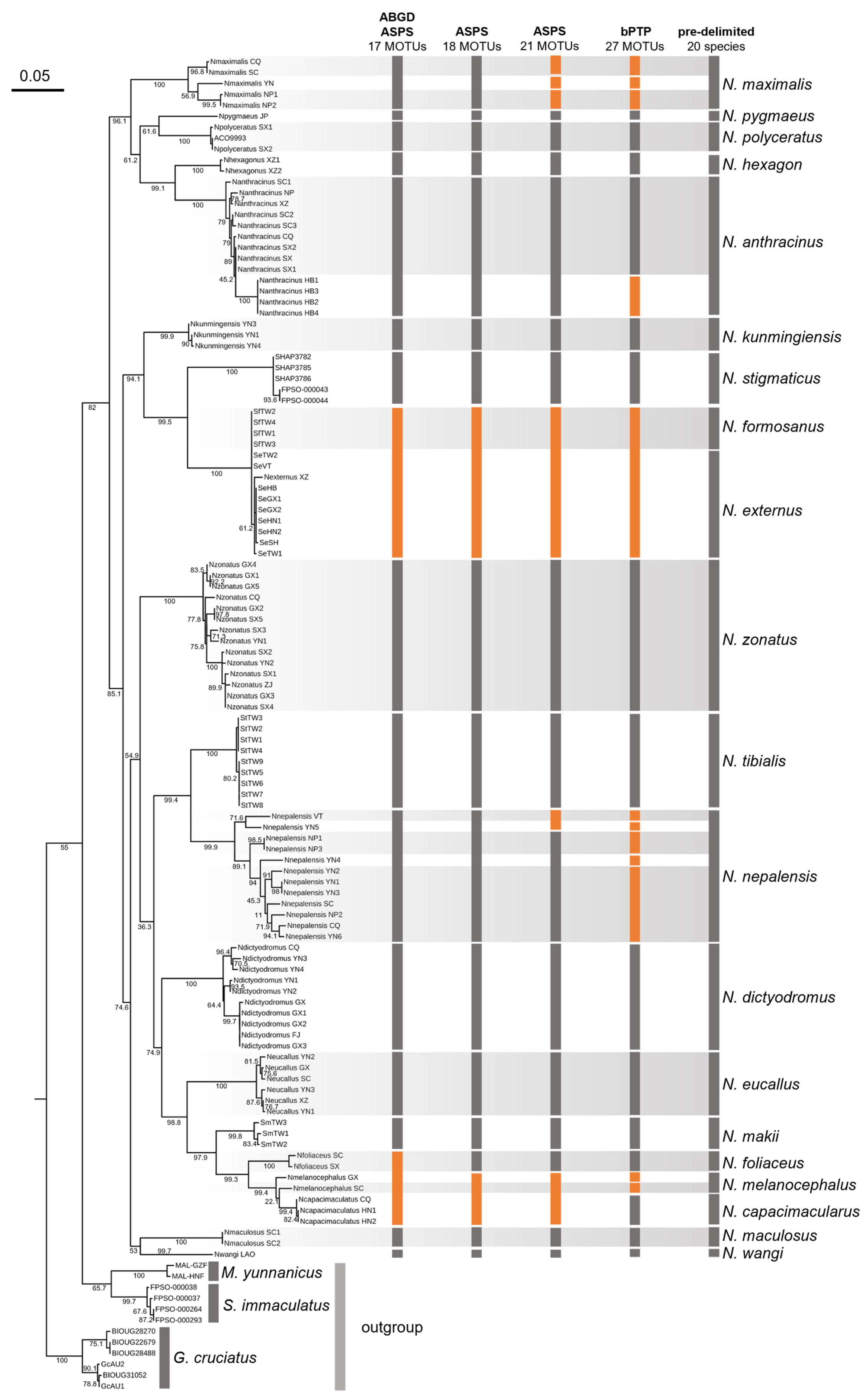
3.2. Systematics
Genus Neostenopsocus Liang & Liu, 2024
Neostenopsocus Liang & Liu, 2024 [
5]: 439. Type species: Stenopsocus externus Banks, 1937 [
38]: 259, by original designation.
Diagnosis: See Liang & Liu [
5].
Remarks: See Liang & Liu [
5].
Distribution: Oriental, Palearctic, and Australian regions.
Stenopsocus anthracinus Li, 1989 [
39]: 36. Type locality: China (Shaanxi: Zhenba).
Stenopsocus angustifurcus Li, 2002 [
2]: 658. Type locality: China (Guizhou: Dushan).
syn. nov. Stenopsocus biconvexus Li, 1997 [
40]: 428. Type locality: China (Hubei: Xingshan).
syn. nov. Stenopsocus bipunctatus Li, 2002 [
2]: 637. Type locality: China (Yunnan: Longling).
syn. nov. Stenopsocus cassideus Li, 1997 [
41]: 313. Type locality: China (Hunan: Sangzhi).
syn. nov. Stenopsocus dichospilus Li, 2002 [
2]: 657. Type locality: China (Anhui: Huangshan).
syn. nov. Stenopsocus flavifrons Li, 1989 [
39]: 35. Type locality: China (Shaanxi: Liuba).
syn. nov. Stenopsocus frontalis Li, 1989 [
39]: 38. Type locality: China (Shaanxi: Zhenba).
syn. nov. Stenopsocus fulivertex Li, 2002 [
2]: 647. Type locality: China (Hubei: Jiugongshan).
syn. nov. Stenopsocus parviforficatus Li, 2002 [
2]: 649. Type locality: China (Ningxia: Liupanshan).
syn. nov. Stenopsocus podorphus Li, 1997 [
