Questionable Species Names for Distinct Species Clusters: An Empirical Test of the BOLD Molecular Identification Engine
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
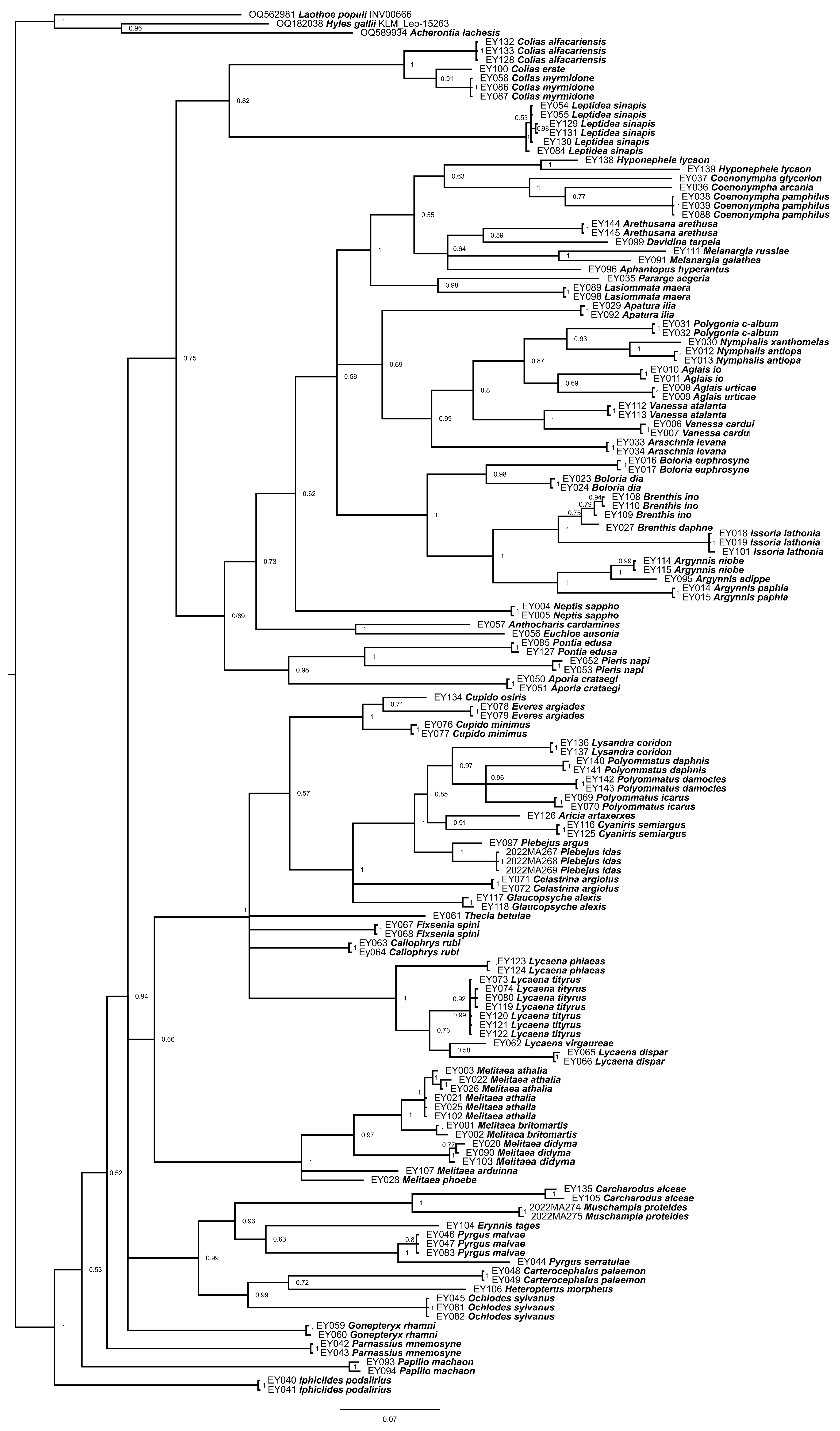
3. Results
4. Discussion
4.1. Species Identification Errors in the BOLD Database (Problems from a Same Genus)
4.2. Possible Contamination/Inaccurate Labeling (Errors in Identification of Genera and Families)
4.3. Undifferentiated DNA Barcodes
4.4. Existing Taxonomy Uncertainty
4.5. Anomalous DNA Barcodes
4.6. Incompleteness of the BOLD Database
5. Conclusions
- Species identification based on DNA barcode comparison, as implemented in the BOLD database (https://boldsystems.org/) (accessed on 6 November 2025), is very effective; however, it needs to be improved. Our study revealed problems with this approach, which partially coincide with the problems of DNA barcoding of the Hemiptera insects [47]. First of all, these problems are associated with errors in the barcode public data as well as with human errors, such as specimen misidentification, sample confusion, and contamination [47]. It is extremely urgent to work on minimizing errors and typos in species identifications in the BOLD database. Gross errors (at the level of genera and families) can be detected and marked by BOLD staff. Correction of identification errors at the species level usually requires access to voucher specimens and can only be performed by experts in specific groups of organisms. In our opinion, such experts should be coordinators and participants of the relevant BOLD projects. The need for manual, semi-automated or automated curation of the DNA barcode databases to improve DNA identifications is well recognized [15,48,49]. However, the curation procedure is not yet integrated into the standard BOLD system algorithm. We recommend creating such an algorithm in BOLD, where, upon detection of conflicting identifications, the system automatically sends a request for clarification to the participants of the corresponding BOLD projects.
- In many cases, even when individuals of different species share DNA barcodes, individuals of local faunas can be unambiguously identified if a geographic correction is introduced. Such a correction can be based on the fact that an identical DNA barcode of another species is found only in a different geographic region. For this reason, DNA barcoding of not only global and regional but also local faunas and floras should be welcomed. However, it should be kept in mind that this approach should be used with caution, since the most complete species lists may contain gaps and species may be found far beyond their known ranges [50]. Unrecognized cryptic species, as well as migrations and range expansions of species, can create additional problems.
- One of the problems of molecular species identification is the identity or high similarity of DNA barcodes in different species. However, as our analysis shows, the identity of DNA barcodes in nominally different species may be a consequence of their real conspecificity. In other words, units formally described as different species may belong to the same evolutionary lineage and actually represent a single species. Recognition of such cases requires further intensive research in the field of classical and integrative taxonomy. Although this type of work is far beyond the scope of the pure DNA barcoding studies, DNA barcoding can be an incentive for them. In cases of poorly studied, formally described taxa, the identity of DNA barcodes may indicate the need for their further analysis and testing for conspecificity.
