Genome-Wide Identification and Characterization of Heat Shock Proteins in the Stored-Product Pest Rhyzopertha dominica (Fabricius): Phylogenetic, Structural, and Stress-Induced Expression Analyses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Genome-Wide Identification of the HSP Genes
2.3. Bioinformatics Analysis of HSP Genes
2.4. Controlled Nitrogen Atmosphere Treatment
2.5. Extreme High Temperature Treatment
2.6. Phosphine Fumigation Treatment
2.7. K-Obiol Grain Protectant Treatment
2.8. Transcriptome Sequencing and Expression Pattern Analysis of HSP Genes
2.9. RT-qPCR Verification
3. Results
3.1. Genome-Wide Identification of HSP Genes in R. dominica
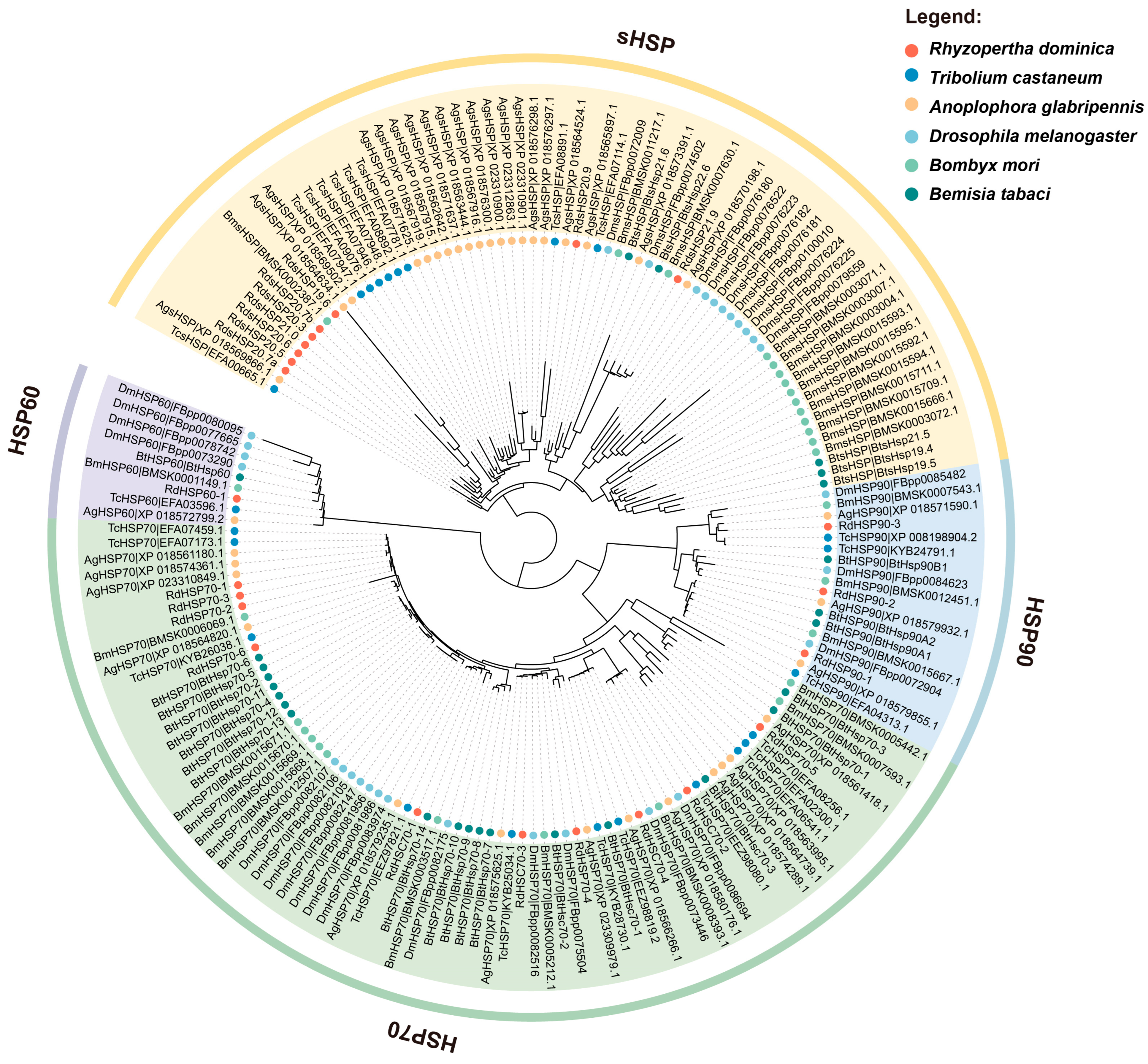
3.2. Phylogenetic Analysis
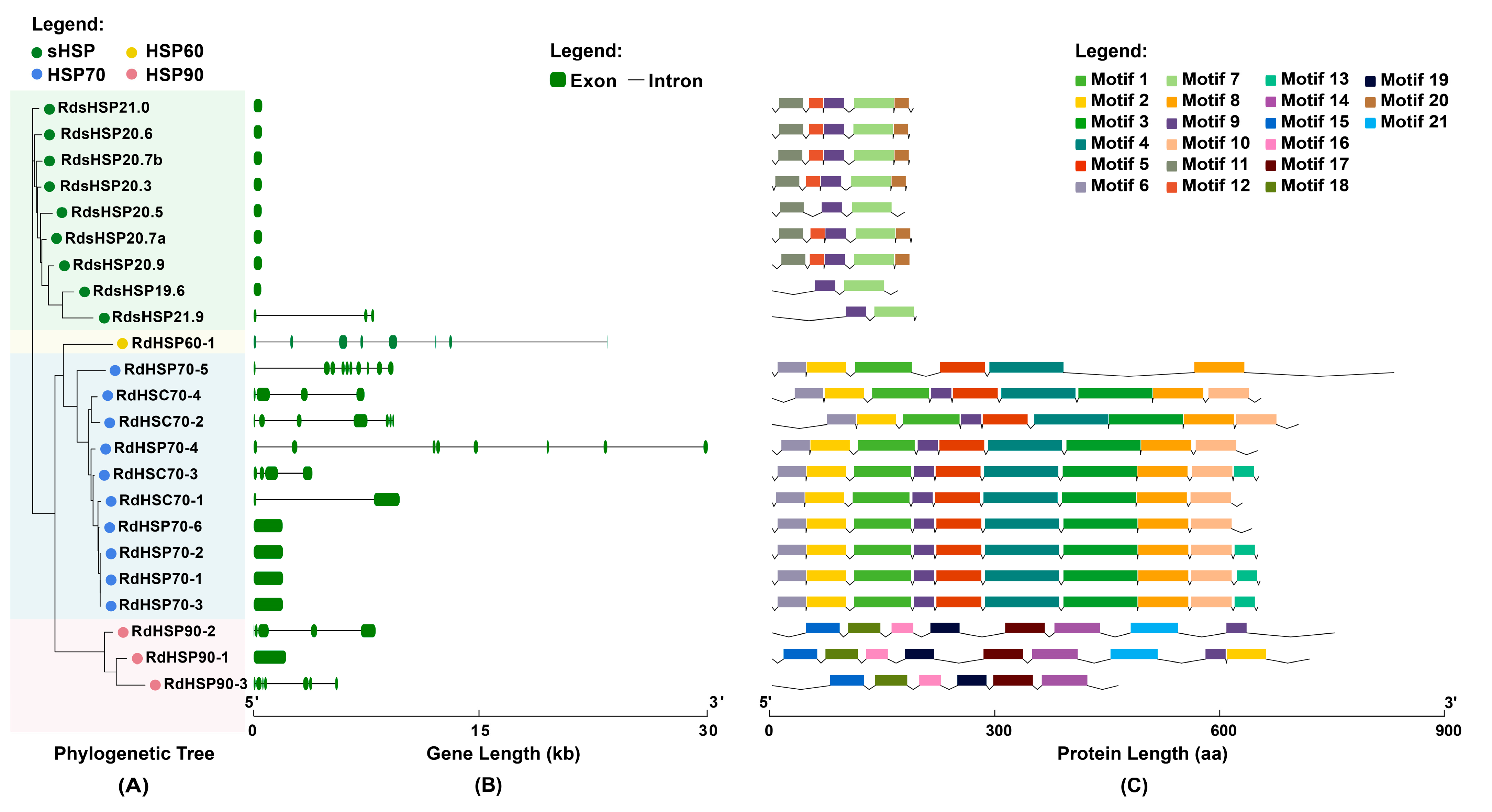
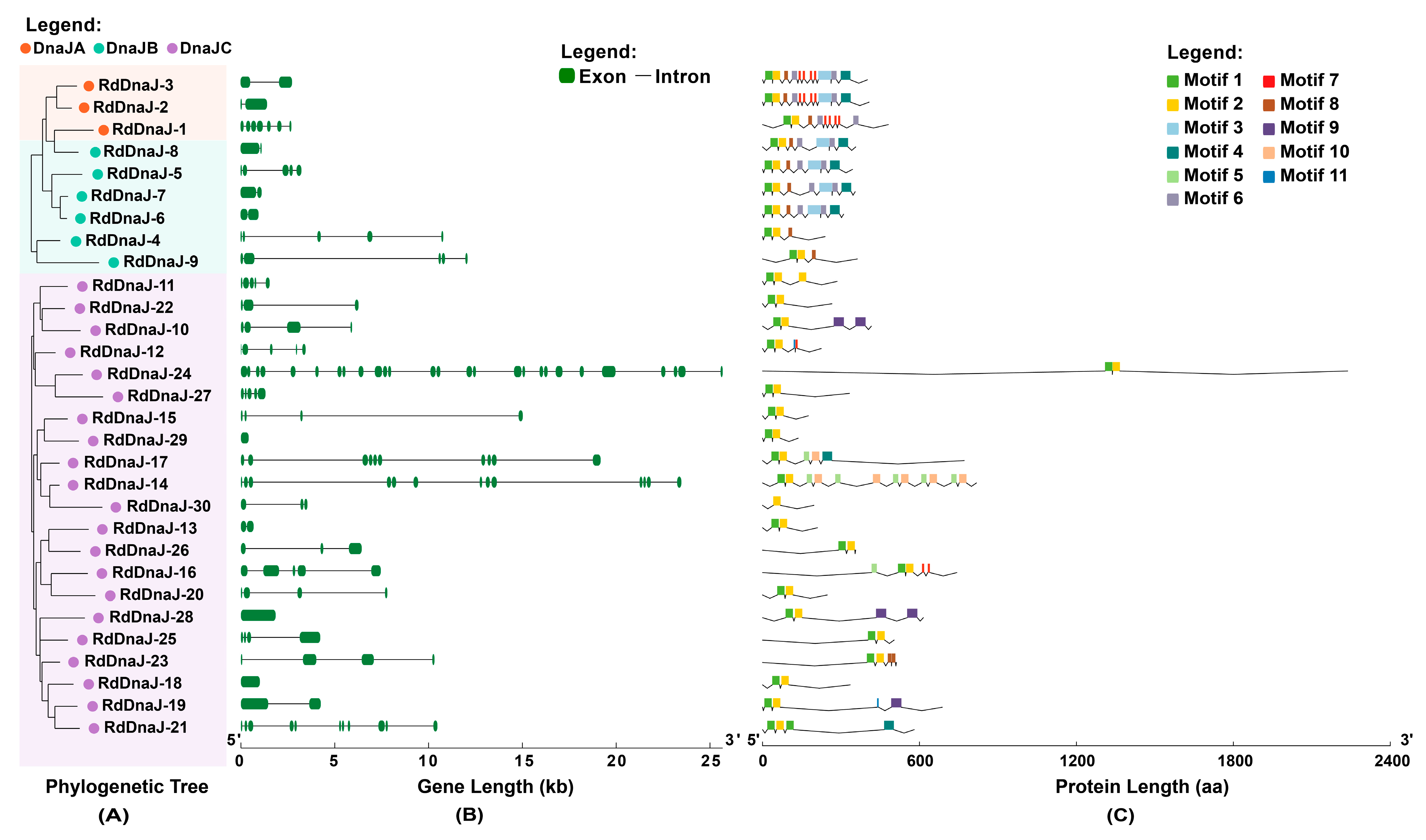
3.3. Gene Structure and Conserved Motif Analysis
3.4. Evaluation of the Lethal Effects of Four Treatments on Rhyzopertha dominica
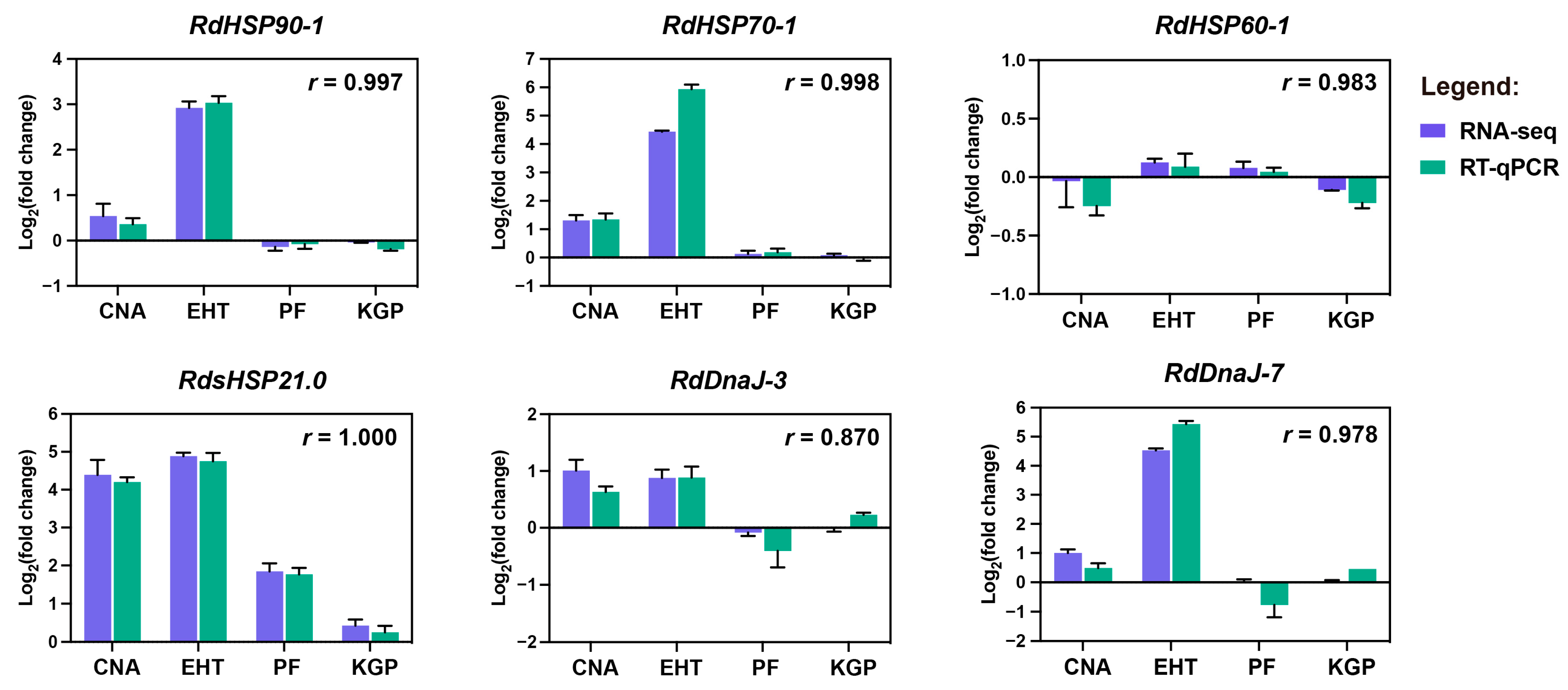
3.5. Transcriptome Sequencing and Expression Patterns of RdHsps
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashburner, M.; Bonner, J.J. The induction of gene activity in Drosophila by heat shock. Cell 1979, 17, 241–254. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; MacRae, T.H. Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 2015, 60, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Chen, H.; Xu, X.L.; Li, Y.P.; Wu, J.X. Characterization of heat shock protein 90, 70 and their transcriptional expression patterns on high temperature in adult of Grapholita molesta (Busck). Insect Sci. 2014, 21, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Feng, Z.; Lu, M.; Du, Y. Chilo Suppressalis heat shock proteins are regulated by heat shock factor 1 during heat stress. Insect Mol. Biol. 2023, 32, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Q.; Zhang, Y.L.; Wang, X.Q.; Dong, H.; Gao, P.; Jia, L.Y. Characterization of multiple heat-shock protein transcripts from Cydia Pomonella: Their response to extreme temperature and insecticide exposure. J. Agric. Food Chem. 2016, 64, 4288–4298. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, W.; Li, R.; Zhao, C.; Pan, L.; Yan, C. A chromosome level genome assembly of Propsilocerus akamusi to understand its response to heavy metal exposure. Mol. Ecol. Resour. 2021, 21, 1996–2012. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Meng, J.Y.; Zhou, L.; Zhang, C.Y. Induced heat shock protein 70 confers biological tolerance in UV-B stress–adapted Myzus persicae (Hemiptera). Int. J. Biol. Macromol. 2022, 220, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Wrońska, A.K.; Boguś, M.I. Heat shock proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (Lepidoptera) hemolymph are affected by infection with Conidiobolus coronatus (Entomophthorales). PLoS ONE 2020, 15, e0228556. [Google Scholar] [CrossRef] [PubMed]
