Genome-Wide Identification and Expression Profiling of Odorant-Binding Protein Genes in the Bean Flower Thrips Megalurothrips usitatus (Bagnall) (Thysanoptera: Thripidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of OBP Genes in M. usitatus
2.2. Chromosomal Location, Phylogenetic Analysis, and Structural Characteristics Analyses of MusiOBPs
2.3. Insect Rearing and Sample Collection
2.4. RNA Extraction and cDNA Synthesis
2.5. Real-Time Quantitative PCR Analysis of MusiOBPs
3. Results
3.1. Genome-Wide Identification of OBP Genes in M. usitatus
3.2. Chromosomal Location of OBP Genes in M. usitatus
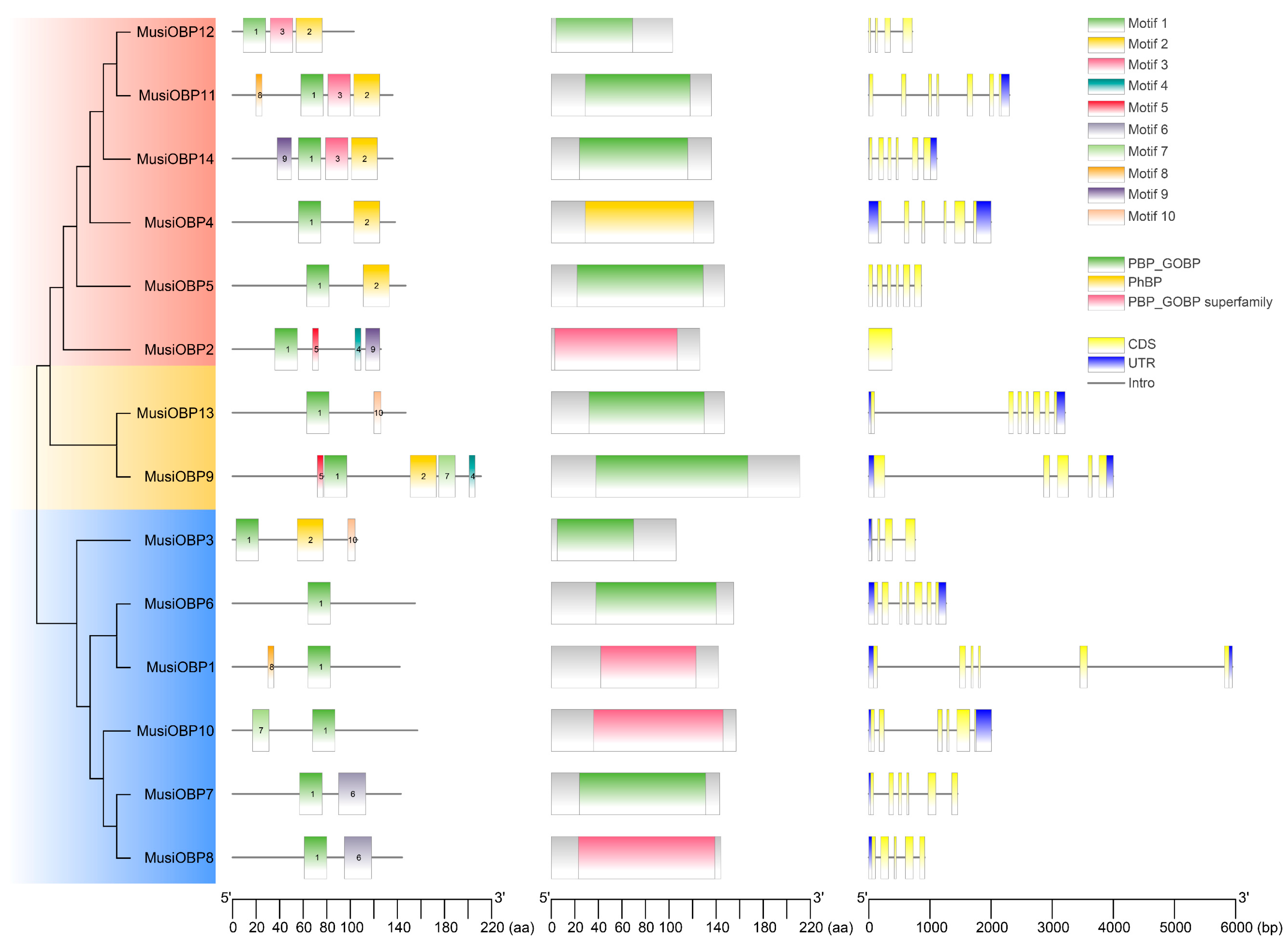
3.3. Gene and Protein Structure of OBPs in M. usitatus
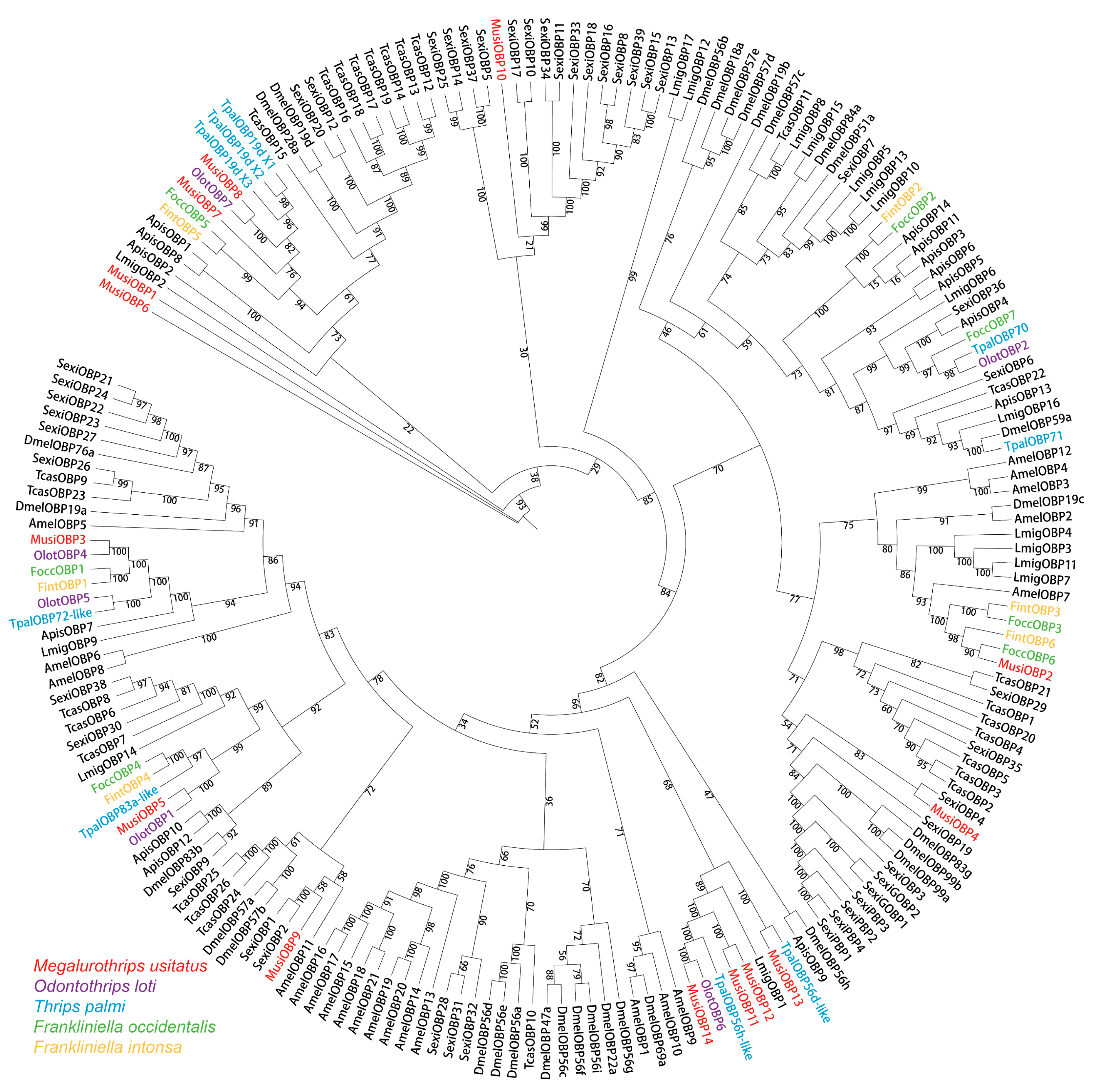
3.4. Phylogenetic Analysis of OBPs
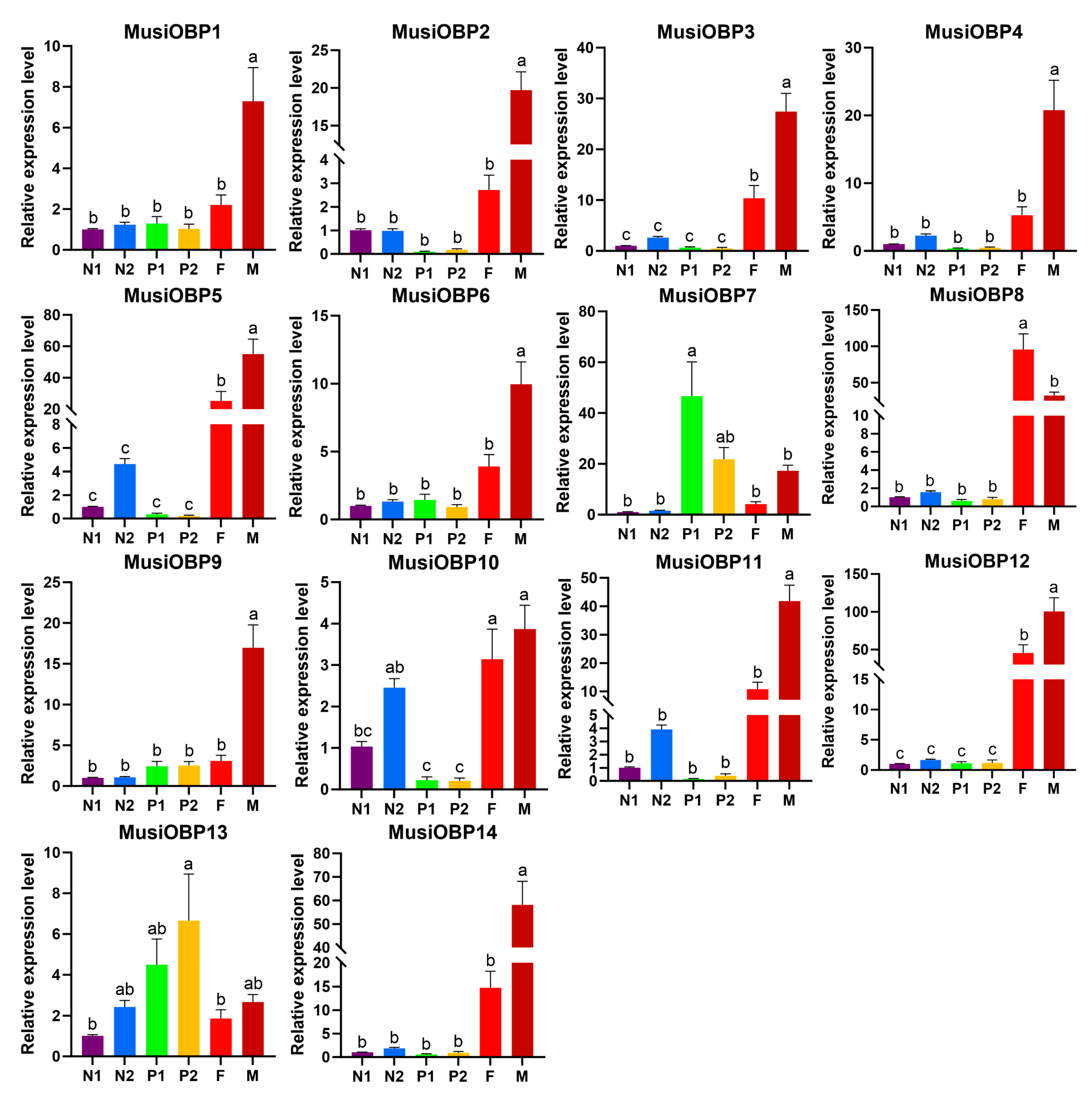
3.5. Expression Patterns of OBPs in Different Developmental Stages of M. usitatus
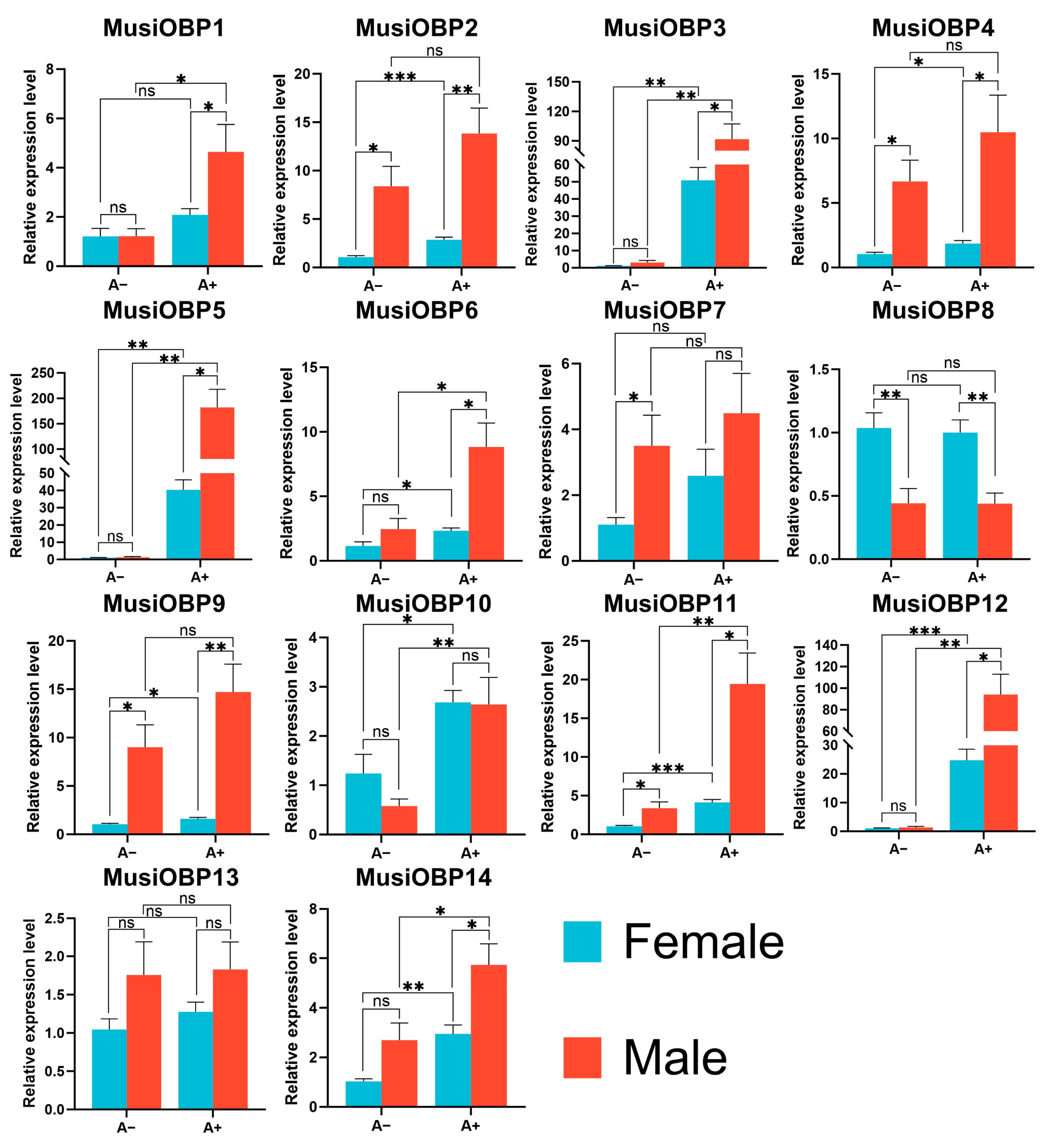
3.6. Tissue- and Sex-Specific Expression Patterns of OBPs in M. usitatus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oparaeke, A.M. The sensitivity of flower bud thrips, Megalurothrips sjostedti Trybom (Thysanoptera: Thripidae), on cowpea to three concentrations and spraying schedules of Piper guineense Schum. & Thonn. extracts. Plant Protect. Sci. 2006, 42, 106–111. [Google Scholar]
- Duraimurugan, P.; Tyagi, K. Pest spectra, succession and its yield losses in mungbean and urdbean under changing climatic scenario. Legume Res. 2014, 37, 212–222. [Google Scholar] [CrossRef]
- Tang, L.D.; Yan, K.L.; Fu, B.L.; Wu, J.H.; Liu, K.; Lu, Y.Y. The life table parameters of Megalurothrips usitatus (Thysanoptera: Thripidae) on four leguminous crops. Fla. Entomol. 2015, 98, 620–625. [Google Scholar] [CrossRef]
- Yasmin, S.; Ali, M.; Rahman, M.M.; Akter, M.S.; Latif, M.A. Biological traits of bean flower thrips, Megalurothrips usitatus (Thysanoptera: Thripidae) reared on mung bean. Herit. Sci. 2021, 5, 29–33. [Google Scholar] [CrossRef]
- Pan, X.; Yang, L.; Jin, H.; Lu, R.; Li, F.; Cao, F.; Wu, S. Research advances in occurrence and control of Megalurothrips usitatus in Hainan. J. Trop. Biol. 2021, 12, 508–513. [Google Scholar]
- Peter, C.; Govindarajulu, V. Management of blossom thrips, Megalurothrips usitatuson pigeonpea. Trop. Pest Manag. 1990, 36, 312–313. [Google Scholar] [CrossRef]