40]: 435. Type locality: China (Hubei: Xingshan).
syn. nov. Stenopsocus qianipullus Li, 2005 [
42]: 92. Type locality: China (Guizhou: Dashahe).
syn. nov. Stenopsocus symipsarous Li, 2002 [
2]: 656. Type locality: China (Guizhou: Guiyang).
syn. nov. Diagnosis. This species exhibits noticeable sexual dimorphism in body coloration. The females are characterized by the antenna, with 9–13 antennomeres whitish, forewing pterostigma with brown marking extending to mid of r-rs, and black–brown genital segments. In contrast, males display paler overall pigmentation, uniformly brown antennae, and forewing pterostigma with brown marking not extending to r-rs.
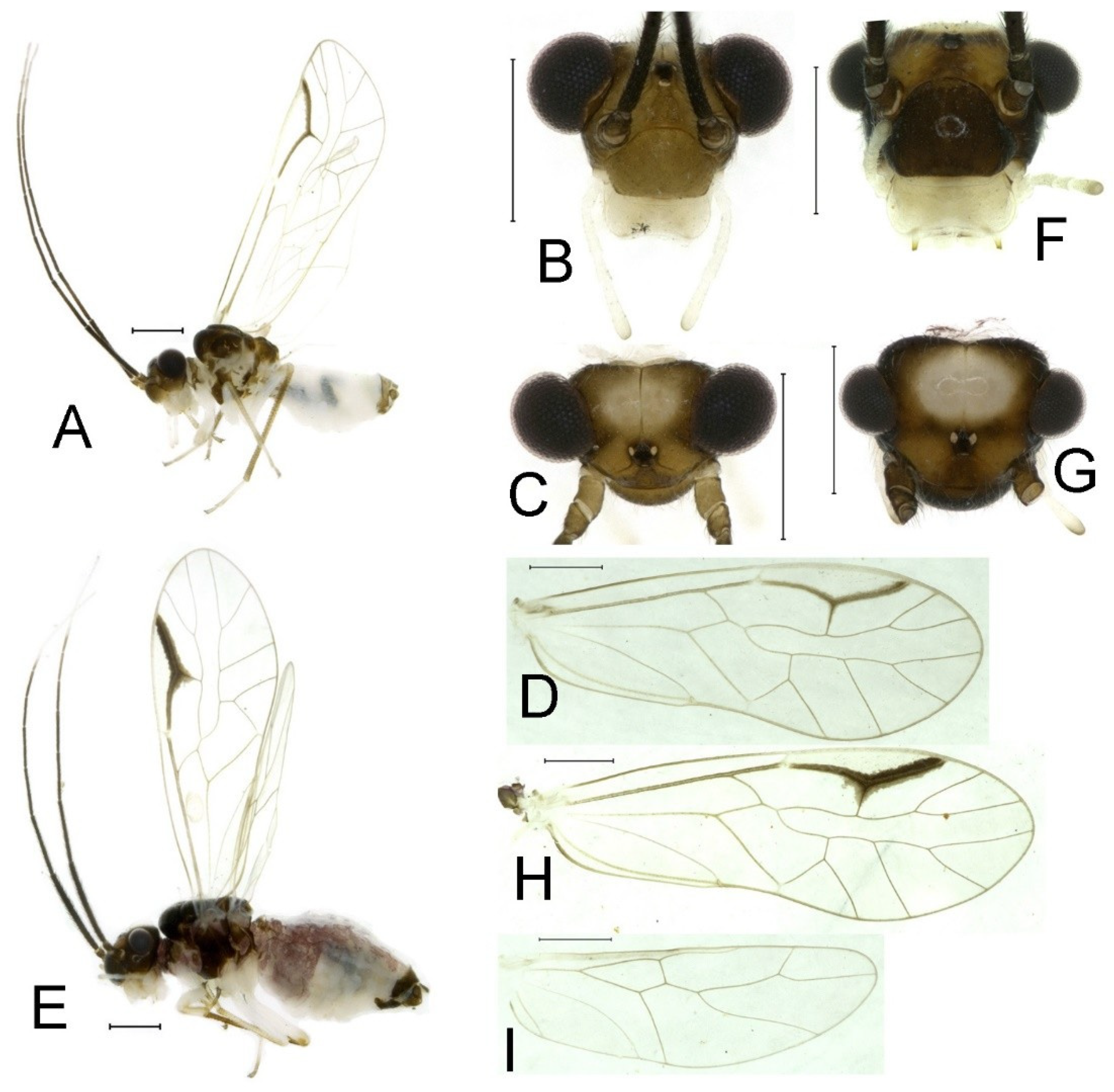
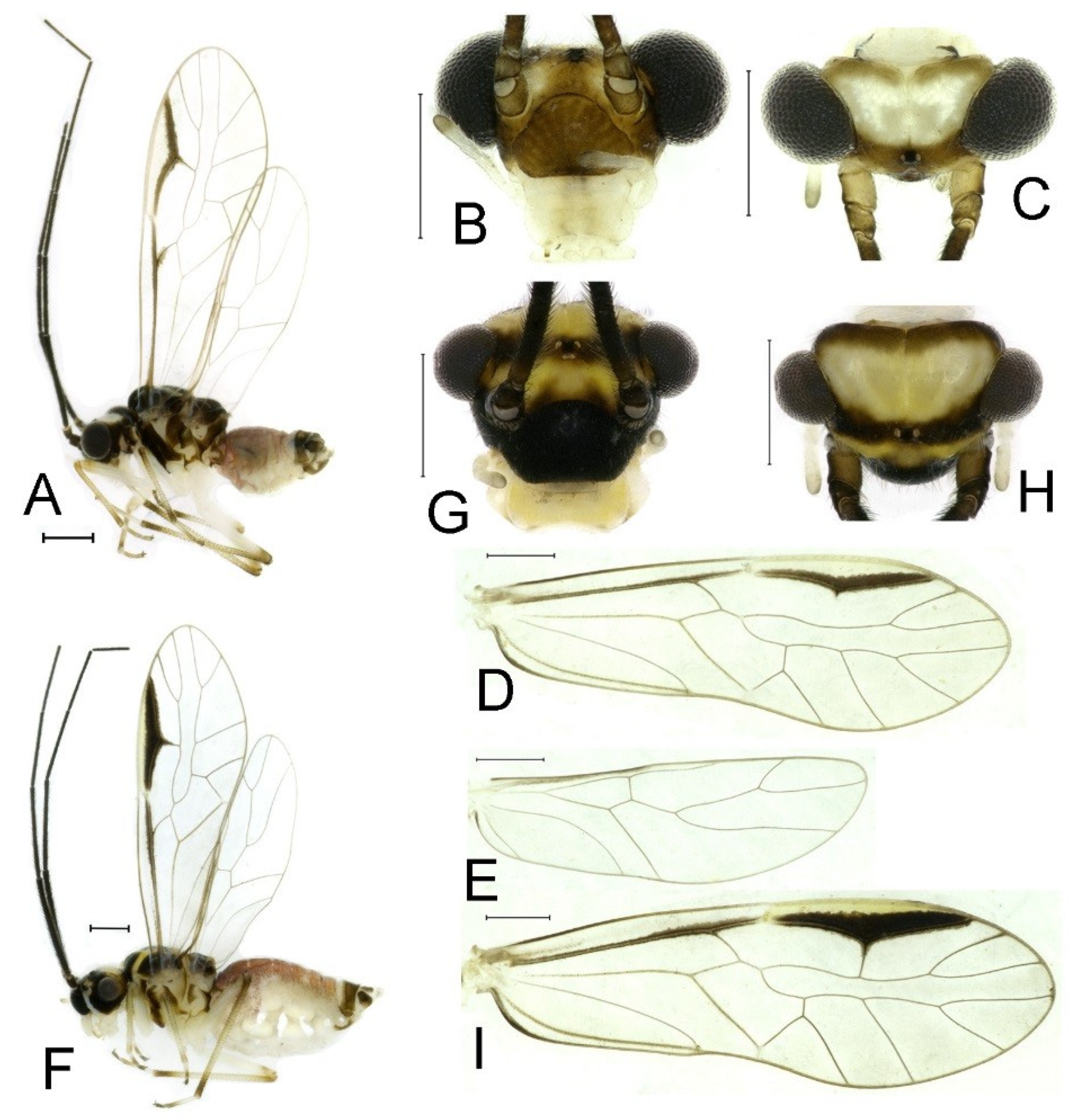
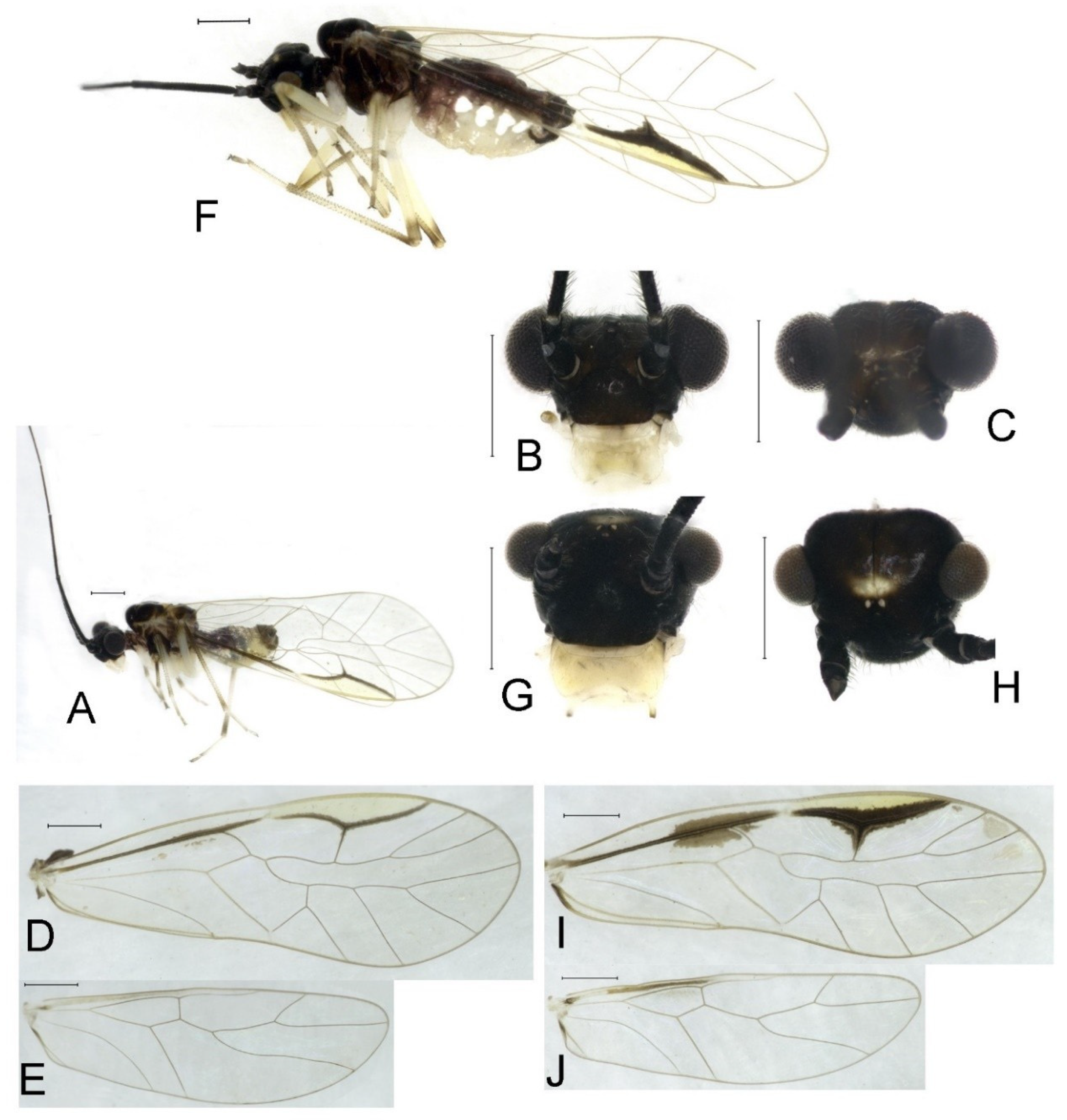
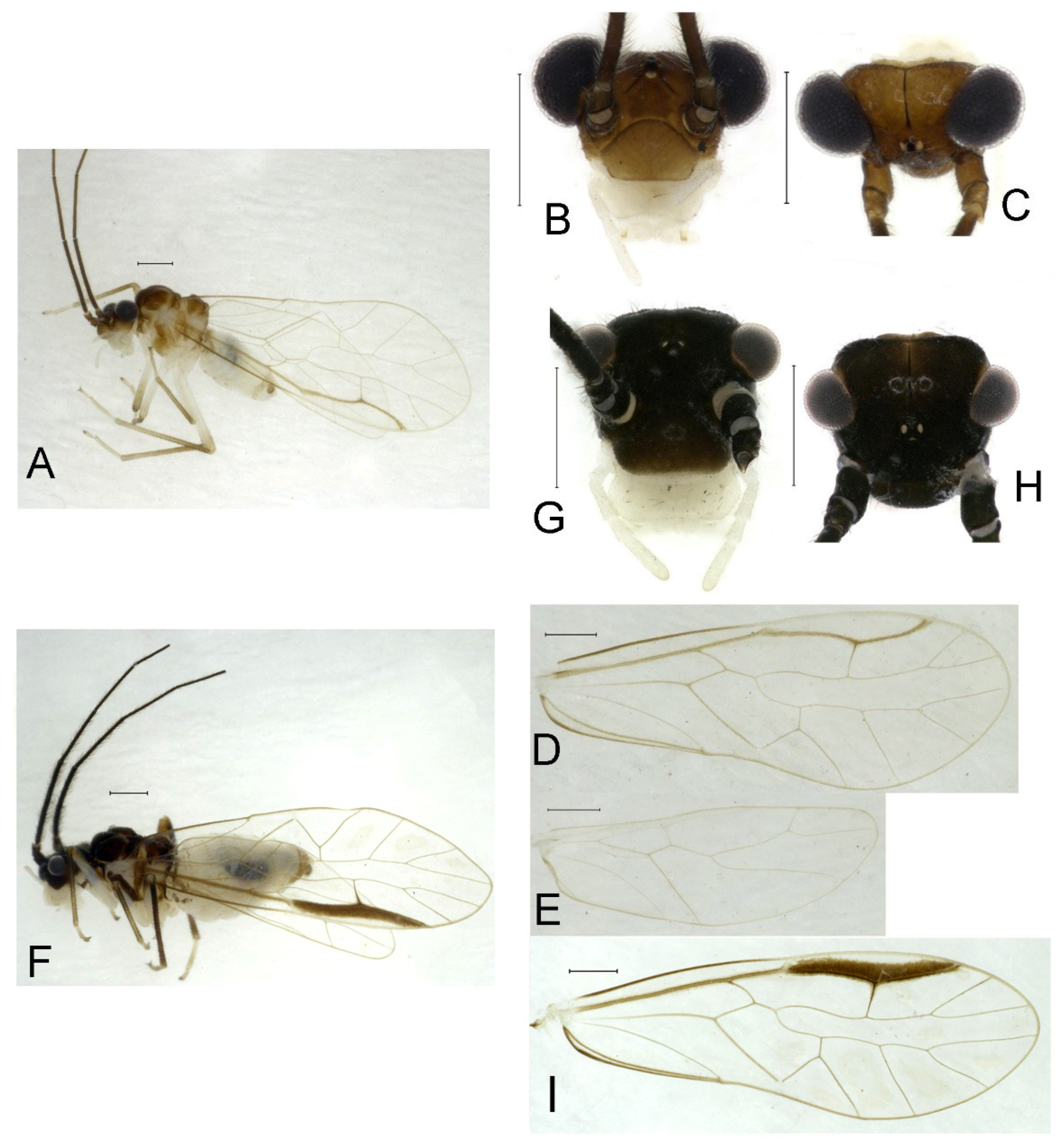
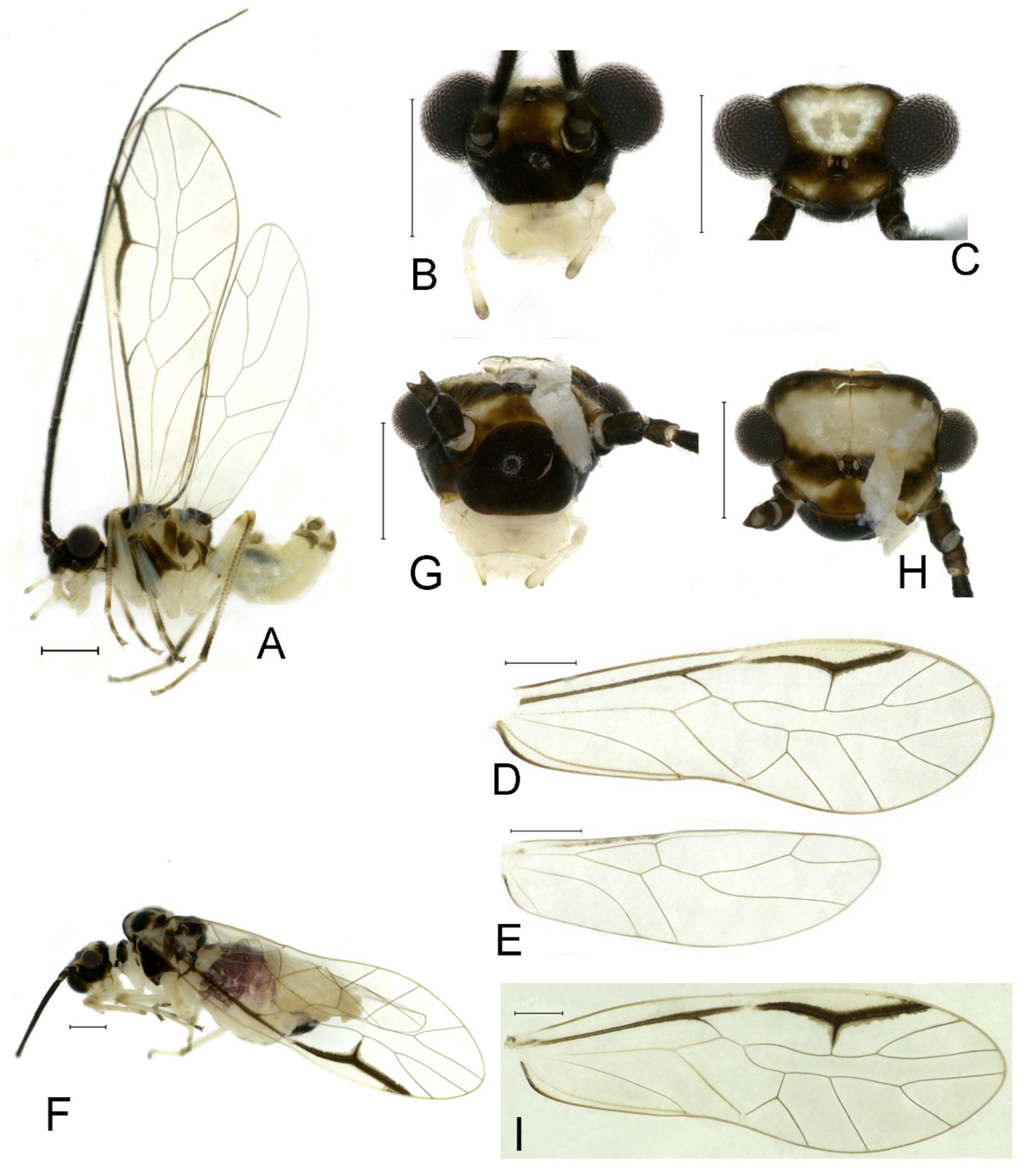
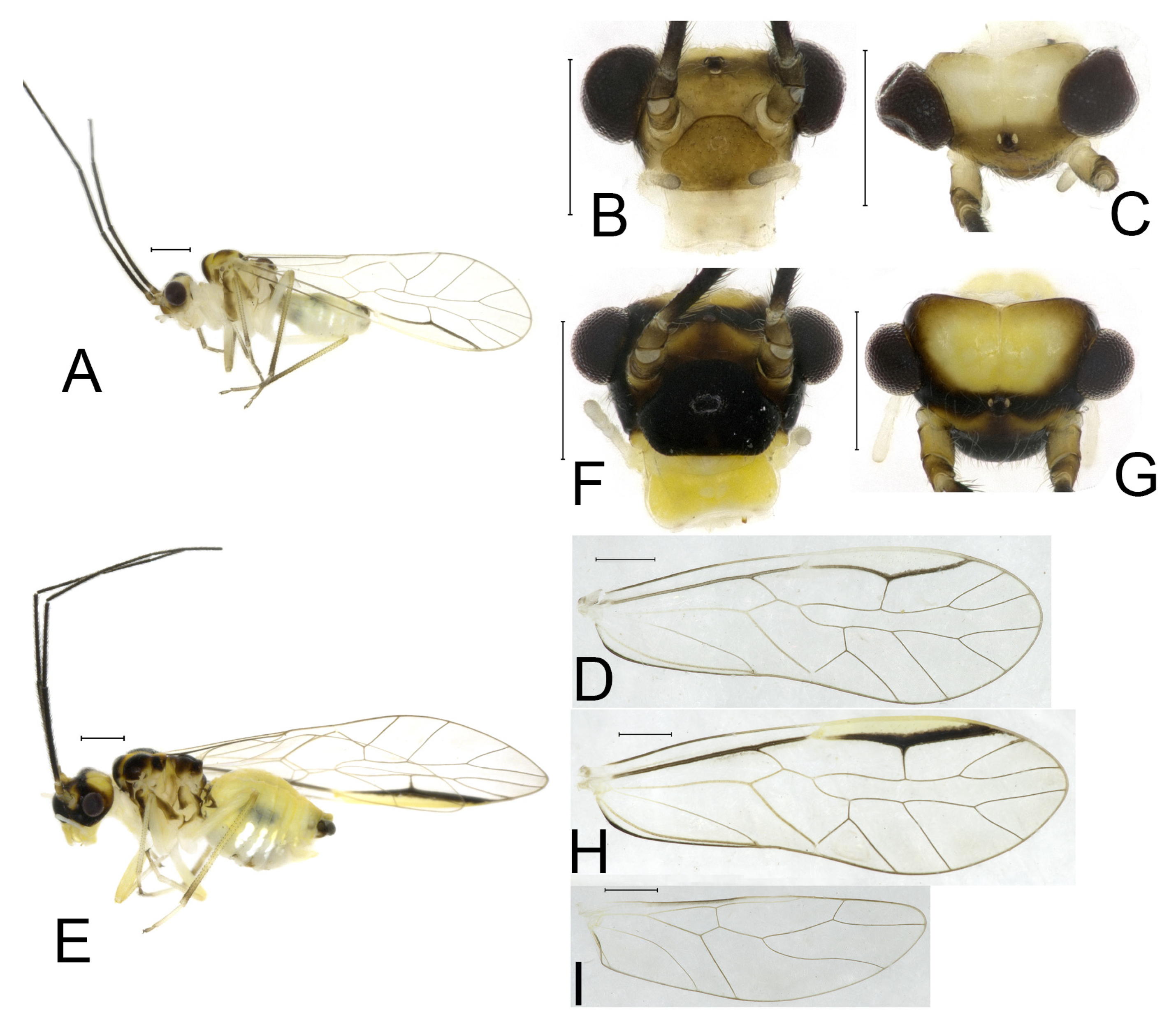
Redescription. Adult male: Body (
Figure 2A) length 2.42 mm, length from postclypeus to wing tip 4.42 mm. IO: 0.29 mm; d: 0.27 mm; IO/d = 1.07; f1: 0.76 mm; f2: 0.59 mm; f3: 0.51 mm; FWL: 3.65 mm; FWW: 1.34 mm; HWL: 2.69 mm; HWW: 0.88 mm; t1: 0.42 mm; t2: 0.15 mm.
Color (in alcohol): Head (
Figure 2B,C) yellowish brown, vertex with a yellowish area. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax dark brown. Leg femur mostly yellowish, tibia brown. Abdomen yellowish white, genital segments brown.