- Teets, N.M.; Peyton, J.T.; Colinet, H.; Renault, D.; Kelley, J.L.; Kawarasaki, Y.; Lee, R.E.; Denlinger, D.L. Gene expression changes governing extreme dehydration tolerance in an Antarctic insect. Proc. Natl. Acad. Sci. USA 2012, 109, 20744–20749. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Feder, M.E.; Kang, L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Mol. Ecol. 2018, 27, 3040–3054. [Google Scholar] [CrossRef]
- Lindquist, S. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-M.; Zhang, Q.; Zhang, Y.-L.; Zhang, G.-Z.; Zhang, Z.; Yu, Q.-Y. Heat shock protein 70 family in response to multiple abiotic stresses in the silkworm. Insects 2021, 12, 928. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Z.-D.; Li, D.-T.; Jiang, M.-X.; Zhang, C.-X. HSP70/DNAJ family of genes in the brown planthopper, Nilaparvata lugens: Diversity and function. Genes 2021, 12, 394. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, K. Constantly updated knowledge of Hsp90. J. Biochem. 2005, 137, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lin, S.; Wu, Y.; Zhao, Z.; Zhou, X.; Sadiq, S.; Zhang, Z.; Guo, X.; Wu, P. Hsp90 could promote BmNPV proliferation by interacting with Actin-4 and enhance its expression. Dev. Comp. Immunol. 2023, 142, 104667. [Google Scholar] [CrossRef]
- Gao, J.; Guo, Y.; Chen, J.; Diao, Q.Y.; Wang, Q.; Dai, P.L.; Zhang, L.; Li, W.M.; Wu, Y.Y. Acute oral toxicity, apoptosis, and immune response in nurse bees (Apis mellifera) induced by flupyradifurone. Front. Physiol. 2023, 14, 1150340. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Shin, Y.; Han, S.; Ha, J.; Tiwari, P.K.; Kim, S.S.; Kang, I. Molecular chaperonin HSP60: Current understanding and future prospects. Int. J. Mol. Sci. 2024, 25, 5483. [Google Scholar] [CrossRef]
- Sundaram, V.K.; Sampathkumar, N.K.; Massaad, C.; Grenier, J. Optimal use of statistical methods to validate reference gene stability in longitudinal studies. PLoS ONE 2019, 14, e0219440. [Google Scholar] [CrossRef] [PubMed]
- Basha, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- de Jong, W.W.; Caspers, G.-J.; Leunissen, J.A.M. Genealogy of the α-crystallin-:small heat-shock protein superfamily. Int. J. Biol. Macromol. 1998, 22, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Turan, M. Genome-wide analysis and characterization of HSP gene families (HSP20, HSP40, HSP60, HSP70, HSP90) in the yellow fever mosquito (Aedes aegypti) (Diptera: Culicidae). J. Insect Sci. 2023, 23, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Abou-Shaara, H.F. The response of heat shock proteins in honey bees to abiotic and biotic stressors. J. Therm. Biol. 2024, 119, 103784. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Xu, K.K.; Cao, Y.; Meng, Y.L.; Liu, Y.; Li, C. Identification and expression analysis of four small heat shock protein genes in cigarette beetle, Lasioderma serricorne (Fabricius). Insects 2019, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Musskopf, M.K.; De Mattos, E.P.; Bergink, S.; Kampinga, H.H. HSP40/DNAJ Chaperones. In Encyclopedia of Life Sciences; John Wiley & Sons: Chichester, UK, 2018. [Google Scholar]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, M.; Caplan, A. Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 1998, 3, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Bashir, T. Reproduction of Rhyzopertha dominica (F.) (Coleoptera: Bostrichidae) on different host-grains. Pak. J. Biol. Sci. 2002, 5, 91–93. [Google Scholar] [CrossRef]