- Hossain, M.A. Efficacy of some insecticides against insect pests of mungbean (Vigna radiata L.). Bangladesh J. Agril. Res. 2015, 40, 657–667. [Google Scholar] [CrossRef][Green Version]
- Maradi, R.M.; Rajashekharappa, K.; Pradhan, K. Evaluation of bio-efficacy of newer molecules of insecticides against thrips, Megalurothrips usitatus in yard long bean, Vigna unguiculata subsp. sesquipedalis. Bioscan 2020, 15, 189–192. [Google Scholar]
- Tang, L.D.; Zhao, H.Y.; Fu, B.L.; Han, Y.; Liu, K.; Wu, J.H. Colored sticky traps to selectively survey thrips in cowpea ecosystem. Neotrop. Entomol. 2016, 45, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Paola, S.; Lin, M.-Y.; Chhun Hy, H.; Sareth, K.; Sor, S. Development and validation of an integrated pest management strategy for the control of major insect pests on yard-long bean in Cambodia. Crop Prot. 2019, 116, 82–91. [Google Scholar] [CrossRef]
- Tang, L.D.; Guo, L.H.; Ali, A.; Desneux, N.; Zang, L.S.; Musser, F. Synergism of adjuvants mixed with spinetoram for the management of bean flower thrips, Megalurothrips usitatus (Thysanoptera: Thripidae) in Cowpeas. J. Econ. Entomol. 2022, 115, 2013–2019. [Google Scholar] [CrossRef]
- Bruce, T.J.A.; Wadhams, L.J.; Woodcock, C.M. Insect host location: A volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef]
- Hansson, B.; Stensmyr, M. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, D.; Larsson, M.C.; Anderson, P. Insect host plant selection in complex environments. Curr. Opin. Insect Sci. 2015, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rützler, M.; Zwiebel, L.J. Molecular biology of insect olfaction: Recent progress and conceptual models. J. Comp. Physiol. A 2005, 191, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Guo, M.; Ban, L.; Song, L.M.; Liu, Y.; Pelosi, P.; Wang, G. Niemann-Pick C2 proteins: A new function for an old family. Front. Physiol. 2018, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Q.; Wang, Q.; Dong, K.; Xiao, Y.; Zhang, Y.J. Identification and characterization of odorant binding proteins in the forelegs of Adelphocoris lineolatus (Goeze). Front. Physiol. 2017, 8, 735. [Google Scholar] [CrossRef]
- Tang, B.; Tai, S.; Dai, W.; Zhang, C. Expression and functional analysis of two odorant-binding proteins from Bradysia odoriphaga (Diptera: Sciaridae). J. Agric. Food Chem. 2019, 67, 3565–3574. [Google Scholar] [CrossRef] [PubMed]