- Although the incompleteness of DNA barcode libraries may be a major limitation for molecular identification in poorly studied groups of organisms, for well-studied groups such as European butterflies, this problem is of least significance. It may arise when analyzing rare species.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BPP | Bayesian posterior probability |
| ID | Identification |
References
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLOS Biol. 2004, 2, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932. [Google Scholar] [CrossRef] [PubMed]
- Huemer, P.; Berggren, K.; Aarvik, L.; Rennwald, E.; Hausmann, A.; Segerer, A.; Staffoni, G.; Aspaas, A.M.; Trichas, A.; Hebert, P.D.N. Extensive DNA barcoding of Lepidoptera of Crete (Greece) reveals significant taxonomic and faunistic gaps and supports the first comprehensive checklist of the island’s fauna. Insects 2025, 16, 438. [Google Scholar] [CrossRef] [PubMed]
- Twyford, A.D.; Beasley, J.; Barnes, I.; Allen, H.; Azzopardi, F.; Bell, D.; Blaxter, M.L.; Broad, G.; Campos-Dominguez, L.; Choonea, D.; et al. A DNA barcoding framework for taxonomic verification in the Darwin Tree of Life Project. Wellcome Open Res. 2024, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Nat. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.D.H.; Hebert, P.D.N. Identifying spiders through DNA barcodes. Can. J. Zool. 2005, 83, 481–491. [Google Scholar] [CrossRef]
- Phillips, J.D.; Griswold, C.K.; Young, R.G.; Hubert, N.; Hanner, R.H. A Measure of the DNA barcode gap for applied and basic research. Methods Mol. Biol. 2024, 2744, 375–390. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcode of life data system. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Raupach, M.J.; Hannig, K.; Morinière, J.; Hendrich, L.A. DNA barcode library for ground beetles of Germany: The genus Pterostichus Bonelli, 1810 and allied taxa (Insecta, Coleoptera, Carabidae). ZooKeys 2020, 980, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Wiemers, M.; Fiedler, K. Does the DNA barcoding gap exist?—A case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 2007, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Janko, Š.; Rok, Š.; Blaž, K.; Danilo, B.; Andrej, G.; Denis, K.; Klemen, Č.; Matjaž, G. DNA barcoding insufficiently identifies European wild bees (Hymenoptera, Anthophila) due to undefined species diversity, genus-specific barcoding gaps and database errors. Mol Ecol Resour. 2024, 24, e13953. [Google Scholar] [CrossRef]
- Krivosheeva, V.; Salnitska, M.; Semerikova, D.; Gebremeskel, A.; Ivanova, A.; Solodovnikov, A. Identification of West Siberian Quedius (Coleoptera, Staphylinidae) by COI barcodes calls for integrative taxonomy and curation of public DNA libraries. Deut. Entomol. Zeit. 2025, 72, 101–117. [Google Scholar] [CrossRef]