Forewing (
Figure 2D) transparent. R dark brown. Posterior margin of pterostigma with narrow brown stripe not extending along r-rs. Hindwing immaculate.
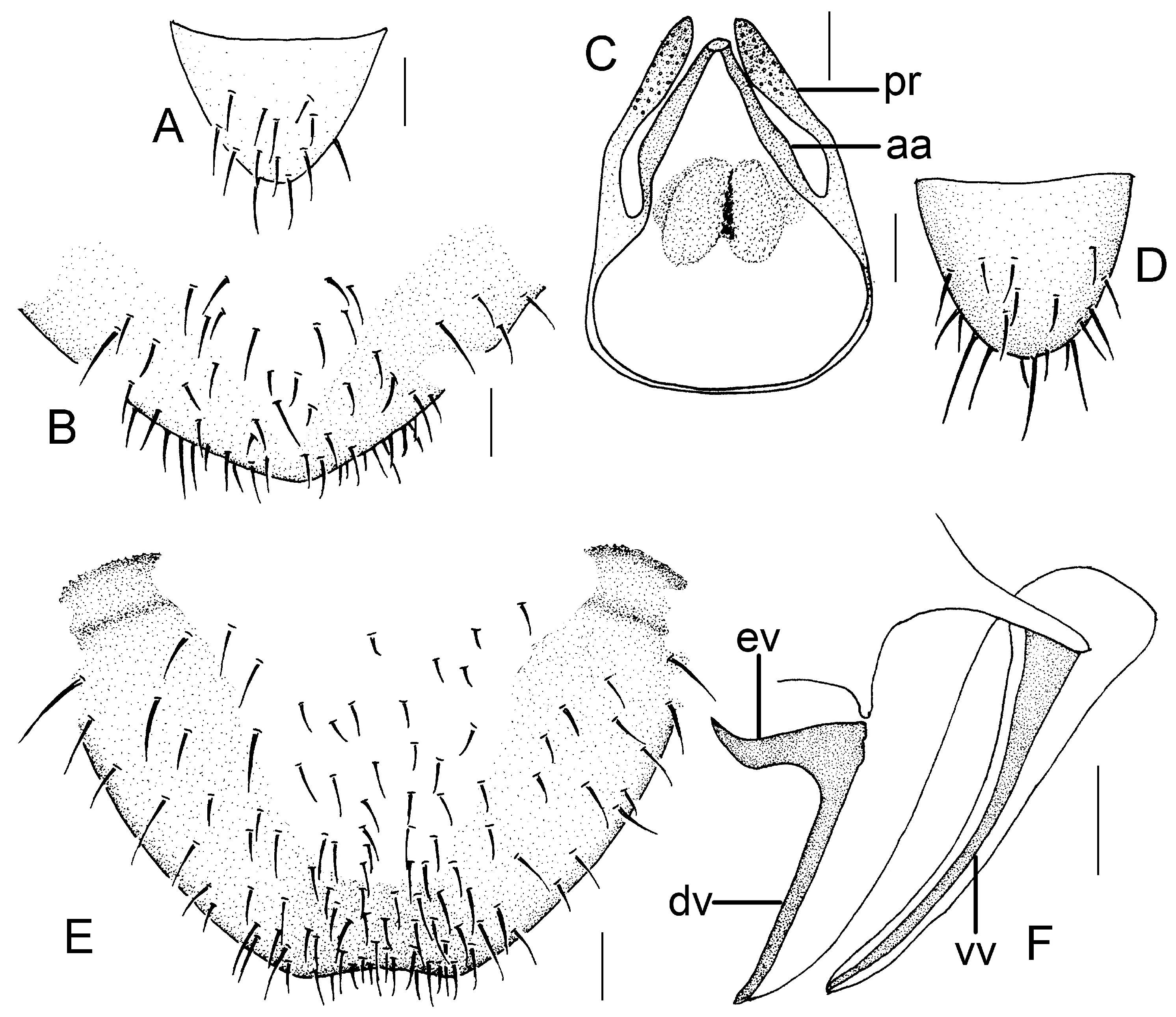
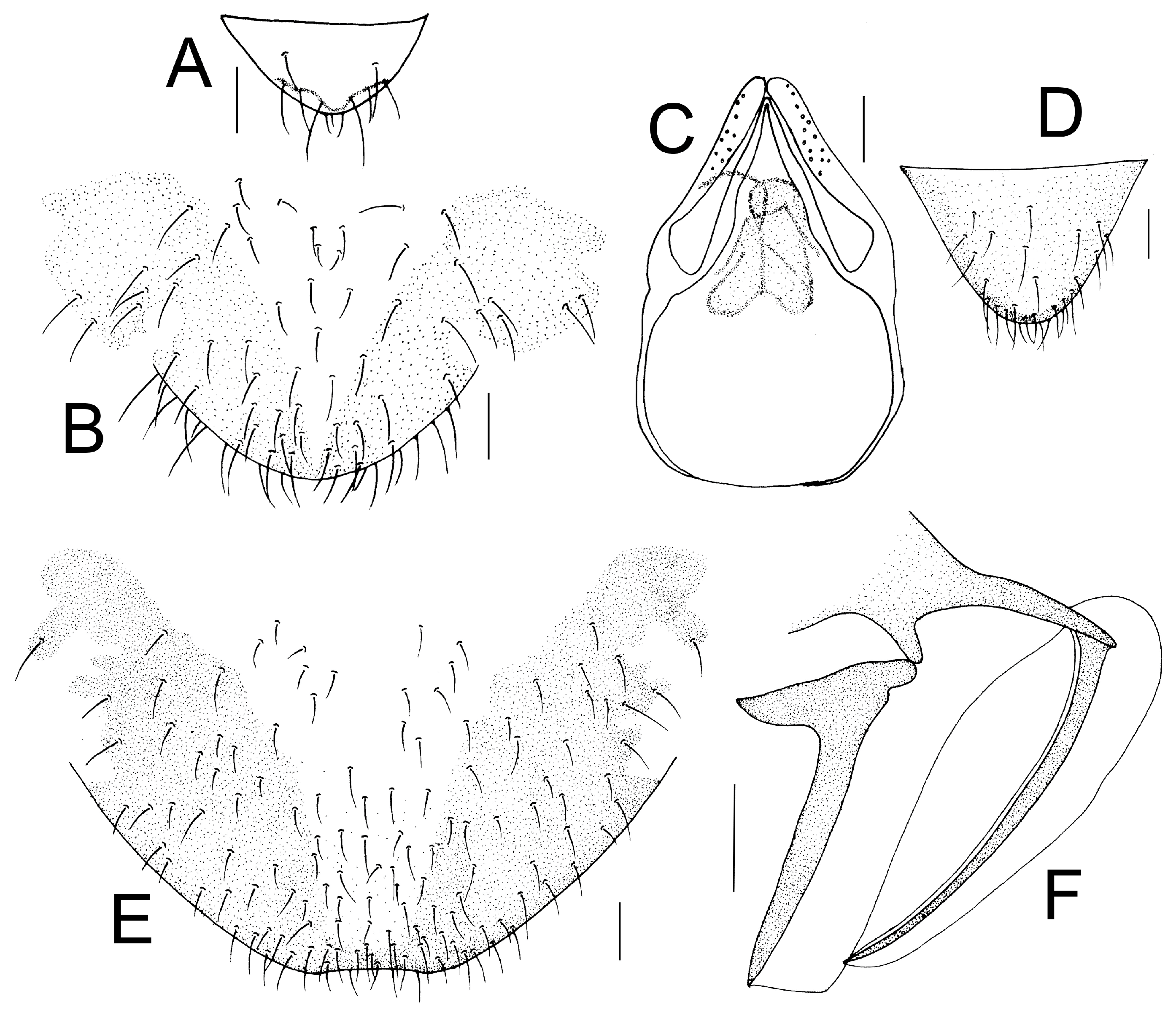
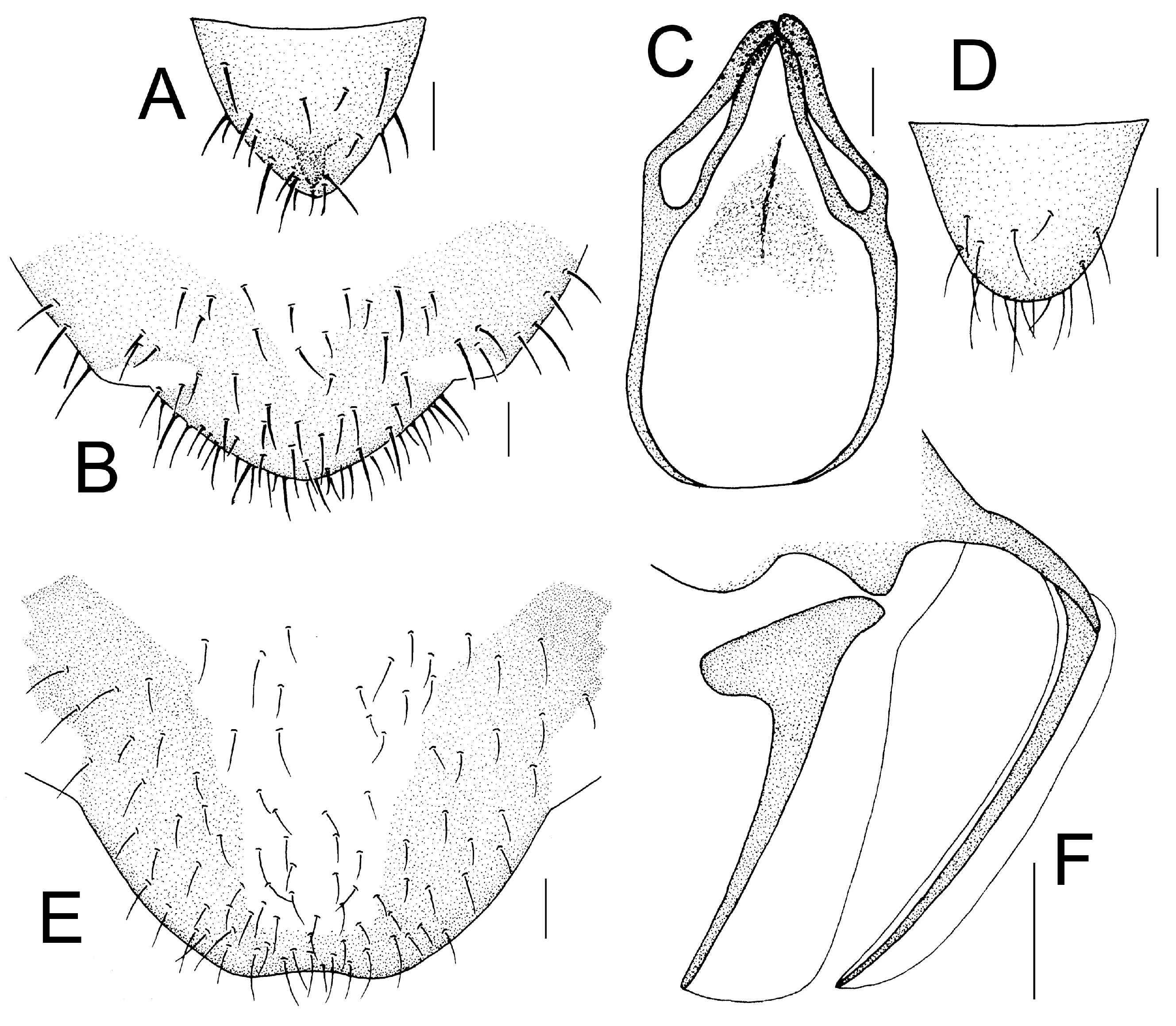
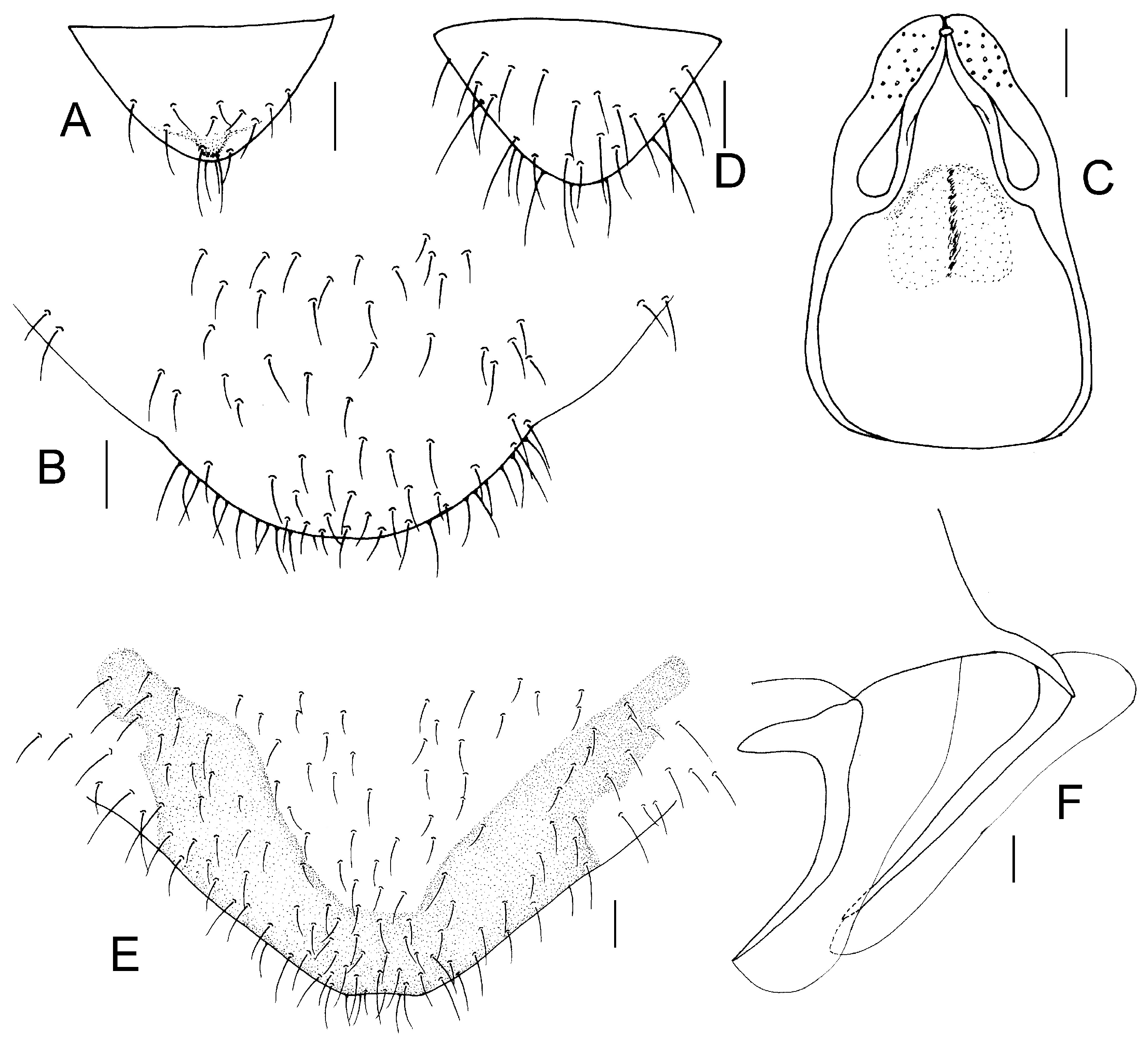
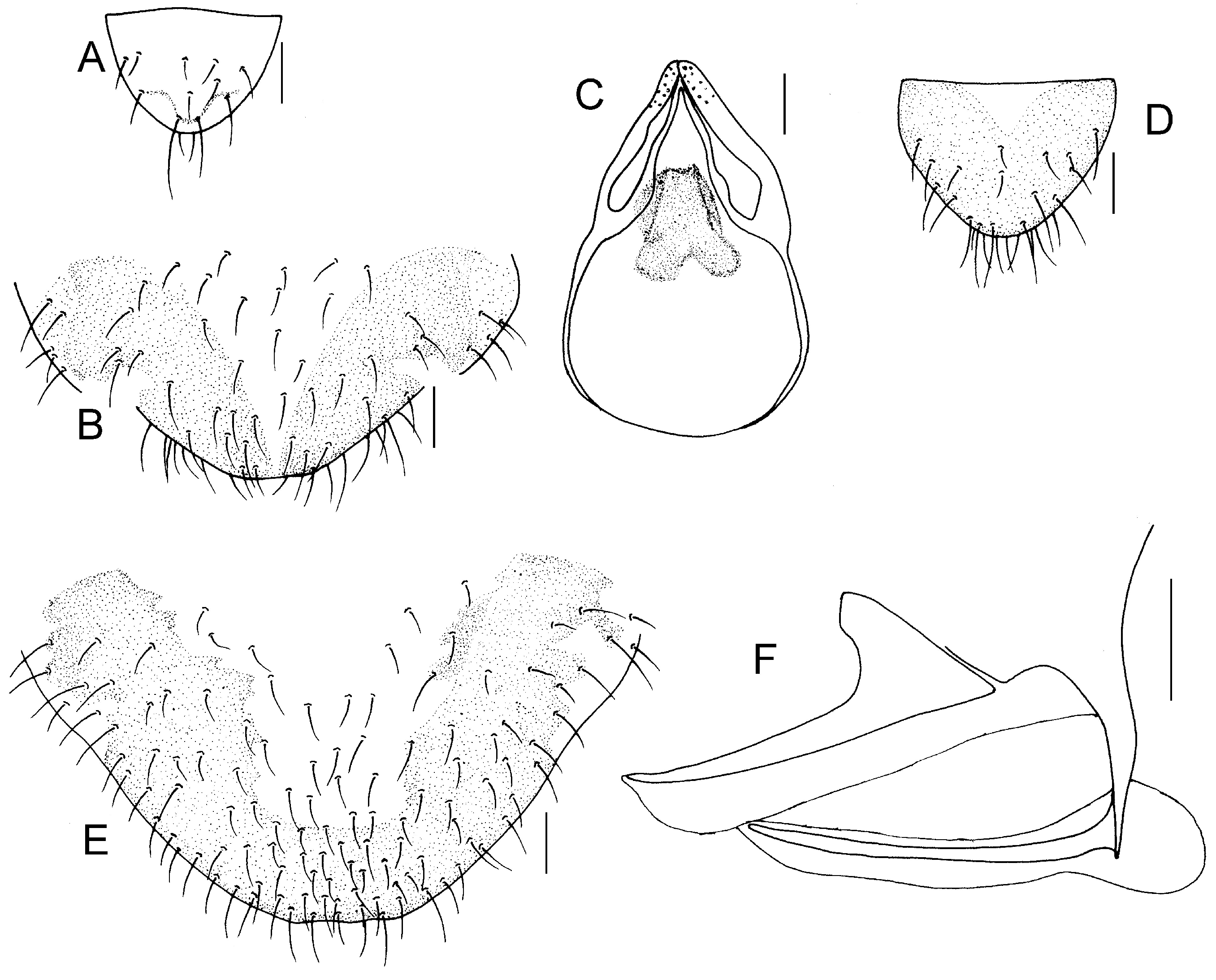
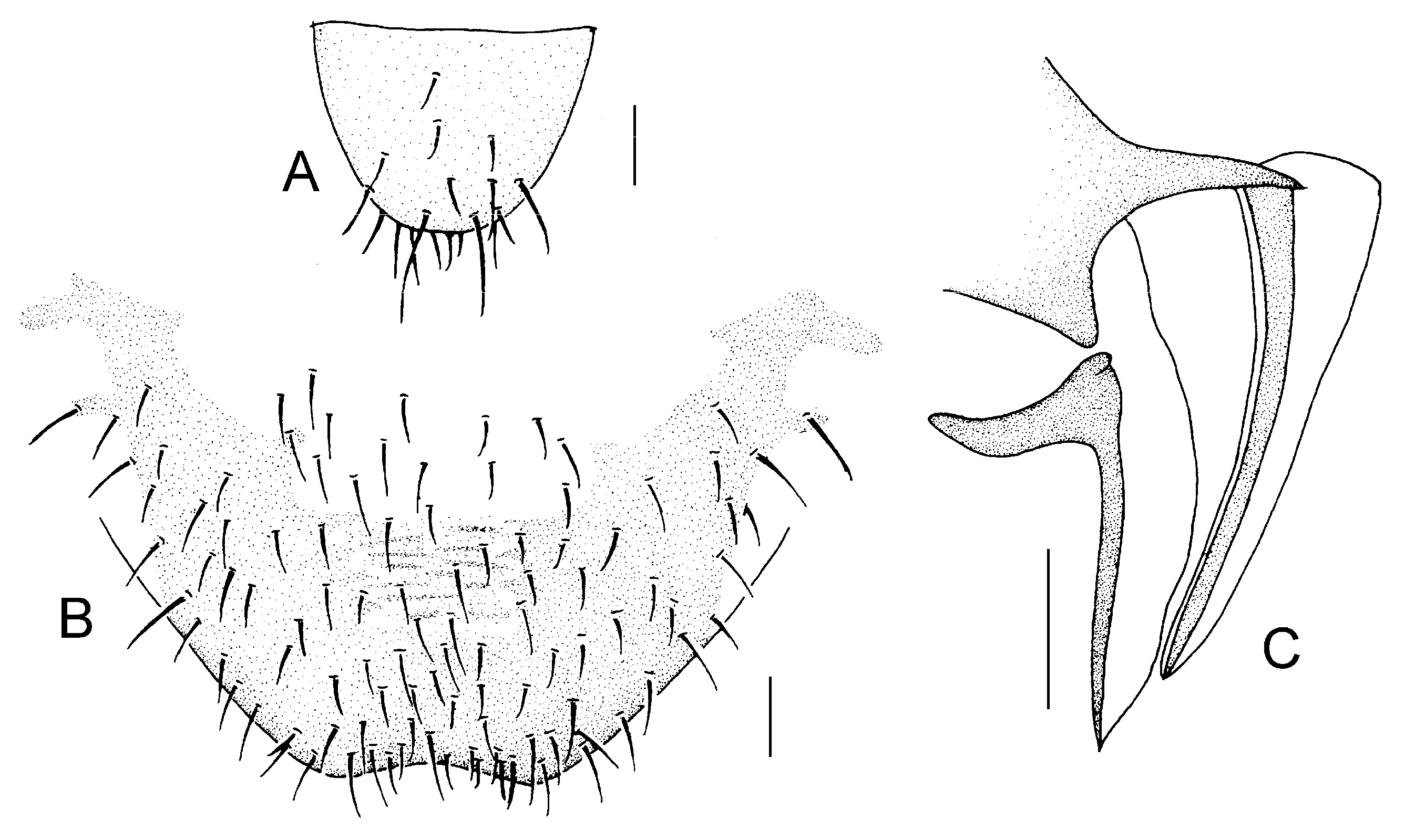
Genital segments strongly sclerotized. Epiproct (
Figure 3A) subtriangular. Paraproct with 22 trichobothria. Hypandrium (
Figure 3B) sclerotized strongly. Endophallus (
Figure 3C) strongly sclerotized, external parameres robust, with some punctures on apex, and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 2E) length 2.68 mm, length from postclypeus to wing tip 5.02 mm. IO: 0.44 mm; d: 0.14 mm; IO/d = 3.14; f1: 0.89 mm; f2: 0.65 mm; f3: 0.45 mm; FWL: 4.08 mm; FWW: 1.41 mm; HWL: 2.86 mm; HWW: 0.91 mm; t1: 0.40 mm; t2: 0.14 mm.
Color similar to male, slightly darker (
Figure 2E–I). Forewing (
Figure 2H) markings much larger than those in male. Antenna with 9–13 antennomeres whitish. Pterostigma with brown marking extending to mid of r-rs. Abdomen purplish red, 4–8 segments ventrally yellowish white. Genital segments dark brown.
Genital segments strongly sclerotized. Epiproct (