- Brito, N.F.; Moreira, M.F.; Melo, A.C.A. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 199–228. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Hekmat-Scafe, D.S.; Scafe, C.R.; Mckinney, A.J.; Tanouye, M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, M.; Ng Fuk Chong, M.; Vaïtinadapoulé, A.; Frumence, E.; Sowdhamini, R.; Offmann, B. Comparative genomics of odorant binding proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 2013, 5, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Billeter, J.C.; Levine, J.D. The role of cVA and the odorant binding protein lush in social and sexual behavior in Drosophila melanogaster. Front. Ecol. Evol. 2015, 3, 75. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, B.; Grossi, G.; Falabella, P.; Liu, Y.; Yan, S.; Lu, J.; Xi, J.; Wang, G. Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 2017, 27, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hu, W.; Hou, Y.M. Host plant recognition by two odorant-binding proteins in Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Pest Manag. Sci. 2023, 79, 4521–4534. [Google Scholar] [CrossRef] [PubMed]
- Rihani, K.; Fraichard, S.; Chauvel, I.; Poirier, N.; Delompré, T.; Neiers, F.; Tanimura, T.; Ferveur, J.F.; Briand, L. A conserved odorant binding protein is required for essential amino acid detection in Drosophila. Commun. Biol. 2019, 2, 245. [Google Scholar] [CrossRef]
- Swarup, S.; Williams, T.I.; Anholt, R.R.H. Functional dissection of odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011, 10, 648–657. [Google Scholar] [CrossRef]
- Sun, J.S.; Larter, N.K.; Chahda, J.S.; Rioux, D.; Gumaste, A.; Carlson, J.R. Humidity response depends on the small soluble protein Obp59a in Drosophila. Elife 2018, 7, e39249. [Google Scholar] [CrossRef] [PubMed]
- Benoit, J.B.; Vigneron, A.; Broderick, N.A.; Wu, Y.; Sun, J.S.; Carlson, J.R.; Aksoy, S.; Weiss, B.L. Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. Elife 2017, 6, e19535. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yin, X.H.; Hu, S.Y.; Miao, H.N.; Wang, Z.; Li, H.; Zhang, Y.J.; Liang, P.; Gu, S.H. Overexpression of two odorant binding proteins confers chlorpyrifos resistance in the green peach aphid Myzus persicae. J. Agric. Food Chem. 2024, 72, 20101–20113. [Google Scholar] [CrossRef]
