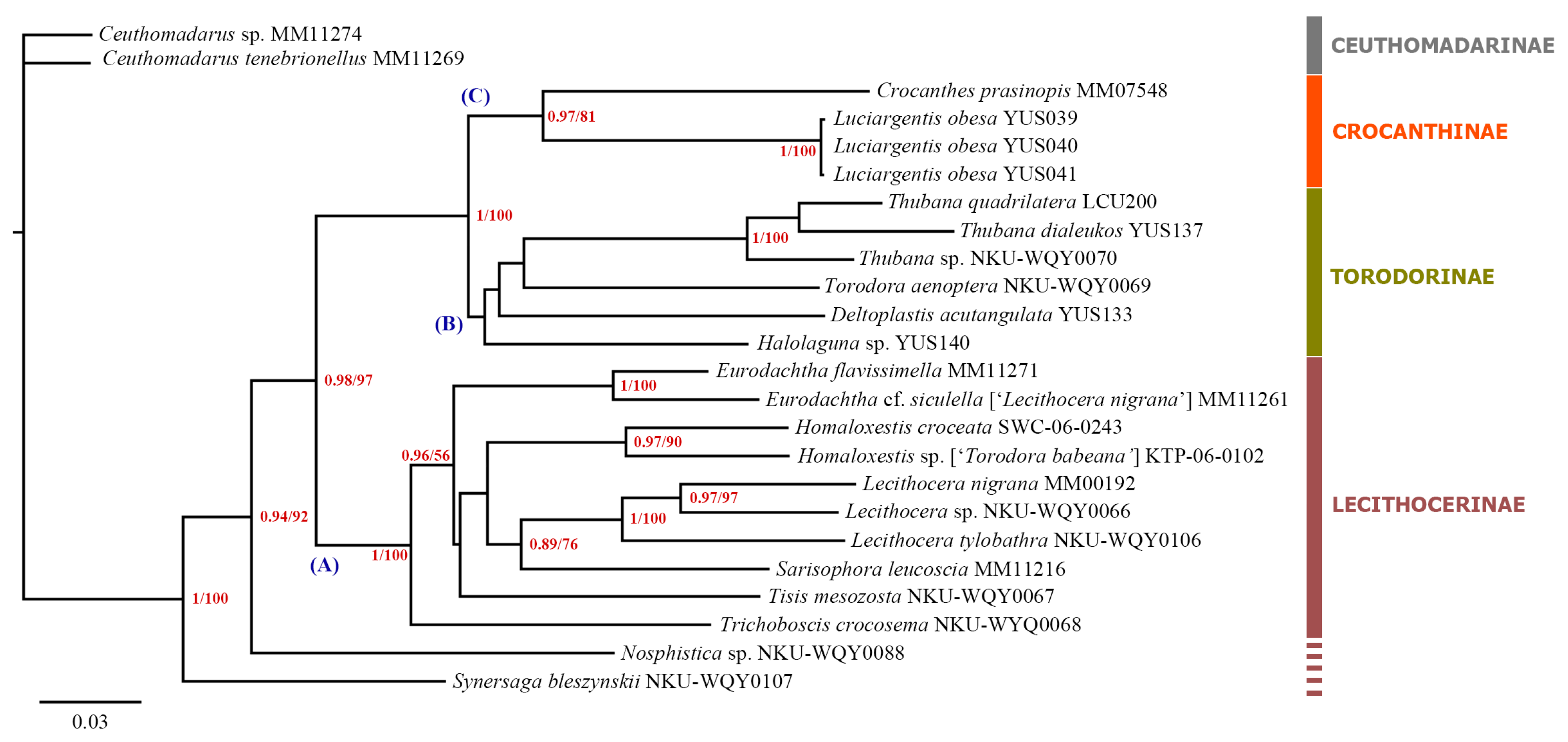
3.1. Molecular Analysis Results
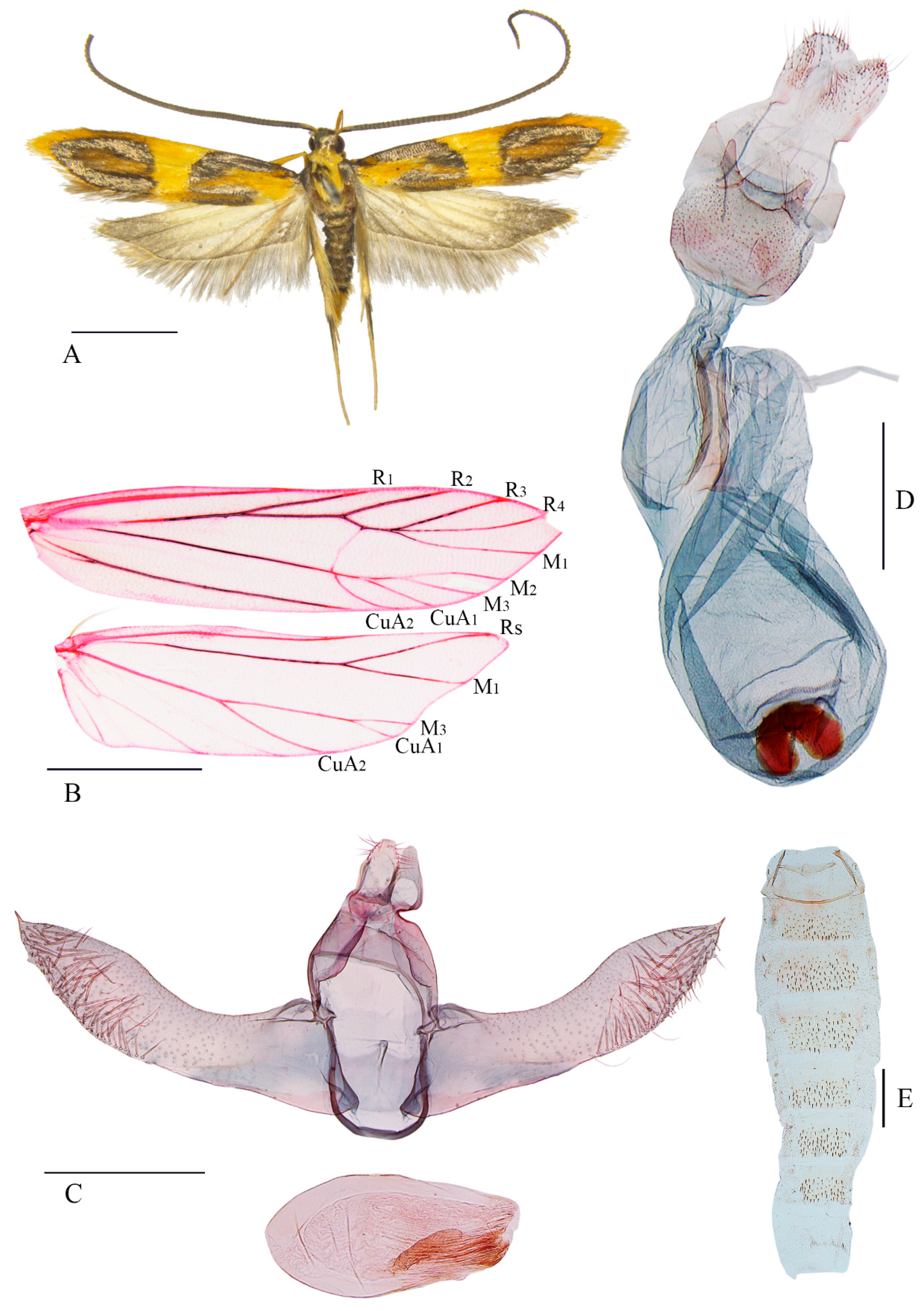
3.2. Morphological Results
3.2.1. Luciargentis Yu and Wang, gen. nov.
- 1.
- Male genitalia with reduced gnathos (i.e., presence of a basal plate and absence of a median process) ……………………………………..…………………………………..... 2
- -
- Male genitalia with gnathos entirely absent …………………………………………… 3
- 2.
- Antenna with long cilia; forewing subrectangular or triangular …...……… Gonaepa
- -
- Antenna smooth; forewing lanceolate ………………………… Luciargentis gen. nov.
- 3.
- Labila palpus with sexual dimorphism ………………………………………………… 4
- -
- Labial palpus with no sexual dimorphism ……………………………………………... 5
- 4.