Figure 3D) subtrapezoidal. Paraproct with 30 trichobothria. Subgenital plate (
Figure 3E) with a broad sclerotized area. External valve short and robust, with a shark tip, almost perpendicular to dorsal valve (
Figure 3F).
Specimens examined. Holotype of
Stenopsocus anthracinus, ♂, China, Shaanxi, Zhenba (800 m), 1985.VII.20, Fasheng Li (CAU) (
Figure S3);
Holotype of
Stenopsocus angustifurcus, ♂, China, Guizhou, Dushan (1000 m), 1981.VI.1, Fasheng Li (CAU) (
Figure S4);
Holotype of
Stenopsocus biconvexus, ♂, China, Sichuan, Wushan, Liziping (1850 m), 1994.VI.1, Fasheng Li (CAU) (
Figure S5);
Holotype of
Stenopsocus bipunctatus, ♀, China, Yunnan, Longling (900 m), 1989.IX.17, Fasheng Li (CAU) (
Figure S6);
Holotype of
Stenopsocus cassideus, China, Hunan, Sangzhi, Mt. Tianping, 1981.IX.17, Xingwang Tong (CAU) (
Figure S7);
Holotype of
Stenopsocus dichospilus, ♂, China, Anhui, Mt. Huangshan, Beihai (2000 m), 1977.VIII.23, Fasheng Li (CAU) (
Figure S8);
Holotype of
Stenopsocus flavifrons, ♀, China, Shaanxi, Foping, Zhangliangmiao (1200 m), 1985.VII.23, Fasheng Li (CAU) (
Figure S9);
Holotype of
Stenopsocus fulivertex, ♂, China, Hubei, Tongshan, Mt. Jiugong, 1984.VI.16, Chikun Yang (CAU) (
Figure S10);
Holotype of
Stenopsocus frontalis, ♀, China, Shaanxi, Foping (1200 m), 1985.VII.16, Fasheng Li (CAU) (
Figure S11);
Holotype of