- Anikin, V.V.; Sachkov, S.A.; Zolotuhin, V.V. Fauna Lepidopterologica Volgo–Uralensis: From P. Pallas to Present Days. In Proceedings of the Museum Witt Munich; Museum Witt: Munich, Germany, 2017. 671p. [Google Scholar]
- Dinca, V.; Zakharov, E.V.; Hebert, P.D.; Vila, R. Complete DNA barcode reference library for a country’s butterfly fauna reveals high performance for temperate Europe. Proc. Biol. Sci. 2011, 278, 347–355. [Google Scholar] [CrossRef]
- Dincă, V.; Dapporto, L.; Somervuo, P.; Voda, R.; Cuvelier, S.; Gascoigne-Pees, M.; Huemer, P.; Mutanen, M.; Hebert, P.; Vila, R. High resolution DNA barcode library for European butterflies reveals continental patterns of mitochondrial genetic diversity. Commun. Biol. 2021, 4, 315. [Google Scholar] [CrossRef]
- Bartonova, A.; Konvicka, M.; Korb, S.; Kramp, K.; Schmitt, T.; Fric, Z. Range dynamics of Palaearctic steppe species under glacial cycles: The phylogeography of Proterebia afra (Lepidoptera: Nymphalidae: Satyrinae). Biol. J. Linn. Soc. 2018, 125, 867–884. [Google Scholar] [CrossRef]
- Krupitsky, A.V.; Shapoval, N.A.; Schepetov, D.M.; Ekimova, I.A.; Lukhtanov, V.A. Phylogeny, species delimitation and biogeography of the endemic Palaearctic tribe Tomarini (Lepidoptera, Lycaenidae). Zool. J. Linn. Soc. 2022, 196, 630–646. [Google Scholar] [CrossRef]
- Dincă, V.; Montagud, S.; Talavera, G.; Hernández-Roldán, J.; Munguira, M.L.; García-Barros, E.; Hebert, P.D.; Vila, R. DNA barcode reference library for Iberian butterflies enables a continental-scale preview of potential cryptic diversity. Sci. Rep. 2015, 2, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Dapporto, L.; Menchetti, M.; Voda, R.; Corbella, C.; Cuvelier, S.; Djemadi, I.; Gascoigne-Pees, M.; Hinojosa, J.; Lam, N.T.; Serracanta, M.; et al. The atlas of mitochondrial genetic diversity for Western Palaearctic butterflies. Glob. Ecol. Biogeogr. 2022, 31, 2184–2190. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Sourakov, A.; Zakharov, E.V.; Hebert, P.D. DNA barcoding Central Asian butterflies: Increasing geographical dimension does not significantly reduce the success of species identification. Mol. Ecol. Resour. 2009, 9, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- D’Ercole, J.; Dapporto, L.; Opler, P.; Schmidt, C.B.; Ho, C.; Menchetti, M.; Zakharov, E.V.; Burns, J.M.; Hebert, P.D.N. A genetic atlas for the butterflies of continental Canada and United States. PLoS ONE 2024, 19, e0300811. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Lvovsky, A.L.; Morgun, D.V. Bulavousye Cheshuekrylye Vostochnoy Evropy [Butterflies of Eastern Europe]; KMK Scientific Press Ltd.: Moscow, Russia, 2007; 452p. [Google Scholar]
- Tuzov, V.K.; Bogdanov, P.V.; Churkin, S.V.; Dantchenko, A.V.; Devyatkin, A.L.; Murzin, V.S.; Samodurov, G.D.; Zhdanko, A.B. Guide to the Butterflies of Russia and Adjacent Territories (Lepidoptera, Rhopalocera). Libytheidae, Danaidae, Nymphalidae, Riodinidae, Lycaenidae; Pensoft: Sofia, Bulgaria; Moscow, Russia, 2000; Volume 2, 580p. [Google Scholar]
- Gaunet, A.; Dincă, V.; Dapporto, L.; Montagud, S.; Voda, R.; Schär, S.; Badiane, A.; Font, E.; Vila, R. Two consecutive Wolbachia-mediated mitochondrial introgressions obscure taxonomy in Palearctic swallowtail butterflies (Lepidoptera, Papilionidae). Zool. Scr. 2019, 48, 507–519. [Google Scholar] [CrossRef]