- Li, X.; Cheng, J.; Chen, L.; Huang, J.; Zhang, Z.; Zhang, J.; Ren, X.; Hafeez, M.; Zhou, S.; Dong, W.; et al. Comparison and functional analysis of odorant-binding proteins and chemosensory proteins in two closely related thrips species, Frankliniella occidentalis and Frankliniella intonsa (Thysanoptera: Thripidae) based on antennal transcriptome analysis. Int. J. Mol. Sci. 2022, 23, 13900. [Google Scholar] [CrossRef]
- Jia, C.; Mohamed, A.; Cattaneo, A.M.; Huang, X.; Keyhani, N.O.; Gu, M.; Zang, L.; Zhang, W. Odorant-binding proteins and chemosensory proteins in Spodoptera frugiperda: From genome-wide identification and developmental stage-related expression analysis to the perception of host plant odors, sex pheromones, and insecticides. Int. J. Mol. Sci. 2023, 24, 5595. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, W.; Wang, X.; Sang, W.; Pan, H.; Ali, S.; Tang, L.; Wu, J. Transcriptome analysis of Megalurothrips usitatus (Bagnall) identifies olfactory genes with ligands binding characteristics of MusiOBP1 and MusiCSP1. Front. Physiol. 2022, 13, 978534. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Q.; Wei, S.; Liu, S.; Tian, L.; Song, F.; Duan, Y.; Cai, W.; Li, H. Chromosome-level genome assembly of bean flower thrips Megalurothrips usitatus (Thysanoptera: Thripidae). Sci. Data 2023, 10, 252. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Mcginnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.; Song, J.; Thanki, N.; Yamashita, R.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Yuan, L.; Jin, H.; Yan, H.; Li, F.; Wu, S. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Megalurothrips usitatus (Thysanoptera: Thripidae). Front. Physiol. 2023, 14, 1161680. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.K.; Kong, X.Y.; Ke, Y.C.; Wang, S.; Li, Q.J.; Fu, Q.W.; Wu, Q.X.; Liu, Y. Research progress on thrips Megalurothrips usitatus (Bagrall). China Veg. 2018, 21–27. [Google Scholar] [CrossRef]
- Zhang, X.; Purba, E.R.; Sun, J.; Zhang, Q.H.; Dong, S.L.; Zhang, Y.N.; He, P.; Mang, D.; Zhang, L. Functional differentiation of two general-odorant binding proteins in Hyphantria cunea (Drury) (Lepidoptera: Erebidae). Pest Manag. Sci. 2023, 79, 3312–3325. [Google Scholar] [CrossRef]