Stenopsocus parviforficatus, ♂, China, Ningxia, Mt. Liupanshan, Erlong River (2100 m), 1980.VIII.4, Fasheng Li (CAU) (
Figure S12);
Holotype of
Stenopsocus podorphus, ♀, China, Hubei, Xingshan, Longmen River (1300 m), 1994.IX.8, Fasheng Li (CAU) (
Figure S13);
Holotype of
Stenopsocus qianipullus, ♀, China, Guizhou, Daozhen, Dashahe Nature Reserve (1400 m), 2004.VIII.17, Maofa Yang (CAU) (
Figure S14);
Holotype of
Stenopsocus symipsarous, ♂, China, Guizhou, Huaxi (1000 m), Guiyang, Guizhou, 1981.V.22, Chikun Yang (CAU) (
Figure S15).
CHINA: 3♀♀, Sichuan, Mt. Emei, Linggongli, 2012.IX.2, Xuankun Li (CAU); 1♀, Sichuan, Mt. Emei, Leidongping, 2012.IX.3, Ding Yang (CAU); 1♀, Sichuan, Mt. Emei, 2012.VI.8-15, Xiao Zhang (CAU); 1♀, Sichuan, Tianshidong (1000 m), Mt. Qingcheng, 1978.V.2, Fasheng Li (CAU); 1♀, Sichuan, Linggongli, Mt. Emei, 2010.VII.15, Junchao Wang (CAU); 3♂, Leidongping, Mt. Emei, Sichuan, 2012.IX.3, Xuankun Li (CAU); 1♀, 1♀, Xishan (2100 m), Kunming, Yunnan, 1981.V.16, Fasheng Li (CAU); 1♂, Yunnan, Nanjian, Mt. Wuliang (2221 m), 2016.VII.16, Qicheng Yang (CAU); 1♀, Chongqing, Yintiaoling, 2022.VII.8, Leran Cao (CAU); 2♂♂1♀, Hubei, Yinshan, Mt. Wujiashan (715 m), Wencheng Chang (CAU); 1♂1♀, Yunnan, Nujiang, Pianma, 2013.VII.6, Ziqiang Sun (CAU); 2♀♀, Yunnan, Pingbian, Mt. Daweishan (1962 m), 2016.VII.15, Yanan Lv (CAU).