- Davlatov, A.M.; Lukhtanov, V.A. Pontia daplidice (Linnaeus, 1758) and Pontia edusa (Fabricius, [1777]) in Tajikistan: One or two species? (Lepidoptera: Pieridae). SHIL. Rev. Lepidopterol. 2025, 53, 333–342. [Google Scholar] [CrossRef]
- Gwiazdowska, A.; Rutkowski, R.; Sielezniew, M. Conservation genetics of the endangered Danube Clouded Yellow butterfly Colias myrmidone (Esper, 1780) in the last Central European stronghold: Diversity, Wolbachia infection and Balkan connections. Insects 2025, 16, 220. [Google Scholar] [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 2004, 5, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.E.; Lopes, J.F.; Kratochwil, C.F. East African cichlid fishes. EvoDevo 2023, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Stein, F.; Gailing, O. Identification of BOLD engine deficiencies and suggestions for improvement based on a curated Tachina (Diptera) record set. PLoS ONE 2025, 20, e0331216. [Google Scholar] [CrossRef] [PubMed]
- Makhov, I.A.; Gorodilova, Y.Y.; Lukhtanov, V.A. Sympatric occurrence of deeply diverged mitochondrial DNA lineages in Siberian geometrid moths (Lepidoptera, Geometridae): Cryptic speciation, mitochondrial introgression, secondary admixture or effect of Wolbachia? Biol. J. Linn. Soc. 2021, 134, 342–365. [Google Scholar] [CrossRef]
- Hinojosa, J.C.; Dapporto, L.; Brockmann, E.; Dincă, V.; Tikhonov, V.; Grishin, N.; Lukhtanov, V.A.; Vila, R. Overlooked cryptic diversity in Muschampia (Lepidoptera: Hesperiidae) adds two species to the European butterfly fauna. Zool. J. Linn. Soc. 2021, 193, 847–859. [Google Scholar] [CrossRef]
- Dubatolov, V.V.; Sergeev, M.G.; Zdanko, A.B. New and little known species of the butterfly genus Hyponephele Muschamp, 1915. Atalanta 1994, 25, 171–177. [Google Scholar]
- Eckweiler, W.; Bozano, G.C. Guide to the Butterflies of the Palearctic Region: Satyrinae Part IV. Tribe Satyrini, Subtribe Maniolina: Maniola, Pyronia, Aphantopus, Hyponephele; Omnes Artes: Milano, Italy, 2011; 134p. [Google Scholar]
- Kramp, K.; Cizek, O.; Madeira, P.M.; Ramos, A.; Konvicka, M.; Castilho, R.; Schmitt, T. Genetic implications of phylogeographical patterns in the conservation of the boreal wetland butterfly Colias palaeno (Pieridae). Biol. J. Linn. Soc. 2016, 119, 1068–1081. [Google Scholar] [CrossRef]
- Pazhenkova, E.A.; Lukhtanov, V.A. Chromosomal and mitochondrial diversity in Melitaea didyma complex (Lepidoptera, Nymphalidae): Eleven deeply diverged DNA barcode groups in one non-monophyletic species? Comp. Cytogenet. 2016, 10, 697–717. [Google Scholar] [CrossRef]
- Charlat, S.; Duplouy, A.; Hornett, E.A.; Dyson, E.A.; Davies, N.; Roderick, G.K.; Wedell, N.; Hurst, G.D. The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina. BMC Evol. Biol. 2009, 9, 64. [Google Scholar] [CrossRef]
- Sucháčková, B.A.; Konvička, M.; Marešová, J.; Wiemers, M.; Ignatev, N.; Wahlberg, N.; Schmitt, T.; Fric, Z.F. Wolbachia affects mitochondrial population structure in two systems of closely related Palaearctic blue butterflies. Sci. Rep. 2021, 11, 3019. [Google Scholar] [CrossRef]
- Ožana, S.; Dolný, A.; Pánek, T. Nuclear copies of mitochondrial DNA as a potential problem for phylogenetic and population genetic studies of Odonata. Syst. Entomol. 2022, 47, 591–602. [Google Scholar] [CrossRef]