- Edde, P.A. A Review of the biology and control of Rhyzopertha dominica (F.) the lesser grain borer. J. Stored Prod. Res. 2012, 48, 1–18. [Google Scholar] [CrossRef]
- Jia, F.; Toews, M.D.; Campbell, J.F.; Ramaswamy, S.B. Survival and reproduction of lesser grain borer, Rhyzopertha dominica (F.) (Coleoptera: Bostrichidae) on flora associated with native habitats in Kansas. J. Stored Prod. Res. 2008, 44, 366–372. [Google Scholar] [CrossRef]
- Oppert, B.; Morgan, T.D. Improved high-throughput bioassay for Rhyzopertha dominica (F.) (Coleoptera: Bostrichidae). J. Stored Prod. Res. 2013, 52, 68–73. [Google Scholar] [CrossRef]
- Elek, J.A. Methods for collecting eggs and monitoring egg-hatch and immature development of Rhyzopertha dominica (F.) (Coleoptera: Bostrichidae). J. Stored Prod. Res. 1994, 30, 261–265. [Google Scholar] [CrossRef]
- Stewart-Jones, A.; Stirrup, T.J.; Hodges, R.J.; Farman, D.I.; Hall, D.R. Analysis of free fatty acids in food substrates and in the dust and frass of stored-product pests: Potential for species discrimination? J. Stored Prod. Res. 2009, 45, 119–124. [Google Scholar] [CrossRef]
- Shah, J.A.; Vendl, T.; Aulicky, R.; Stejskal, V. Frass produced by the primary pest Rhyzopertha dominica supports the population growth of the secondary stored product pests Oryzaephilus surinamensis, Tribolium castaneum, and T. confusum. Bull. Entomol. Res. 2021, 111, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Nansen, C.; Phillips, T.W.; Palmer, M.W. Analysis of the insect community in a stored-maize facility. Ecol. Res. 2004, 19, 197–207. [Google Scholar] [CrossRef]
- Park, S.-H.; Arthur, F.H.; Bean, S.R.; Schober, T.J. Impact of differing population levels of Rhyzopertha dominica (F.) on milling and physicochemical properties of sorghum kernel and flour. J. Stored Prod. Res. 2008, 44, 322–327. [Google Scholar] [CrossRef]
- Ortega, D.S.; Bacca, T.; Nascimento, S.A.P.; Canal, N.A.; Haddi, K. Control failure and insecticides resistance in populations of Rhyzopertha dominica (Coleoptera: Bostrichidae) from Colombia. J. Stored Prod. Res. 2021, 92, 101802. [Google Scholar] [CrossRef]
- Daglish, G.J. Prevalence of resistance to deltamethrin in Rhyzopertha dominica (F.) in eastern Australia. J. Stored Prod. Res. 2018, 78, 45–49. [Google Scholar] [CrossRef]
- Opit, G.P.; Phillips, T.W.; Aikins, M.J.; Hasan, M.M. Phosphine resistance in Tribolium castaneum and Rhyzopertha dominica from stored wheat in Oklahoma. J. Econ. Entomol. 2012, 105, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, I.G.; Oliveira, J.; Mexia, A.; Barros, G.; Almeida, C.; Brazinha, C.; Vega, A.; Brites, C. Advances in environmentally friendly techniques and circular economy approaches for insect infestation management in stored rice grains. Foods 2023, 12, 511. [Google Scholar] [CrossRef]