Distribution. China (Shaanxi, Yunnan, Guizhou, Sichuan, Hubei, Hunan, Anhui, Guangxi, Zhejiang, Ningxia, Gansu).
Remarks. Li [
2,
39,
40,
41,
42] described
N. anthracinus,
N. flavifrons, and
N. frontalis from Shaanxi;
N. angustifurcus,
N. qianpullus, and
N. symipsarous from Guizhou;
N. biconvexus from Sichuan;
N. bipunctatus from Yunnan;
N. cassideus from Hunan;
N. dichospilus from Anhui;
N. fulivertex and
N. podorphus from Hubei; and
N. parviforficatus from Ningxia. Some of the above species were proposed based on a single male or female specimen. After examining the type specimens of each species and comparing them with the specimens collected in recent years, the latter 13 species are considered synonyms of
N. anthracinus based on the yellowish vertex, the similar color of antenna, and the marking pattern of forewing. We matched the male and female of this species based on the DNA barcoding in the present study. The males and females of this species exhibit distinct differences in their pterostigma markings. Females possess a brown band along the posterior margin of the pterostigma, while males have an extremely narrow and inconspicuous brown band.
N. anthracinus shares a similar external morphology with
N. tibialis. In the NJ tree, these two species clustered within one clade. We consider these two species as valid based on the interspecific K2P distance of 0.1631. Moreover,
N. anthracinus is distributed in the southern Chinese mainland, while
N. tibialis is only distributed in Taiwan Island.
Stenopsocus capacimacularus Li, 1993 [
43]: 347. Type locality: China (Guangdong: Chebaling).
Stenopsocus tamdaoi Georgiev & Quang-Manh, 2024 [
44]: 4. Type locality: Vietnam (Vinh Phuc: Tam Dao).
syn. nov. Diagnosis. This species is characterized by the unique forewing markings near the pterostigma area.
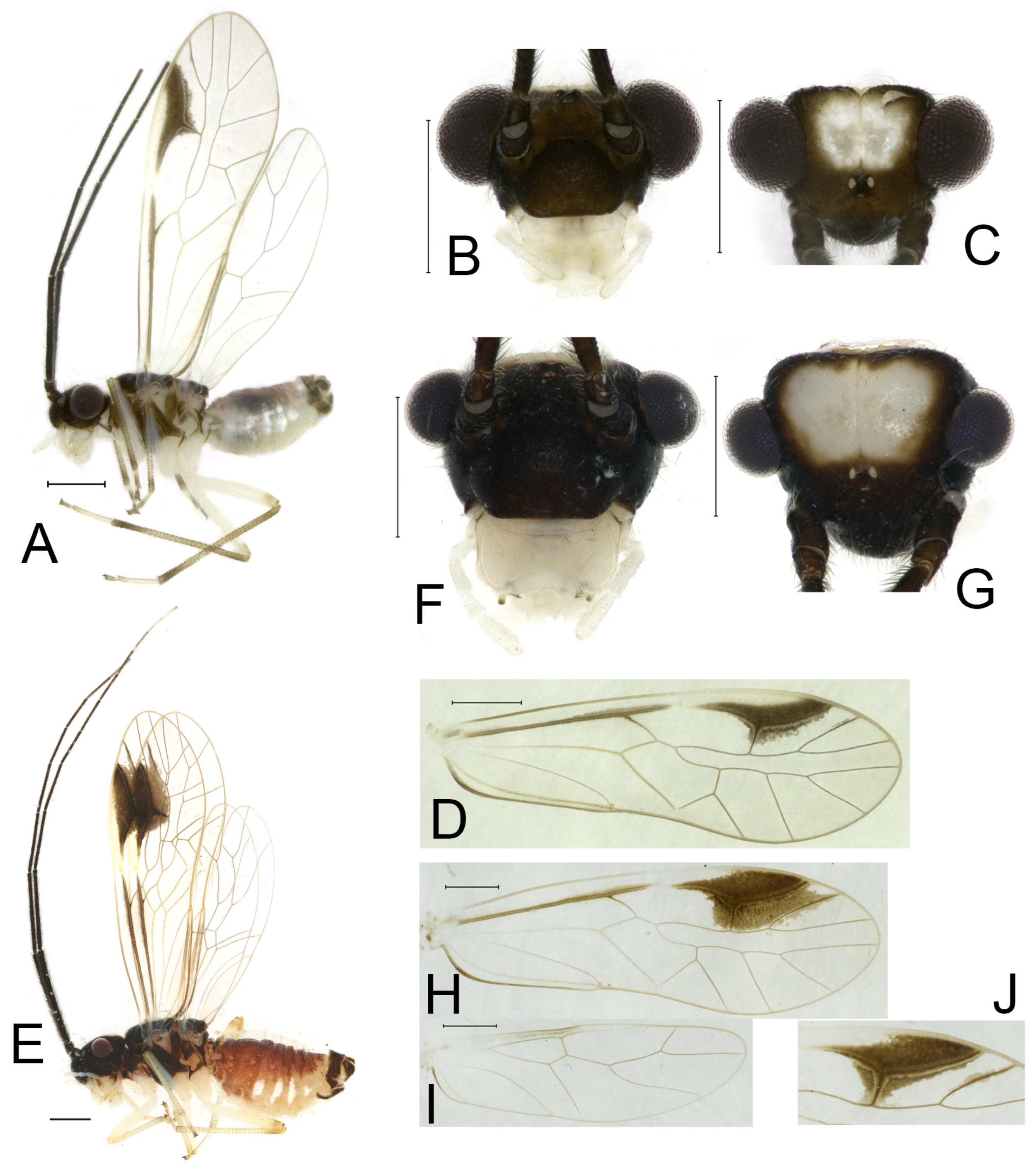
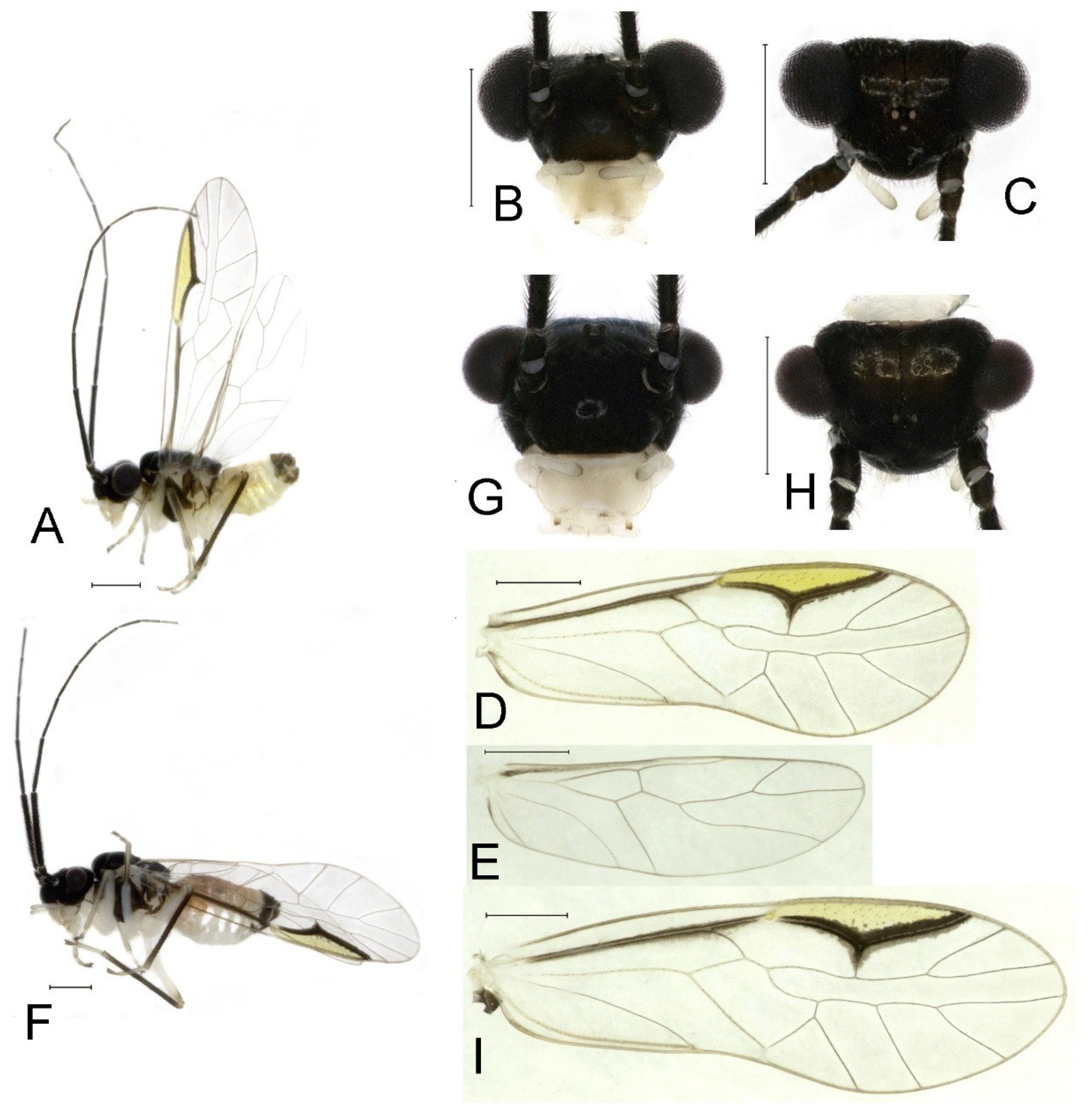
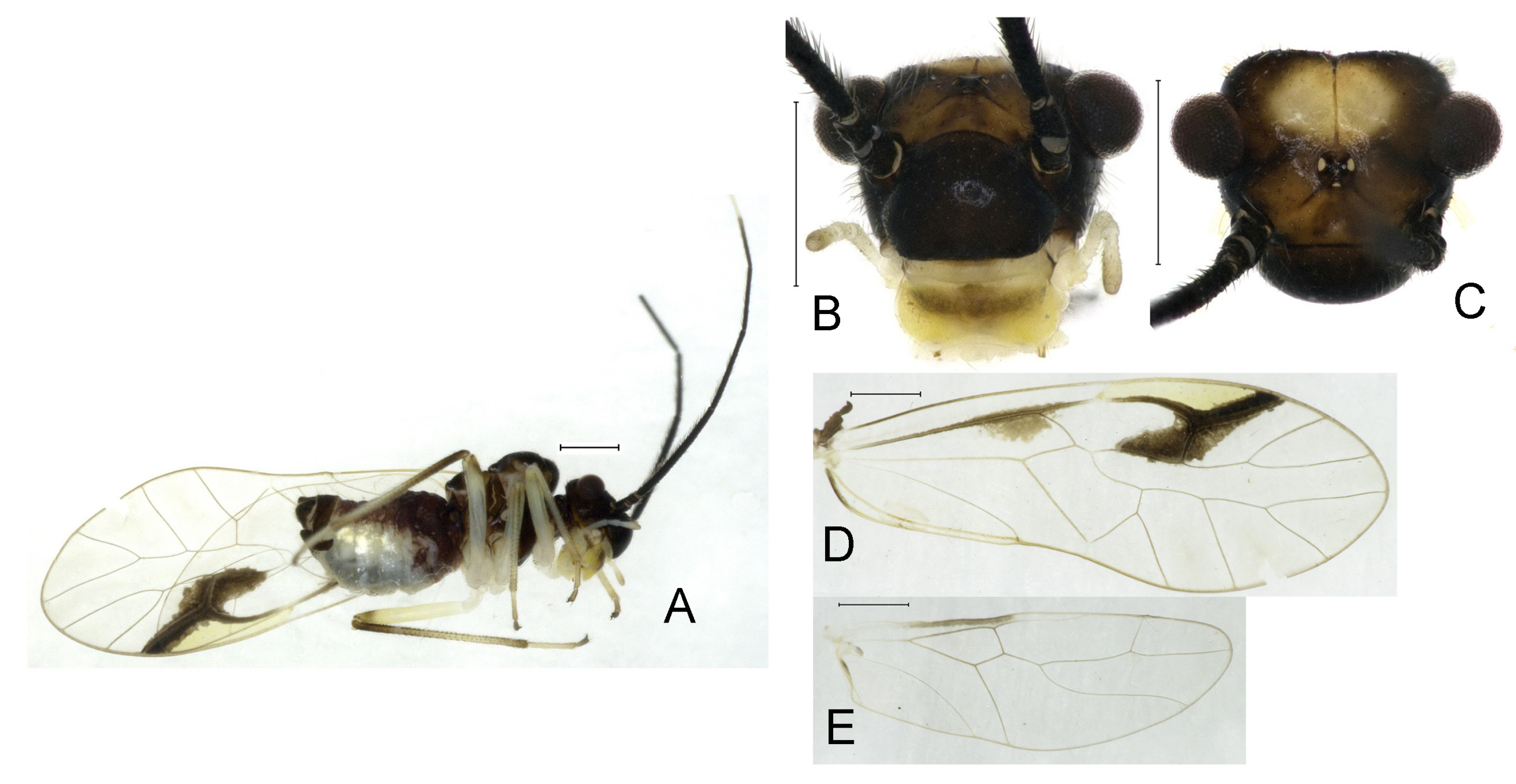
Redescription. Adult male: Body (
Figure 4A) length 2.55 mm, length from postclypeus to wing tip 4.00 mm. IO: 0.31 mm; d: 0.27 mm; IO/d = 1.15; f1: 0.94 mm; f2: 0.83 mm; f3: 0.64 mm; FWL: 3.38 mm; FWW: 1.11 mm; HWL: 2.55 mm; HWW: 0.75 mm; t1: 0.37 mm; t2: 0.12 mm.