- Chen, X.F.; Xu, L.; Zhang, Y.X.; Wei, D.; Wang, J.J.; Jiang, H.B. Genome-wide identification and expression profiling of odorant-binding proteins in the oriental fruit fly, Bactrocera dorsalis. Comp. Biochem. Phys. D 2019, 31, 100605. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Xia, Q.Y.; Xiang, Z.H. The Odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Dippel, S.; Oberhofer, G.; Kahnt, J.; Gerischer, L.; Opitz, L.; Schachtner, J.; Stanke, M.; Schütz, S.; Wimmer, E.A.; Angeli, S. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genom. 2014, 15, 1141. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, Y.; Du, L.; Ban, L. Antennal transcriptome analysis of olfactory genes and characterization of odorant binding proteins in Odontothrips loti (Thysanoptera: Thripidae). Int. J. Mol. Sci. 2023, 24, 5284. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Fu, B.; Tang, L.; Chen, J.; Liu, K. Occurrence regularity of thrips in cowpea and evaluation of insecticides. Chin. Agric. Sci. Bull. 2017, 33, 138–142. [Google Scholar]
- Huang, C.; Ou, X.; Wang, Y.; Zhou, Y.; Zhang, G.; Liu, W.; Wan, F.; Jiang, H.; Zhang, Y. Genome-wide identification, evolution, and female-biased expression analysis of odorant receptors in Tuta absoluta (Lepidoptera: Gelechiidae). Life 2024, 14, 872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, M.; Xu, Y.; Zhao, Y.; Niu, Y.; Zong, S.; Tao, J. Genome-wide identification of the odorant receptor gene family and revealing key genes involved in sexual communication in Anoplophora glabripennis. Int. J. Mol. Sci. 2023, 24, 1625. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yang, Y.T.; Wu, Q.J.; Wang, S.L.; Xie, W.; Zhang, Y.J. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 2018, 26, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Yin, M.Z.; Yao, W.C.; Ma, S.; Dewer, Y.; Liu, X.Z.; Wang, Y.Y.; Wang, C.W.; Li, B.P.; Zhu, X.Y. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the bean bug Riptortus pedestris. Front. Physiol. 2022, 13, 949607. [Google Scholar] [CrossRef] [PubMed]
- Pracana, R.; Levantis, I.; Martínez-Ruiz, C.; Stolle, E.; Priyam, A.; Wurm, Y. Fire ant social chromosomes: Differences in number, sequence and expression of odorant binding proteins. Evol. Lett. 2017, 1, 199–210. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Crabbe, M.J.C.; Ren, Z. Genome-wide identification and characterization of the chemosensory relative protein genes in Rhus gall aphid Schlechtendalia chinensis. BMC Genom. 2023, 24, 222. [Google Scholar] [CrossRef]