- Tshikolovets, V. Butterflies of Europe and the Mediterranean Area; Tshikolovets Publications: Pardubice, Czech Republic, 2011; 544p. [Google Scholar]
- Cheng, Z.; Li, Q.; Deng, J.; Liu, Q.; Huang, X. The devil is in the details: Problems in DNA barcoding practices indicated by systematic evaluation of insect barcodes. Front. Ecol. Evol. 2023, 11, 1149839. [Google Scholar] [CrossRef]
- Quaresma, A.; Ankenbrand, M.J.; Garcia, C.A.Y.; Rufino, J.; Honrado, M.; Amaral, J.; Brodschneider, R.; Brusbardis, V.; Gratzer, K.; Hatjina, F.; et al. Semi-automated sequence curation for reliable reference datasets in ITS2 vascular plant DNA (meta-)barcoding. Sci. Data 2024, 11, 129. [Google Scholar] [CrossRef]
- Keller, A.; Hebert, P. Automating the curation of DNA barcode databases for vascular plants. Environ. DNA 2025, 7, e70125. [Google Scholar] [CrossRef]
- Kosterin, O.E.; Knyazev, S.A. Plusiodonta casta (Butler, 1878) (Lepidoptera, Erebidae, Calpinae) found in West Sayan (Central Siberia, Russia). Acta Biol. Sibir. 2024, 10, 1727–1732. [Google Scholar] [CrossRef]
| Query and Its ID Based on Morphology | BOLD ID for the Majority of the Most Similar Barcodes | Other BOLD IDs for the Most Similar Barcodes (Similarity with the Query, %) | Final BOLD ID for the Query Based on the 25 Closest Barcodes | Final BOLD ID Confidence, % | Reasons for BOLD ID Uncertainty | Geographic Correction * |
|---|---|---|---|---|---|---|
| Aglais io EY010 | Aglais io | Vanessa cardui (100) | Aglais (genus only) | 83 (for the genus) | possible contamination/inaccurate taxon labeling | N/A |
| Apatura ilia EY029 | Apatura ilia | A. metis (99.53) | Apatura (genus only) | 100 (for the genus) | undifferentiated barcodes | No |
| Apatura ilia EY092 | Apatura ilia | A. metis (99.69) | Apatura (genus only) | 100 (for the genus) | undifferentiated barcodes | No |
| Aphantopus hyperantusEY096 | Aphantopus hyperantus | A. sp. (100), A. bieti (99.68) | Aphantopus (genus only) | 100% (for the genus) | undifferentiated barcodes and/or identification problems | N/A |
| Araschnia levana EY033 | Araschnia levana | Boloria selene (100), Polygonia c-album (100) | Araschnia levana | 92 | possible contamination/inaccurate taxon labeling | N/A |
| Argynnis paphia EY014 | Argynnis paphia | Polyommatus icarus (100) | Argynnis paphia | 96 | possible contamination/inaccurate taxon labeling | N/A |
| Aricia artaxerxes EY126 | Aricia artaxerxes | Plebejus argus (100%) | Aricia artaxerxes | 96 | possible contamination/inaccurate taxon labeling | N/A |
| Brenthis ino EY108 | Brenthis ino | Melitaea athalia (99.84%), B. daphne (99.68%) | Brenthis ino | 92 | possible contamination/inaccurate taxon labeling; identification problems | N/A |
| Callophrys rubi EY063 | Callophrys rubi | C. chalybeitincta (100), Novosatsuma collosa (99.84) | Callophrys (genus only) | 83 (for the genus) | undifferentiated barcodes | No |
| Carcharodus alceae EY105 | Carcharodus alceae | Erynnis tages (99.53) | Carcharodus alceae | 96 | possible contamination/inaccurate taxon labeling | N/A |
| Carcharodus alceae EY135 | Carcharodus alceae | C. floccifera (100), C. stauderi (100), C. sp. (100) | Carcharodus (genus only) | 100 (for the genus) | identification problems | N/A |