- Moirangthem, T.T.; Baik, O.-D. Disinfestation of stored grains using non-chemical technologies—A review. Trends Food Sci. Technol. 2021, 107, 299–308. [Google Scholar] [CrossRef]
- Wang, J. Induced tolerance of the psocid, Liposcelis bostrychophila Badonnel (Psocoptera: Liposcelididae), to controlled atmosphere. Int. J. Pest Manag. 1999, 45, 75–79. [Google Scholar] [CrossRef]
- Zheng, H.-Y.; Qin, P.-H.; Yang, K.; Liu, T.-X.; Zhang, Y.-J.; Chu, D. Genome-wide identification and analysis of the heat-shock protein gene superfamily in Bemisia tabaci and expression pattern analysis under heat shock. Insects 2022, 13, 570. [Google Scholar] [CrossRef]
- Wang, X.R.; Wang, C.; Ban, F.X.; Zhu, D.T.; Liu, S.S.; Wang, X.W. Genome-wide identification and characterization of hsp gene superfamily in whitefly (Bemisia tabaci) and expression profiling analysis under temperature stress: HSP gene superfamily of B. tabaci. Insect Sci. 2019, 26, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Si, F.L.; Qiao, L.; He, Q.Y.; Zhou, Y.; Yan, Z.T.; Chen, B. HSP superfamily of genes in the malaria vector Anopheles sinensis: Diversity, phylogenetics and association with pyrethroid resistance. Malar. J. 2019, 18, 132. [Google Scholar] [CrossRef] [PubMed]
- Mau, Y.S.; Collins, P.J.; Daglish, G.J.; Nayak, M.K.; Ebert, P.R. The rph2 gene is responsible for high level resistance to phosphine in independent field strains of Rhyzopertha dominica. PLoS ONE 2012, 7, e34027. [Google Scholar] [CrossRef]
- Schlipalius, D.I.; Chen, W.; Collins, P.J.; Nguyen, T.; Reilly, P.E.B.; Ebert, P.R. Gene interactions constrain the course of evolution of phosphine resistance in the lesser grain borer, Rhyzopertha dominica. Heredity 2008, 100, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Muszewska, A.; Steczkiewicz, K.; Šatović-Vukšić, E.; Plohl, M.; Fabrick, J.; Vinokurov, K.; Koloniuk, I.; Johnston, J.; Smith, T.; et al. The genome of Rhyzopertha dominica (Fab.) (Coleoptera: Bostrichidae): Adaptation for success. Genes 2022, 13, 446. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, H.; Huang, S.; Li, F. OMIGA: Optimized maker-based insect genome annotation. Mol. Genet. Genomics 2014, 289, 567–573. [Google Scholar] [CrossRef]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, F.; Li, Y.; Zong, S.; Tao, J. Genome-wide identification and expression analysis of the hsp gene superfamily in asian long-horned beetle (Anoplophora glabripennis). Int. J. Biol. Macromol. 2022, 200, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bu, C.; Li, T.; Wang, S.; Jiang, F.; Yi, Y.; Yang, H.; Zhang, Z. Cloning and analysis of DnaJ family members in the silkworm, Bombyx mori. Gene 2016, 576, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Ultrafast one-pass fastq data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Kim, D. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare Tools: A zero-code interactive online platform for biological data analysis and visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Feng, H.Y.; Chen, Z.D.; Jiang, S.D.; Miao, Z.Q.; Wang, J.J.; Wei, D.D. Characterization and expression of heat shock protein 70s in Liposcelis bostrychophila: Insights into their roles in insecticidal stress response. J. Stored Prod. Res. 2024, 106, 102289. [Google Scholar] [CrossRef]