- Frei, A.; Gu, H.; Bueno, J.M.; Cardona, C.; Dorn, S. Antixenosis and antibiosis of common beans to Thrips palmi Karny (Thysanoptera: Thripidae). J. Econ. Entomol. 2003, 96, 1577–1584. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, F.; Sun, A.; Song, J.; Shan, S.; Zhang, Y.; Wang, S. Expressional and functional comparisons of five clustered odorant binding proteins in the brown marmorated stink bug Halyomorpha halys. Int. J. Biol. Macromol. 2022, 206, 759–767. [Google Scholar] [CrossRef]
- Liu, P.; Qin, Z.; Feng, M.; Zhang, L.; Huang, X.; Shi, W. The male-produced aggregation pheromone of the bean flower thrips Megalurothrips usitatus in China: Identification and attraction of conspecifics in the laboratory and field. Pest Manag. Sci. 2020, 76, 2986–2993. [Google Scholar] [CrossRef] [PubMed]
- Xiu, C.; Pan, H.; Zhang, F.; Luo, Z.; Bian, L.; Li, Z.; Fu, N.; Zhou, L.; Magsi, F.H.; Cai, X.; et al. Identification of aggregation pheromones released by the stick tea thrips (Dendrothrips minowai) larvae and their application for controlling thrips in tea plantations. Pest Manag. Sci. 2024, 80, 2528–2538. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; He, X.; Liu, Y.; Liu, M.; Liu, X.; Lu, M. Odorant binding protein 18 increases the pathogen resistance of the imported willow leaf beetle, Plagiodera versicolora. Front. Cell. Infect. Mi. 2024, 14, 1360680. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Yang, H.H.; Gu, N.; Li, J.Q.; Zhu, X.Y.; Zhang, Y.N. Identification of attractants for adult Spodoptera litura based on the interaction between odorant-binding protein 34 and host volatiles. Pestic. Biochem. Physiol. 2024, 203, 106005. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Tang, R.; Lin, L.; Zhao, W.; Wei, S.; Zhang, F.; Uddin, M.K.; Xie, M.; Chen, H. RpedOBP1 plays key roles in aggregation pheromones reception of the Riptortus pedestris. Pestic. Biochem. Physiol. 2024, 204, 106073. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Mang, D.; Purba, E.R.; Ye, J.; Qian, J.; Rao, F.; Wang, H.; Wu, Z.; Zhang, W.; Zheng, Y.; et al. Identification and functional analysis of odorant binding proteins in Apriona germari (Hope). J. Agric. Food Chem. 2024, 72, 17248–17259. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Han, K.R.; Jing, X.F.; Liu, T.X.; Zhang, S.Z. Odorant-binding protein CrufOBP1 in Cotesia ruficrus females plays a pivotal role in the detection of Spodoptera frugiperda larvae. Int. J. Biol. Macromol. 