| Carterocephalus palaemon EY048 | Carterocephalus palaemon | C. silvicola (99.52) | Carterocephalus palaemon | 92 | identification problems | N/A |
| Coenonympha arcania EY036 | Coenonympha arcania | C. leander (99.68), C. orientalis (99.36) | Coenonympha arcania | 96 | undifferentiated barcodes | No |
| Coenonympha pamphilus EY038 | Coenonympha pamphilus | C. lyllus (100) | Coenonympha pamphilus | 92 | existing taxonomy uncertainty | N/A |
| Colias erate EY100 | Colias crocea, C. erate | C. crocea (100), C. marnoana (100), C. poliographus (99.84) | Colias (genus only) | 100 (for the genus) | undifferentiated barcodes | No |
| Colias myrmidone EY058 | Colias myrmidone | C. caucasica (100) | Colias (genus only) | 100 (for the genus) | undifferentiated barcodes | Yes |
| Cupido minimus EY076 | Cupido minimus | C. tusovi (99.84), C. osiris (99.84), Glaucopsyche lycormas (99.69) | Cupido (genus only) | 100% (for the genus) | identification problems; existing taxonomy uncertainty | N/A |
| Cupido osiris EY134 | Cupido osiris | C. staudingeri (98.87) | Cupido (genus only) | 100 (for the genus) | undifferentiated barcodes | N/A |
| Cyaniris semiargus EY116 | Cyaniris semiargus | Glaucopsyche lycormas (100) | Cyaniris semiargus | 92 | identification problems or possible contamination/inaccurate taxon labeling | N/A |
| Davidina tarpeia EY099 | Davidina tarpeia | D. dzhulukuli (100), D. lederi (99.66) | Davidina (genus only) | 100 (for the genus) | undifferentiated barcodes | Yes |
| Erynnis tages EY104 | Erynnis tages | Colias alfacariensis (100) | Erynnis tages | 96 | possible contamination/inaccurate taxon labeling | N/A |
| Euchloe ausonia EY056 | Euchloe ausonia | E. pulverata (99.69), E. persica (99.67), E. ochracea (98.44) | Euchloe (genus only) | 100 (for the genus) | existing taxonomy uncertainty and/or identification problems | N/A |
| Fabriciana niobe EY115 | Fabriciana niobe | F. adippe (100), F. xipe (100) | Fabriciana (genus only) | 100 (for the genus) | identification problems | N/A |
| Hyponephele lycaon EY138 | Hyponephele lycaon | H. przhewalskyi (100), Coenonympha pamphilus (99.84) | Hyponephele lycaon | 92 | existing taxonomy uncertainty; possible contamination/inaccurate taxon labeling | N/A |
| Hyponephele lycaon EY139 | Different taxa | different taxa with a low level of similarity | Satyrinae (subfamily only) | 100 (for the subfamily) | anomalous barcode | N/A |
| Iphiclides podalirius EY040 | Iphiclides podalirius | I. feistahameli (100) | Iphiclides (genus only) | 100 (for the genus) | undifferentiated barcodes | Yes |
| Leptidea sinapis EY054 | Leptidea sinapis | L. descimoni (99.84) | Leptidea sinapis | 92 | identification problems | N/A |
| Lysandra coridon EY136 | Lysandra coridon | Hipparchia semele (100) | Lysandra coridon | 96 | possible contamination/inaccurate taxon labeling | N/A |
| Melitaea arduinna EY107 | Melitaea arduinna | M. cinxia (100) | Melitaea arduinna | 91 | identification problems | N/A |
| Melitaea britomartisEU002 | Melitaea britomartis | M. sp. (100), M. aurelia (99.84) | Melitaea (genus only) | 100 (for the genus) | identification problems | N/A |
| Melitaea phoebe EY028 | Melitaea phoebe | M. sibina (99.84), M. ornata (99.69) | Melitaea (genus only) | 100 (for the genus) | undifferentiated barcodes | No |