- Li, Z.W.; Li, X.; Yu, Q.Y.; Xiang, Z.H.; Kishino, H.; Zhang, Z. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol. Biol. 2009, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Leask, M.; Lovegrove, M.; Walker, A.; Duncan, E.; Dearden, P. Evolution and genomic organization of the insect sHSP gene cluster and coordinate regulation in phenotypic plasticity. BMC Ecol. Evol. 2021, 21, 154. [Google Scholar] [CrossRef]
- Comeron, J.M. Selective and mutational patterns associated with gene expression in humans: : Influences on synonymous composition and intron presence. Genetics 2004, 167, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yin, Z.; Li, H.; Guo, J. HSP gene superfamily in Aspongopus chinensis Dallas: Unravelling identification, characterisation and expression patterns during diapause and non-diapause stages. Bull. Entomol. Res. 2024, 114, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xu, K.; Zhu, X.; Bai, Y.; Yang, W.; Li, C. Role of modified atmosphere in pest control and mechanism of its effect on insects. Front. Physiol. 2019, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Michaud, M.R.; Teets, N.M.; Peyton, J.T.; Blobner, B.M.; Denlinger, D.L. Heat shock response to hypoxia and its attenuation during recovery in the flesh fly, Sarcophaga crassipalpis. J. Insect Physiol. 2011, 57, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y. Effects of anoxia on survival and gene expression in Bactrocera dorsalis. J. Insect Physiol. 2018, 107, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Nayak, M.K.; Daglish, G.J.; Phillips, T.W.; Ebert, P.R. Resistance to the fumigant phosphine and its management in insect pests of stored products: A global perspective. Annu. Rev. Entomol. 2020, 65, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Price, N.R.; Dance, S.J. Some biochemical aspects of phosphine action and resistance in three species of stored product beetles. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1983, 76, 277–281. [Google Scholar] [CrossRef]
- Alzahrani, S.M.; Ebert, P.R. Pesticidal toxicity of phosphine and its interaction with other pest control treatments. Curr. Issues Mol. Biol. 2023, 45, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Erban, T.; Harant, K.; Chalupnikova, J.; Kocourek, F.; Stara, J. Beyond the survival and death of the deltamethrin-threatened pollen beetle Meligethes aeneus: An in-depth proteomic study employing a transcriptome database. J. Proteomics 2017, 150, 281–289. [Google Scholar] [CrossRef]
- Lin, T.; Wu, H.; Luo, L. The gene expression profile of Monochamus alternatus in response to deltamethrin exposure. J. Asia-Pac. Entomol. 2014, 17, 893–899. [Google Scholar] [CrossRef]
- Yang, Q.; Lu, Y. Heat shock protein 70 genes are involved in the thermal tolerance of Hippodamia variegata. Insects 2024, 15, 678. [Google Scholar] [CrossRef] [PubMed]