Color (in alcohol): Head (
Figure 4B) dark brown, vertex (
Figure 4C) with a subtrapezoidal yellowish area. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax dark brown. Leg femur mostly yellowish, tibia brown, 1st tarsomeres and entire 2nd tarsomere brown. Abdomen dorsally pale purplish, ventrally yellowish white, genital segments brown.
Forewing transparent (
Figure 4D). R dark brown. Pterostigma yellowish, with brown marking which do not extend along r-rs. Hindwing with pale brown markings present between Sc and R.
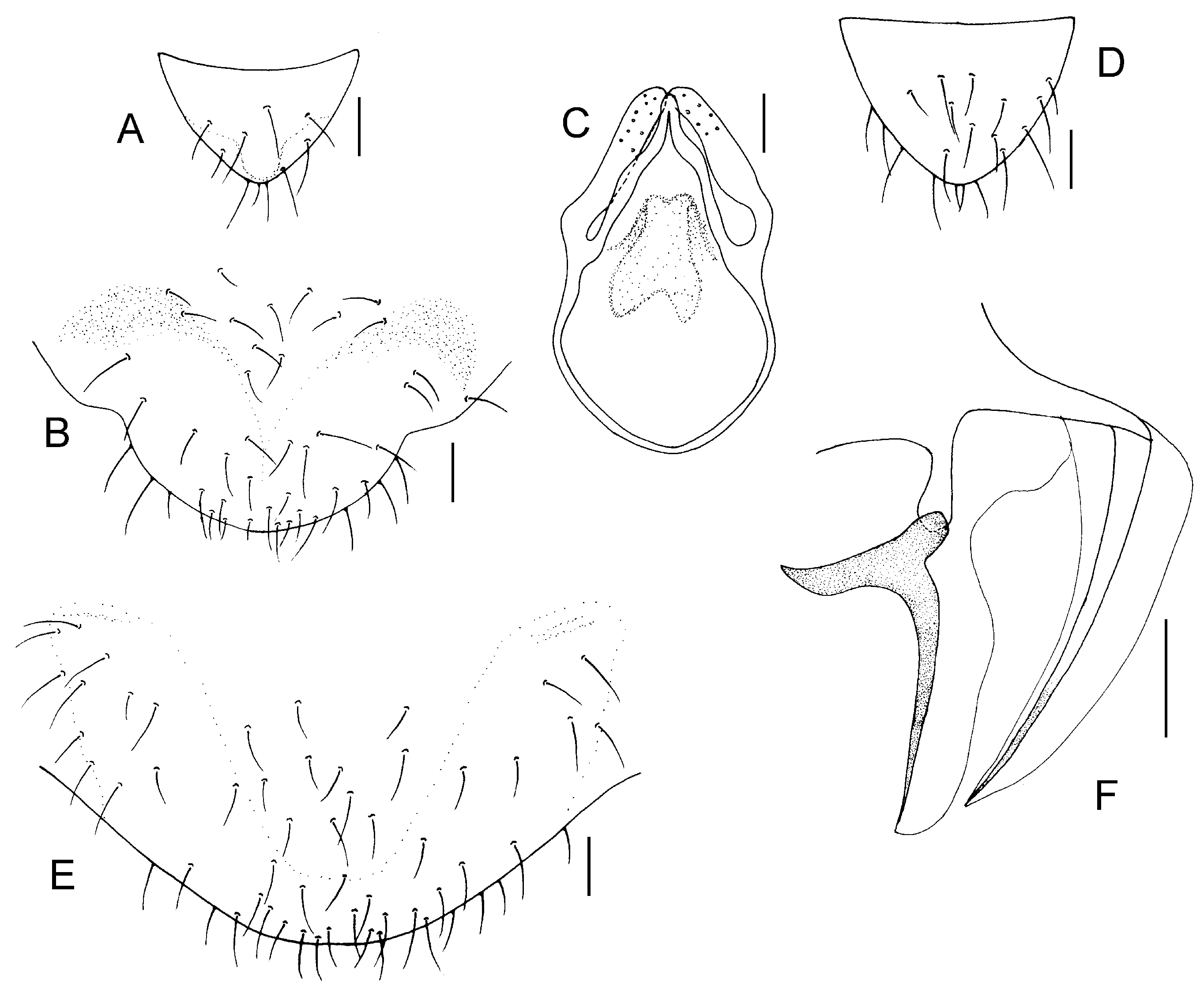
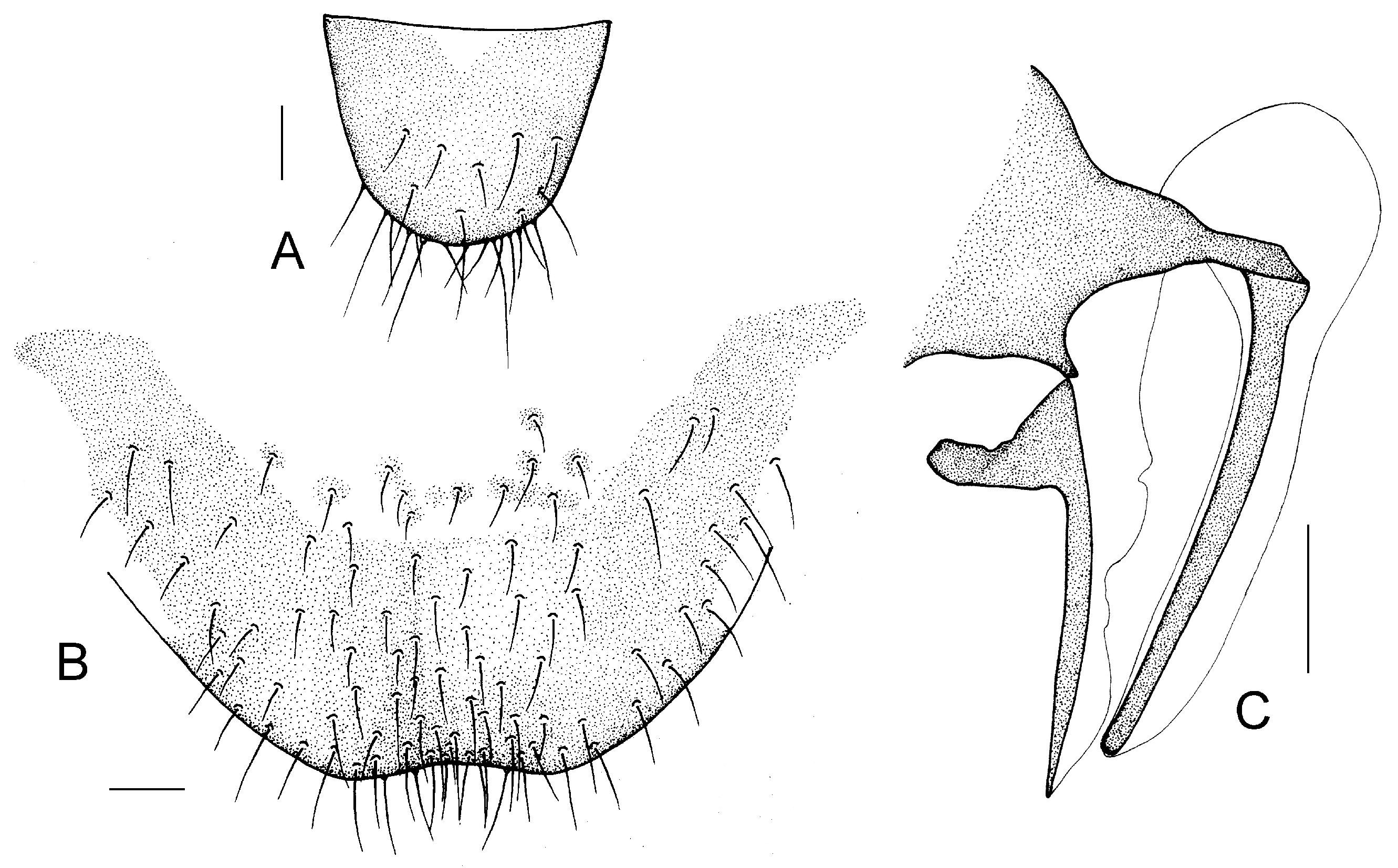
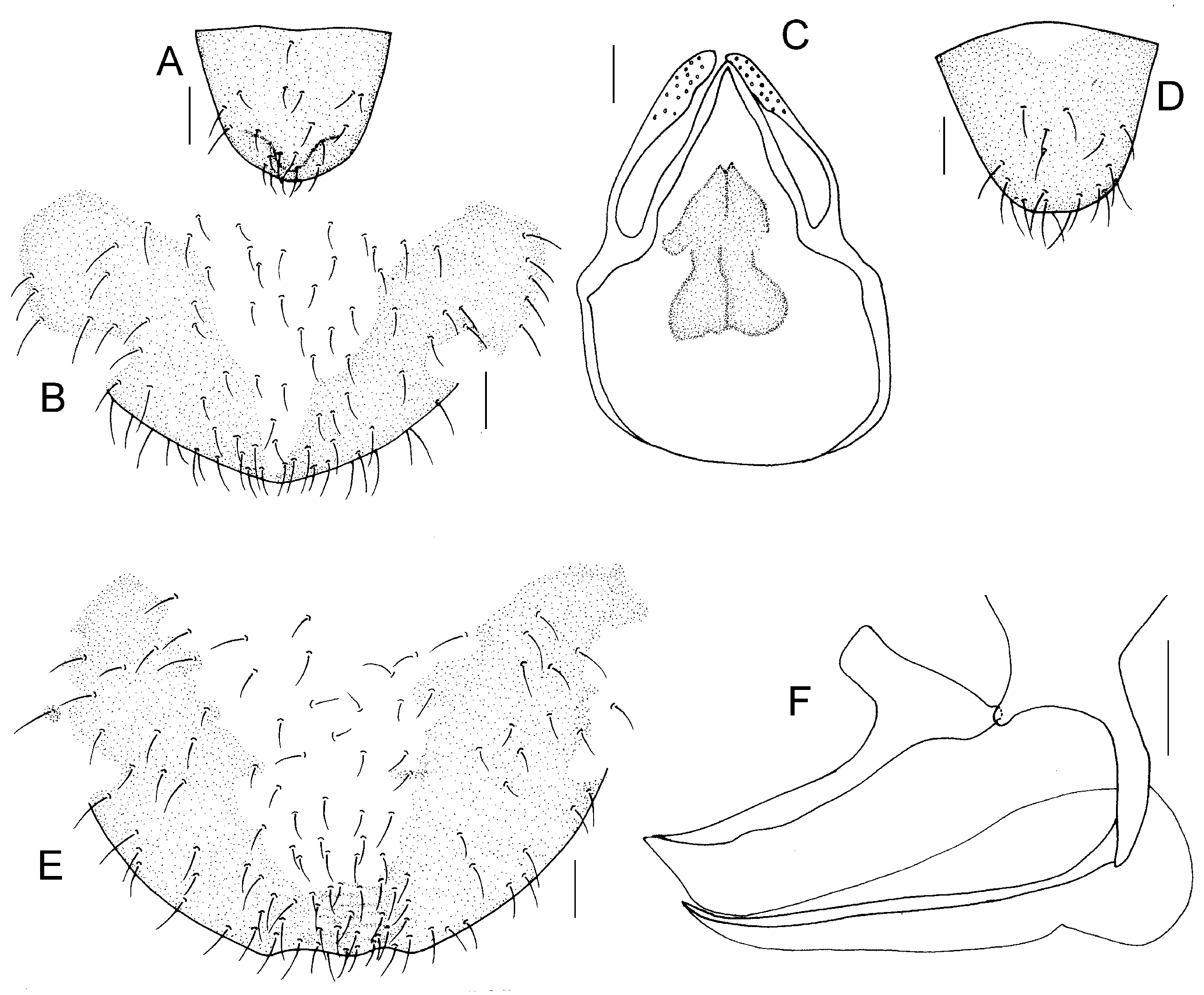
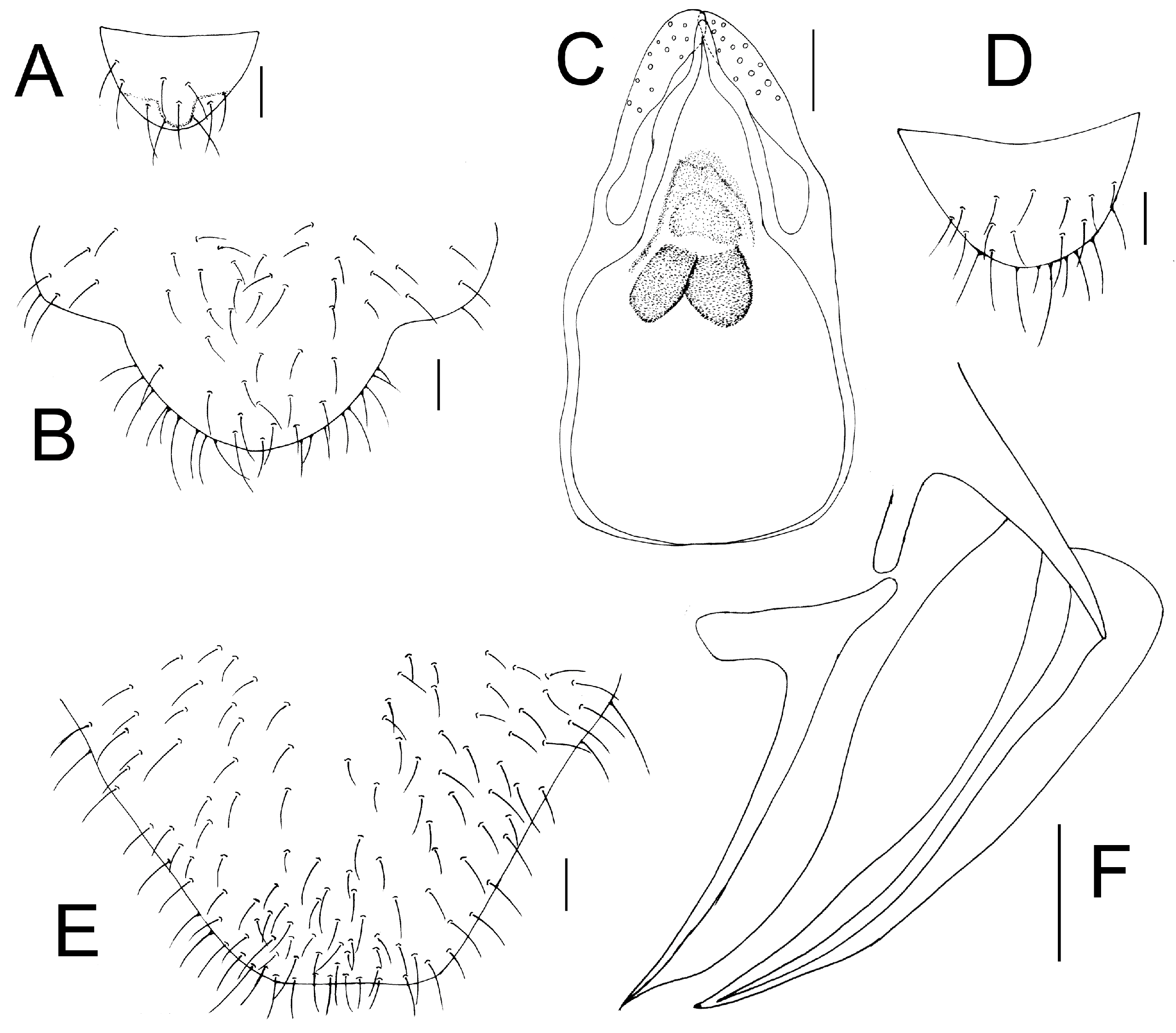
Genital segments strongly sclerotized. Epiproct (
Figure 5A) subtriangular. Paraproct with 27 trichobothria. Hypandrium (
Figure 5B) sclerotized strongly. Endophallus (
Figure 5C) strongly sclerotized, external parameres robust, with some punctures on broad end toward apex, and exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 4E) length 4.10 mm, length from postclypeus to wing tip 5.33 mm. IO: 0.60 mm; d: 0.25 mm; IO/d = 2.4; f1: 1.29 mm; f2: 1.12 mm; f3: 0.84 mm; FWL: 4.17 mm; FWW: 1.41 mm; HWL: 3.07 mm; HWW: 0.92 mm; t1: 0.38 mm; t2: 0.14 mm.
Color similar to male, slightly darker. Forewing (
Figure 4H) markings much larger than those in male. More than 2/3 area of pterostigma with a big brown marking. Abdomen purplish red, with 4–8 segments ventrally yellowish white.
Genital segments strongly sclerotized. Epiproct (
Figure 5D) subtrapezoidal. Paraproct with 30 trichobothria. Subgenital plate (
Figure 5E) with a broad sclerotized area. External valve short and robust, with a shark tip, almost perpendicular to dorsal valve (
Figure 5F).
Specimens examined. Holotype of
Stenopsocus capacimacularus, ♀, Chebaling Nature Reserve, Shixing, Guangdong, 1991.IV.23, Fasheng Li (CAU) (
Figure S16).
CHINA: 5♀♀, Zhejiang, Lin’an, Mt. Tianmushan, 2012.IX.4, Hong Wu (Malaise trap) (CAU); 2♂♂2♀♀, Mt. Daweishan, Liuyang, Hunan, 2019.V.28, Feiyang Liang (HNUST); ♀, Guangxi, Nanning, Wuming, Mt. Damingshan, 2013.IX.23, Xingyue Liu (CAU); 2♀♀, Fujian, Nanping, Tongmu, 2009.VII.6, Xiushuai Yang (CAU); 9♀♀, Zhejiang, Lin’an, Mt. Tianmushan (300–1000 m), 2016.IV.27, Feiyang Liang (CAU); 1♂3♀♀, Guangxi, Jinxiu, Yinshan Park (1150 m), 2016.V.22, Feiyang Liang (CAU); 4♂♂, Guangxi, Jinxiu, Lianhuashan Park (950 m), 2016.V.22, Xingyue Liu (CAU); 1♀, Guangxi, Tianlin, Mt. Cenwanglaoshan, Dalongping Station, 2016.V.8, Xingyue Liu (CAU); 1♀, Zhejiang, Qingliangfeng, Tianchi, 2012.X, Malaise trap (CAU).