2024, 274, 133491. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, W.; Wang, S.; Miao, W.; Liu, Z.; Wu, F.; Wang, J.; Sheng, S. Binding characteristics and structural dynamics of two general odorant-binding proteins with plant volatiles in the olfactory recognition of Glyphodes pyloalis. Insect Biochem. Mol. Biol. 2024, 173, 104177. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence (5′-3′) | Length (bp) |
|---|---|---|
| qMusiOBP1 | F: AAAAGCCCAAGATGCAGGTG | 162 |
| R: TGCACTTTTCAATCCCAGCC | ||
| qMusiOBP2 | F: GCAAGGAGATGATGAAGGAC | 182 |
| R: CTTGTACTCCTCGGACCAG | ||
| qMusiOBP3 | F: AGAACAATAAAGTGGACGCC | 160 |
| R: TTTACGCAGCGAATGAAGTT | ||
| qMusiOBP4 | F: CGAGCTGACCCACATCAA | 155 |
| R: TCAACTTGTCCTCTGGCG | ||
| qMusiOBP5 | F: GAGCTCACAGAGGACCAGAA | 153 |
| R: CTGGTACACGCACTTCATGT | ||
| qMusiOBP6 | F: ACGATCCCATGATGAAGGCA | 160 |
| R: AATCAGCAGCTATGTCCGGT | ||
| qMusiOBP7 | F: AGGCGAGGGAAATCAAGAGG | 174 |
| R: TCGACCCTTCCTTGGAAAACT | ||
| qMusiOBP8 | F: TAAAGAAGCACGCGATGGAG | 158 |
| R: CTCGTCCAGCACTTTGAAGG | ||
| qMusiOBP9 | F: CGACGATGTGTGCGAGAAAT | 188 |
| R: ATGACCTTGCATGCCATTGG | ||
| qMusiOBP10 | F: AAGAACATGCTGGCCTGT | 211 |
| R: ACATGTTCTTGGCCCGTT | ||
| qMusiOBP11 | F: CACCACCTCCAAAACCAGC | 160 |
| R: CCATCTTCGGACAACATGCC | ||
| qMusiOBP12 | F: GCAACTTCAAGTGCATCATG | 100 |
| R: GCGTGCATTTTCTCAGGT | ||
| qMusiOBP13 | F: CCGACAATTTGGAGGCCTAC | 153 |
| R: ACTTTGTGGCAATGTCTCGC | ||
| qMusiOBP14 | F: TGCAAATGGATATGGACGCC | 162 |
| R: CGAGTAGTAGGTGACCGTCC | ||
| qMusiACT | F: ACGACGTACAACTCCATCAT | 125 |
| R: GTAATCTCCTTCTGCATCCTGT | ||
| qMusiRPL | F: ACATCGAGCTGGGTACTG | 122 |
| R: CACCACCATTTACTGAGCAT |
| Gene Name | Gene ID | ORF (aa) | Complete ORF | Signal Peptide | Homology Search with Known Proteins | ||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Acc. Number | E-Value | Identity | Coverage | |||||
| MusiOBP1 | ONE63_000073 | 142 | Yes | 1–20 | Nezara viridula | QCZ25102.1 | 0.01 | 48.98% | 34% |
| MusiOBP2 | ONE63_005245 | 126 | Yes | No | Frankliniella intonsa | WBW64304.1 | 5 × 10−75 | 88.71% | 98% |
| MusiOBP3 | ONE63_006116 | 106 | Yes | No | Odontothrips loti | WBU77197.1 | 6 × 10−43 | 87.65% | 76% |
| MusiOBP4 | ONE63_006437 | 138 | Yes | 1–20 | Frankliniella fusca | KAK3931445.1 | 5 × 10−32 | 50.00% | 92% |