- Ashbrook, A.R.; Feder, J.L.; Scharf, M.E.; Bennett, G.W.; Gondhalekar, A.D. Characterization of heat exposure-associated escape behaviors and HSP gene expression in bed bugs (Cimex lectularius L.). Pest Manag. Sci. 2022, 78, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.T.; Zhai, Z.; Liu, Z.Y.; Zhou, L.; Yang, H.; Chen, L.; Zhang, Y.S.; Yu, H. Expressional respones of hsp70 genes against abiotic and entomopathgenic stresses in four different noctuid larval species (Lepidoptera: Noctidae). Pestic. Biochem. Physiol. 2024, 204, 106059. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Meng, J.Y.; Zhou, L.; Yao, M.S.; Zhang, C.Y. Identification of five small heat shock protein genes in Spodoptera frugiperda and expression analysis in response to different environmental stressors. Cell Stress Chaperones 2021, 26, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta 2012, 1823, 607–613. [Google Scholar] [CrossRef] [PubMed]



| Family | Gene Name | Scaffold | Strand | Protein Length (aa) | Molecular Weight (kDa) | PI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| HSP90 | RdHSP90-1 | QCXZ01000042.1 | − | 718 | 82.3 | 5.01 | Cytoplasmic |
| RdHSP90-2 | QCXZ01000138.1 | + | 752 | 86.1 | 4.90 | Cytoplasmic/Extracellular | |
| RdHSP90-3 (partial) | QCXZ01000137.1 | + | 471 | - | - | - | |
| HSP70 | RdHSP70-1 | QCXZ01000003.1 | + | 652 | 71.1 | 5.43 | Cytoplasmic |
| RdHSP70-2 | QCXZ01000003.1 | − | 649 | 70.8 | 5.44 | Cytoplasmic | |
| RdHSP70-3 | QCXZ01000003.1 | − | 649 | 70.7 | 5.44 | Cytoplasmic | |
| RdHSP70-4 | QCXZ01000003.1 | + | 649 | 71.4 | 5.28 | Cytoplasmic | |
| RdHSP70-5 | QCXZ01000041.1 | − | 831 | 93.0 | 5.69 | Nuclear | |
| RdHSP70-6 | QCXZ01000053.1 | + | 641 | 70.4 | 5.82 | Cytoplasmic | |
| RdHSC70-1 | QCXZ01000137.1 | + | 629 | 69.1 | 5.45 | Cytoplasmic | |
| RdHSC70-2 | QCXZ01000137.1 | − | 703 | 77.0 | 5.88 | Mitochondrial | |
| RdHSC70-3 | QCXZ01000137.1 | − | 650 | 71.1 | 5.33 | Cytoplasmic | |
| RdHSC70-4 | QCXZ01000137.1 | − | 653 | 72.8 | 5.17 | Endoplasmic Reticulum | |
| HSP60 | RdHSP60-1 | QCXZ01000001.1 | − | 591 | 63.4 | 5.52 | Mitochondrial |
| sHSP | RdsHSP21.0 | QCXZ01000001.1 | + | 191 | 21.0 | 5.62 | Mitochondrial/Nuclear/Extracellular |
| RdsHSP20.6 | QCXZ01000001.1 | + | 186 | 20.6 | 6.85 | Cytoplasmic/Mitochondrial | |
| RdsHSP20.5 | QCXZ01000001.1 | + | 178 | 20.5 | 6.52 | Nuclear/Cytoplasmic | |
| RdsHSP19.6 | QCXZ01000003.1 | − | 167 | 19.6 | 6.14 | Nuclear/Cytoplasmic | |
| RdsHSP20.7a | QCXZ01000041.1 | + | 186 | 20.7 | 5.86 | Extracellular/Cytoplasmic/Nuclear | |
| RdsHSP20.7b | QCXZ01000064.1 | − | 183 | 20.7 | 6.60 | Mitochondrial/Cytoplasmic | |
| RdsHSP20.9 | QCXZ01000064.1 | − | 183 | 20.9 | 6.52 | Nuclear | |
| RdsHSP20.3 | QCXZ01000064.1 | + | 179 | 20.3 | 7.03 | Extracellular | |
| RdsHSP21.9 | QCXZ01000137.1 | − | 192 | 21.9 | 5.61 | Nuclear |
| Subfamily | Gene Name | Scaffold | Strand | Protein Length (aa) | Molecular Weight (kDa) | PI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| DnaJA | RdDnaJ-1 | QCXZ01000041.1 | + | 481 | 52.9 | 9.29 | Mitochondrial |
| RdDnaJ-2 | QCXZ01000042.1 | − | 407 | 45.5 | 6.29 | Nuclear | |