- Hindwing unicolorous …………………………………………………..……… Lamprista
- -
- Hindwing with similar maculation as forewing ………………………………… Hanara
- 5.
- Forewing with R2 usually free; hindwing usually unicolor ……………….... Crocanthes
- -
- Forewing with R2 usually stalked with R3+4; hindwing with similar maculation as forewing ……………………………………………………………………………………. 6
- 6.
- Male labial palpus with second palpomere normal, third palpomere long or diversified ……………………………………………………………………………... Aprosoesta
- -
- Male labial palpus with second palpomere extremely elongate, third palpomere very short or absent …………………………………………………………………… Pacificulla
- The dichotomous key was partially adapted from Park [1].
3.2.2. Luciargentis obesa Yu and Wang, sp. nov.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, K.T.; Cho, S.; Koo, J.M. The Subfamily Torodorinae of the World (Lepidoptera: Lecithoceridae); National Institute of Biological Resources: Incheon, Republic of Korea, 2022; 584p. [Google Scholar]
- Wang, Q.Y.; Li, H.H. Phylogeny of the superfamily Gelechioidea (Lepidoptera: Obtectomera), with an exploratory application on geometric morphometrics. Zool. Scr. 2020, 49, 307–328. [Google Scholar] [CrossRef]
- Gozmány, L. Lecithoceridae. In Microlepidoptera Palaearctica; Amsel, H.G., Reisser, H., Gregor, F., Eds.; Georg Fromme & Co.: Vienna, Austria, 1978; Volume 5, pp. 1–306. [Google Scholar]
- Common, I.F.B. Moths of Australia; Melbourne University Press: Melbourne, VIC, Australia, 1990; 544p. [Google Scholar]
- Komai, F.; Yoshiyasu, Y.; Nasu, Y.; Saito, T. A Guide to the Lepidoptera of Japan; Tokai University Press: Tokyo, Japan, 2011; 1308p. [Google Scholar]
- Park, K.T.; Mey, W. A review of the genus Lecithocera Herrich-Schäffer, 1853 in the Philippines, with descriptions of seven new species (Lepidoptera: Lecithoceridae). SHILAP Revta. lepid. 2016, 33, 339–352. [Google Scholar] [CrossRef]
- Park, K.T. A new subfamily Crocanthinae based on the genus Crocanthes Meyrick and its related genera, with a world catalog of the subfamily (Lepidoptera, Lecithoceridae). J. Asia-Pac. Biodivers. 2015, 8, 251–286. [Google Scholar] [CrossRef]
- Mi, X.C.; Feng, G.; Hu, Y.; Zhang, J.; Chen, L.; Corlett, R.T.; Hughes, A.C.; Pimm, S.; Schmid, B.; Shi, S.; et al. The global significance of biodiversity science in China: An overview. Natl. Sci. Rev. 2021, 8, nwab032. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H. The Gelechiidae of China (I) (Lepidoptera: Gelechioidea); Nankai University Press: Tianjin, China, 2002; 538p. [Google Scholar]
- Folmer, O.; Black, M.B.; Hoch, W.; Lutz, R.A.; Vrijehock, R.C. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Cho, S.W.; Mitchell, A.; Regier, J.C.; Mitter, C.; Poole, R.W.; Friedlander, T.P.; Zhao, S.W. A highly conserved nuclear gene for low-level phylogenetics: Elongation factor-1α recovers morphology-based tree for heliothine moths. Mol. Biol. Evol. 1995, 12, 650–656. [Google Scholar] [PubMed]
- Brower, A.V.Z.; DeSalle, R. Patterns of mitochondrial versus nuclear DNA sequence divergence among nymphalid butterflies: The utility of wingless as a source of characters for phylogenetic inference. Insect Mol. Biol. 1998, 7, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Wahlberg, N.; Wheat, C.H. Genomic outposts serve the phylogenetic pioneers: Designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 2008, 57, 231–242. [Google Scholar] [CrossRef]