| MusiOBP5 | ONE63_006734 | 147 | Yes | 1–19 | Odontothrips loti | WBU77195.1 | 1 × 10−77 | 88.64% | 89% |
| MusiOBP6 | ONE63_006862 | 155 | Yes | 1–20 | Macrosteles quadrilineatus | XP_054263813.1 | 0.22 | 25.64% | 71% |
| MusiOBP7 | ONE63_006863 | 143 | Yes | 1–17 | Frankliniella occidentalis | XP_026276672.1 | 5 × 10−74 | 79.56% | 95% |
| MusiOBP8 | ONE63_006864 | 144 | Yes | 1–19 | Odontothrips loti | WBU77200.1 | 1 × 10−54 | 80.33% | 84% |
| MusiOBP9 | ONE63_006989 | 211 | Yes | 1–29 | Frankliniella fusca | KAK3918161.1 | 2 × 10−90 | 67.51% | 91% |
| MusiOBP10 | ONE63_007105 | 157 | Yes | 1–17 | Zophobas morio | XP_063907370.1 | 0.86 | 29.00% | 63% |
| MusiOBP11 | ONE63_008094 | 136 | Yes | 1-22 | Frankliniella fusca | KAK3914559.1 | 6 × 10−39 | 48.12% | 97% |
| MusiOBP12 | ONE63_009400 | 103 | Yes | No | Frankliniella fusca | KAK3914559.1 | 5 × 10−33 | 66.27% | 80% |
| MusiOBP13 | ONE63_001264 | 147 | Yes | 1–22 | Thrips palmi | XP_034232292.1 | 1 × 10−31 | 50.85% | 78% |
| MusiOBP14 | ONE63_002028 | 136 | Yes | 1–18 | Odontothrips loti | WBU77199.1 | 2 × 10−81 | 86.03% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, G.; Yang, L.; Li, B.; Wang, Q.; Huang, L.; Tian, X.; Zhang, G. Genome-Wide Identification and Expression Profiling of Odorant-Binding Protein Genes in the Bean Flower Thrips Megalurothrips usitatus (Bagnall) (Thysanoptera: Thripidae). Insects 2025, 16, 212. https://doi.org/10.3390/insects16020212
Xia G, Yang L, Li B, Wang Q, Huang L, Tian X, Zhang G. Genome-Wide Identification and Expression Profiling of Odorant-Binding Protein Genes in the Bean Flower Thrips Megalurothrips usitatus (Bagnall) (Thysanoptera: Thripidae). Insects. 2025; 16(2):212. https://doi.org/10.3390/insects16020212
Chicago/Turabian StyleXia, Gen, Lang Yang, Boliao Li, Qinli Wang, Lifei Huang, Xiaoli Tian, and Guohui Zhang. 2025. "Genome-Wide Identification and Expression Profiling of Odorant-Binding Protein Genes in the Bean Flower Thrips Megalurothrips usitatus (Bagnall) (Thysanoptera: Thripidae)" Insects 16, no. 2: 212. https://doi.org/10.3390/insects16020212
APA StyleXia, G., Yang, L., Li, B., Wang, Q., Huang, L., Tian, X., & Zhang, G. (2025). Genome-Wide Identification and Expression Profiling of Odorant-Binding Protein Genes in the Bean Flower Thrips Megalurothrips usitatus (Bagnall) (Thysanoptera: Thripidae). Insects, 16(2), 212. https://doi.org/10.3390/insects16020212