| RdDnaJ-3 | QCXZ01000137.1 | − | 401 | 45.0 | 6.44 | Nuclear | |
| DnaJB | RdDnaJ-4 | QCXZ01000138.1 | + | 239 | 27.3 | 9.20 | Nuclear |
| RdDnaJ-5 | QCXZ01000042.1 | − | 344 | 38.9 | 5.88 | Cytoplasmic | |
| RdDnaJ-6 | QCXZ01000138.1 | − | 310 | 35.0 | 8.77 | Cytoplasmic | |
| RdDnaJ-7 | QCXZ01000086.1 | + | 354 | 38.9 | 9.23 | Cytoplasmic | |
| RdDnaJ-8 | QCXZ01000001.1 | − | 356 | 40.3 | 5.66 | Nuclear | |
| RdDnaJ-9 | QCXZ01000003.1 | − | 362 | 41.7 | 8.88 | Nuclear | |
| DnaJC | RdDnaJ-10 | QCXZ01000064.1 | − | 416 | 48.1 | 8.10 | Nuclear |
| RdDnaJ-11 | QCXZ01000003.1 | + | 285 | 33.0 | 7.67 | Nuclear | |
| RdDnaJ-12 | QCXZ01000075.1 | − | 224 | 24.6 | 7.45 | Extracellular | |
| RdDnaJ-13 | QCXZ01000075.1 | + | 210 | 24.2 | 9.52 | Mitochondrial | |
| RdDnaJ-14 | QCXZ01000137.1 | − | 818 | 94.0 | 5.91 | Cytoplasmic | |
| RdDnaJ-15 | QCXZ01000137.1 | + | 175 | 19.5 | 6.97 | Nuclear | |
| RdDnaJ-16 | QCXZ01000138.1 | − | 743 | 84.8 | 9.46 | Nuclear | |
| RdDnaJ-17 | QCXZ01000138.1 | − | 771 | 89.8 | 8.78 | Plasma Membrane | |
| RdDnaJ-18 | QCXZ01000137.1 | + | 335 | 40.5 | 8.74 | Cytoplasmic | |
| RdDnaJ-19 | QCXZ01000064.1 | + | 687 | 80.0 | 8.00 | Nuclear | |
| RdDnaJ-20 | QCXZ01000137.1 | − | 247 | 29.2 | 9.24 | Nuclear | |
| RdDnaJ-21 | QCXZ01000001.1 | − | 580 | 65.7 | 7.66 | Nuclear/Cytoplasmic/Mitochondrial | |
| RdDnaJ-22 | QCXZ01000137.1 | − | 265 | 31.8 | 8.47 | Nuclear | |
| RdDnaJ-23 | QCXZ01000001.1 | − | 512 | 58.5 | 8.44 | Nuclear | |
| RdDnaJ-24 | QCXZ01000064.1 | − | 2238 | 254.7 | 6.78 | Nuclear | |
| RdDnaJ-25 | QCXZ01000042.1 | − | 503 | 58.1 | 5.95 | Cytoplasmic | |
| RdDnaJ-26 | QCXZ01000003.1 | − | 356 | 41.7 | 8.96 | Plasma Membrane | |
| RdDnaJ-27 | QCXZ01000053.1 | + | 332 | 38.9 | 8.57 | Cytoplasmic | |
| RdDnaJ-28 | QCXZ01000064.1 | − | 615 | 71.2 | 8.84 | Nuclear | |
| RdDnaJ-29 | QCXZ01000041.1 | − | 136 | 15.9 | 4.99 | Nuclear | |
| RdDnaJ-30 | QCXZ01000041.1 | + | 196 | 22.8 | 9.39 | Mitochondrial/Plasma Membrane |
| Treatment | LC/LT10 | LC/LT50 | LC/LT90 | Fit Model | R2 |
|---|---|---|---|---|---|
| Controlled Nitrogen Atmosphere | 40.792 h | 80.291 h | 158.035 h | Logistic | 0.999 |
| Extreme High Temperature | 46.823 °C | 47.471 °C | 48.127 °C | Logistic | 0.995 |
| Phosphine Fumigation | 0.102 g/m3 | 0.551 g/m3 | 2.978 g/m3 | Probit | 0.965 |
| K-Obiol Grain Protectant | 0.090 μg/cm2 | 0.318 μg/cm2 | 1.125 μg/cm2 | Probit | 0.984 |
| Group Name | Sample Name | Treatment Details | Raw Bases | Q20 (%) | Q30 (%) | Clean Reads | Total Mapping Reads (Rates) |
|---|---|---|---|---|---|---|---|
| CNA | N1 | Controlled nitrogen atmosphere treatment (99% N2 for 80 h) | 8.41 Gb | 98.42 | 95.35 | 52,279,194 | 43,583,325 (83.37%) |
| N2 | 6.94 Gb | 98.73 | 96.15 | 43,780,126 | 36,996,673 (84.51%) | ||
| N3 | 7.59 Gb | 98.36 | 95.14 | 48,838,362 | 41,920,620 (85.84%) | ||
| CN1 | Control for controlled nitrogen atmosphere treatment (78% N2 for 80 h) | 7.24 Gb | 98.55 | 95.65 | 46,305,876 | 38,547,717 (83.25%) | |
| CN2 | 7.20 Gb | 98.55 | 95.68 | 44,932,154 | 37,046,999 (82.45%) | ||
| CN3 | 8.02 Gb | 98.26 | 94.97 | 52,145,254 | 41,805,673 (80.17%) | ||