- Kaila, L.; Mutanen, M.; Nyman, T. Phylogeny of the mega-diverse Gelechioidea (Lepidoptera): Adaptations and determinants of success. Mol. Phylogenet. Evol. 2011, 61, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Regier, J.C.; Mitter, C.; Solis, M.A.; Hayden, J.E.; Landry, B.; Nuss, M.; Simonsen, T.J.; Yen, S.H.; Zwick, A.; Cummings, M.P. A molecular phylogeny for the Pyraloid moths (Lepidoptera: Pyraloidea) and its implications for higher-level classification. Syst. Entomol. 2012, 37, 635–656. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Walker, F. List of specimens of Lepidopterous insects in the collection of the British Museum. Tineites 1866, 35, 1535–2040. [Google Scholar]
- Meyrick, E. Lepidoptera Heterocera. fam. Gelechiadae. In Genera Insectorum; Wytsman, P., Ed.; Louis Desmet-Verteneuil: Bruxelles, Belgium, 1925; Volume 184, pp. 1–290. [Google Scholar]
- Park, K.T. The Subfamily Crocanthinae of the World (Lepidoptera, Lecithoceridae); LAP LAMBERT Academic Publishing: Saarbrücken, Germany, 2017; 135p. [Google Scholar]
- Wu, C.S. Fauna Sinica. Insecta. Lepidoptera: Lecithoceridae; Science Press: Beijing, China, 1997; 306p. [Google Scholar]
- Yu, S.; Zhu, Y.M.; Wang, S.X. Eighteen new species and fifteen new records of the genus Torodora Meyrick (Lepidoptera: Lecithoceridae) from China. Zootaxa 2022, 5133, 1–39. [Google Scholar] [CrossRef]
- Sterling, M.J.; Lees, D.C.; Grundy, D. Xenotorodor stygioxanthus gen. nov., sp. nov. (Lepidoptera, Lecithoceridae, Torodorinae), described from an established population in Spain with discussion of taxonomic placement. Nota Lepi. 2023, 46, 103–123. [Google Scholar] [CrossRef]
| Gene Region | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | ||
|---|---|---|---|---|
| EF-1α/elongation factor-1a | EF-1α-F | CCYGCCAAYATCACCACTGAAG | EF-1α-R | AGAGGHGGGAACTCYTGGAAGGA |
| GAPDH/Glyceraldehyde-3-phosphate dehydrogenase | GAPDH-F | TCACTTGGAVGGTGGHGCCAAGAA | GAPDH-R | AGAGAGATACCAGCDGCAGCATC |
| CAD/carbamoyl phosphate synthetase domain protein | CAD-F | AGTTTRGACTACTGTGTAGTTAAAATA | CAD-R | TGATAAAATAACGCCATCAGGA |
| MDH/cytosolic malate dehydrogenase | MDH-F | TGTTGTCATGGAGCTTGCAGATT | MDH-R | CCCATATAACAACATTCTTWACATCC |
| RpS5/ribosomal protein S5 | RpS5-F | GCAGCATGGCCGTCGATAACAT | RpS5-R | TTGATGAACCCTTGGCAGCATTAAT |
| wingless | wingless-F | TGCACAGTGAAAACTTGCTGGAT | wingless-R | GTTACACCTTTCCACAACGAACATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Li, H.; Wang, S. A New Genus, Luciargentis gen. nov. Revealed by Morphological and Phylogenetic Evidence in the Family Lecithoceridae from Tibet, China. Insects 2025, 16, 242. https://doi.org/10.3390/insects16030242
Yu S, Li H, Wang S. A New Genus, Luciargentis gen. nov. Revealed by Morphological and Phylogenetic Evidence in the Family Lecithoceridae from Tibet, China. Insects. 2025; 16(3):242. https://doi.org/10.3390/insects16030242
Chicago/Turabian StyleYu, Shuai, Haotian Li, and Shuxia Wang. 2025. "A New Genus, Luciargentis gen. nov. Revealed by Morphological and Phylogenetic Evidence in the Family Lecithoceridae from Tibet, China" Insects 16, no. 3: 242. https://doi.org/10.3390/insects16030242
APA StyleYu, S., Li, H., & Wang, S. (2025). A New Genus, Luciargentis gen. nov. Revealed by Morphological and Phylogenetic Evidence in the Family Lecithoceridae from Tibet, China. Insects, 16(3), 242. https://doi.org/10.3390/insects16030242