| Muschampia proteides MA274 | Muschampia proteides | M. proto (100), M. sovietica (100), M. sp. (100) | Muschampia (genus only) | 100 (for the genus) | existing taxonomy uncertainty | N/A |
| Muschampia proteides MA275 | Muschampia proteides | M. proto (100), M. sovietica (100), M. sp. (100) | Muschampia (genus only) | 100 (for the genus) | existing taxonomy uncertainty | N/A |
| Neptis sappho EY004 | Neptis sappho | Araschnia levana (100) | Neptis sappho | 94 | possible contamination/inaccurate taxon labeling | N/A |
| Ochlodes sylvanus EY045 | Ochlodes sylvanus | O. hyrcana (100), O. faunus | Ochlodes (genus only) | 100 (for the genus) | existing taxonomy uncertainty | N/A |
| Papilio machaon EY093 | Papilio machaon | P. saharae (99.52) | Papilio machaon | 92 | undifferentiated barcodes | Yes |
| Pararge aegeria EY035 | Pararge aegeria | Argynnis paphia (100) | Pararge aegeria | 95 | possible contamination/inaccurate taxon labeling | N/A |
| Pieris napi EY052 | Pieris napi | P. rapae (100) | Pieris napi | 92 | identification problems | N/A |
| Plebejus idas 2022MA269s | Plebejus idas | P. argus (100), P. sp. (100), P. bellieri (99.84) | Plebejus (genus only) | 100 (for the genus) | identification problems | N/A |
| Polygonia c-album EY031 | Polygonia c-album | Polygonia interposita (100), Aglais io (100) | Polygonia (genus only) | 80 (for the genus) | existing taxonomy uncertainty; possible contamination/inaccurate taxon labeling | N/A |
| Polyommatus damocles EY142 | different taxa | different taxa with a low (<99%) level of similarity | Polyommatus (genus only) | 100 (for the genus) | incompleteness of the BOLD database | N/A |
| Polyommatus icarus EY069 | Polyommatus icarus | P. juno (99.69) | Polyommatus icarus | 96 | existing taxonomy uncertainty | Yes |
| Pontia edusa EY085 | Pontia edusa | P. daplidice (100) | Pontia (genus only) | 100 (for the genus) | identification problems | N/A |
| Pontia edusa EY127 | Pontia edusa | P. daplidice (100) | Pontia (genus only) | 100 (for the genus) | identification problems | N/A |
| Pyrgus malvae EY046 | Pyrgus malvae | P. sp. (100), P. malvoides (99.22) | Pyrgus (genus only) | 100 (for the genus) | undifferentiated barcodes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakimenko, E.V.; Romanovich, A.E.; Lukhtanov, V.A. Questionable Species Names for Distinct Species Clusters: An Empirical Test of the BOLD Molecular Identification Engine. Insects 2025, 16, 1172. https://doi.org/10.3390/insects16111172
Yakimenko EV, Romanovich AE, Lukhtanov VA. Questionable Species Names for Distinct Species Clusters: An Empirical Test of the BOLD Molecular Identification Engine. Insects. 2025; 16(11):1172. https://doi.org/10.3390/insects16111172
Chicago/Turabian StyleYakimenko, Elisaveta V., Anna E. Romanovich, and Vladimir A. Lukhtanov. 2025. "Questionable Species Names for Distinct Species Clusters: An Empirical Test of the BOLD Molecular Identification Engine" Insects 16, no. 11: 1172. https://doi.org/10.3390/insects16111172
APA StyleYakimenko, E. V., Romanovich, A. E., & Lukhtanov, V. A. (2025). Questionable Species Names for Distinct Species Clusters: An Empirical Test of the BOLD Molecular Identification Engine. Insects, 16(11), 1172. https://doi.org/10.3390/insects16111172