| EHT | H1 | Extreme high temperature treatment (46 °C for 3 h) | 6.89 Gb | 98.98 | 96.95 | 41,938,936 | 34,799,525 (82.98%) |
| H2 | 5.98 Gb | 98.93 | 96.76 | 36,007,342 | 30,014,415 (83.36%) | ||
| H3 | 6.49 Gb | 98.98 | 96.89 | 42,156,452 | 34,320,294 (81.41%) | ||
| CH1 | Control for extreme high temperature treatment (30 °C for 3 h) | 6.66 Gb | 98.91 | 96.74 | 40,544,948 | 34,467,807 (85.01%) | |
| CH2 | 6.61 Gb | 98.92 | 96.75 | 40,519,056 | 34,437,334 (84.99%) | ||
| CH3 | 5.30 Gb | 98.91 | 96.69 | 35,313,172 | 29,893,814 (84.65%) | ||
| PF | P1 | Phosphine fumigation treatment (0.55 g/m3 of phosphine gas for 20 h) | 8.49 Gb | 98.42 | 95.37 | 54,428,094 | 45,038,584 (82.75%) |
| P2 | 7.48 Gb | 98.25 | 94.88 | 47,541,036 | 39,146,791 (82.34%) | ||
| P3 | 7.87 Gb | 98.26 | 94.96 | 50,162,810 | 40,906,312 (81.55%) | ||
| CP1 | Control for phosphine fumigation treatment (0 g/m3 of phosphine gas for 20 h) | 7.26 Gb | 98.37 | 95.13 | 46,007,646 | 37,299,197 (81.07%) | |
| CP2 | 6.99 Gb | 98.54 | 95.64 | 43,525,938 | 35,446,750 (81.44%) | ||
| CP3 | 6.45 Gb | 98.29 | 94.98 | 40,529,910 | 34,389,670 (84.85%) | ||
| KGP | K1 | K-Obiol grain protectant treatment (0.32 μg/cm2 of deltamethrin for 24 h) | 7.11 Gb | 98.20 | 94.68 | 46,005,272 | 39,206,013 (85.22%) |
| K2 | 6.90 Gb | 98.36 | 95.16 | 44,714,288 | 37,767,301 (84.46%) | ||
| K3 | 6.90 Gb | 98.29 | 95.00 | 44,494,096 | 38,208,768 (85.87%) | ||
| CK1 | Control for K-Obiol grain protectant treatment (0 μg/cm2 of deltamethrin for 24 h) | 7.56 Gb | 98.38 | 95.19 | 48,579,578 | 40,769,074 (83.92%) | |
| CK2 | 6.98 Gb | 98.27 | 94.88 | 44,928,050 | 37,932,915 (84.43%) | ||
| CK3 | 6.88 Gb | 98.29 | 94.97 | 43,128,218 | 36,892,070 (85.54%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Xie, Y.; Yao, J.; Zeng, F.; Wang, D. Genome-Wide Identification and Characterization of Heat Shock Proteins in the Stored-Product Pest Rhyzopertha dominica (Fabricius): Phylogenetic, Structural, and Stress-Induced Expression Analyses. Insects 2025, 16, 127. https://doi.org/10.3390/insects16020127
Bai Y, Xie Y, Yao J, Zeng F, Wang D. Genome-Wide Identification and Characterization of Heat Shock Proteins in the Stored-Product Pest Rhyzopertha dominica (Fabricius): Phylogenetic, Structural, and Stress-Induced Expression Analyses. Insects. 2025; 16(2):127. https://doi.org/10.3390/insects16020127
Chicago/Turabian StyleBai, Yueliang, Yanzhu Xie, Junji Yao, Fangfang Zeng, and Dianxuan Wang. 2025. "Genome-Wide Identification and Characterization of Heat Shock Proteins in the Stored-Product Pest Rhyzopertha dominica (Fabricius): Phylogenetic, Structural, and Stress-Induced Expression Analyses" Insects 16, no. 2: 127. https://doi.org/10.3390/insects16020127
APA StyleBai, Y., Xie, Y., Yao, J., Zeng, F., & Wang, D. (2025). Genome-Wide Identification and Characterization of Heat Shock Proteins in the Stored-Product Pest Rhyzopertha dominica (Fabricius): Phylogenetic, Structural, and Stress-Induced Expression Analyses. Insects, 16(2), 127. https://doi.org/10.3390/insects16020127