Distribution. China (Guangxi, Guangdong, Hunan, Zhejiang) and Vietnam (Vinh Phuc).
Remarks. Li [
43] described
S. capacimacularus based on a single female specimen from Chebaling Nature Reserve, Guangdong. Although no specimens were collected from the type locality during our study, recent examinations have identified several individuals from Guangxi, Hunan, and Zhejiang as conspecific based on the vertex coloration and forewing marking patterns. This species exhibits notable intraspecific variation in the forewing markings. The female specimens from Mt. Tianmu (Zhejiang) revealed two distinct marking patterns: a large-marked form (
Figure 4H) and a small-marked form (
Figure 4J). Additionally, male individuals sampled from Guangxi and Hunan were found to display forewing markings similar to the small-marked female form, though the size of these markings was relatively diminished.
S. tamdaoi described from Tam Dao, northern Vietnam, near China, is herein synonymized with
N. capacimacularus based on shared diagnostic characters: a uniformly yellowish vertex and an extensive brown forewing marking of the latter species.
Stenopsocus dictyodromus Li, 1993 [
43]: 349. Type locality: China (Guangdong: Chebaling).
Stenopsocus beroni Georgiev & Quang-Manh, 2024 [
44]: 2. Type locality: Vietnam (Vinh Phuc: Tam Dao).
syn. nov. Stenopsocus longitudinalis Li, 2002 [
2]: 623. Type locality: China (Guangxi: Tianlin).
syn. nov. Stenopsocus trisetus Li, 2002 [
2]: 638. Type locality: China (Guangxi: Pingxiang).
syn. nov. Diagnosis. This species is characterized by a yellowish vertex, a yellowish frontal area with brown markings, posterior margin of pterostigma with brown stripe, and sclerotized genitalia.
Redescription. Adult male: Body (
Figure 6A) length 2.51 mm, length from postclypeus to wing tip 3.89 mm. IO: 0.38 mm; d: 0.34 mm; IO/d = 1.12; f1: 0.99 mm; f2: 0.72 mm; f3: 0.65 mm; FWL: 3.96 mm; FWW: 1.32 mm; HWL: 3.01 mm; HWW: 0.91 mm; t1: 0.47 mm; t2: 0.15 mm.
Color (in alcohol): Head (
Figure 6B) yellowish brown, vertex (
Figure 6C) yellowish white, mid of frontal area with a brown marking. Antenna dark brown. Mouthparts yellowish white, with brown postclypeus. Thorax dark brown. Leg yellowish white, hindleg with brown tibia. Abdomen purplish red, with 4–7 segments ventrally yellowish white, genital segments brown.
Forewing (
Figure 6D) transparent. Posterior margin of pterostigma with narrowly brown marking. Hindwing (
Figure 6E) immaculate.
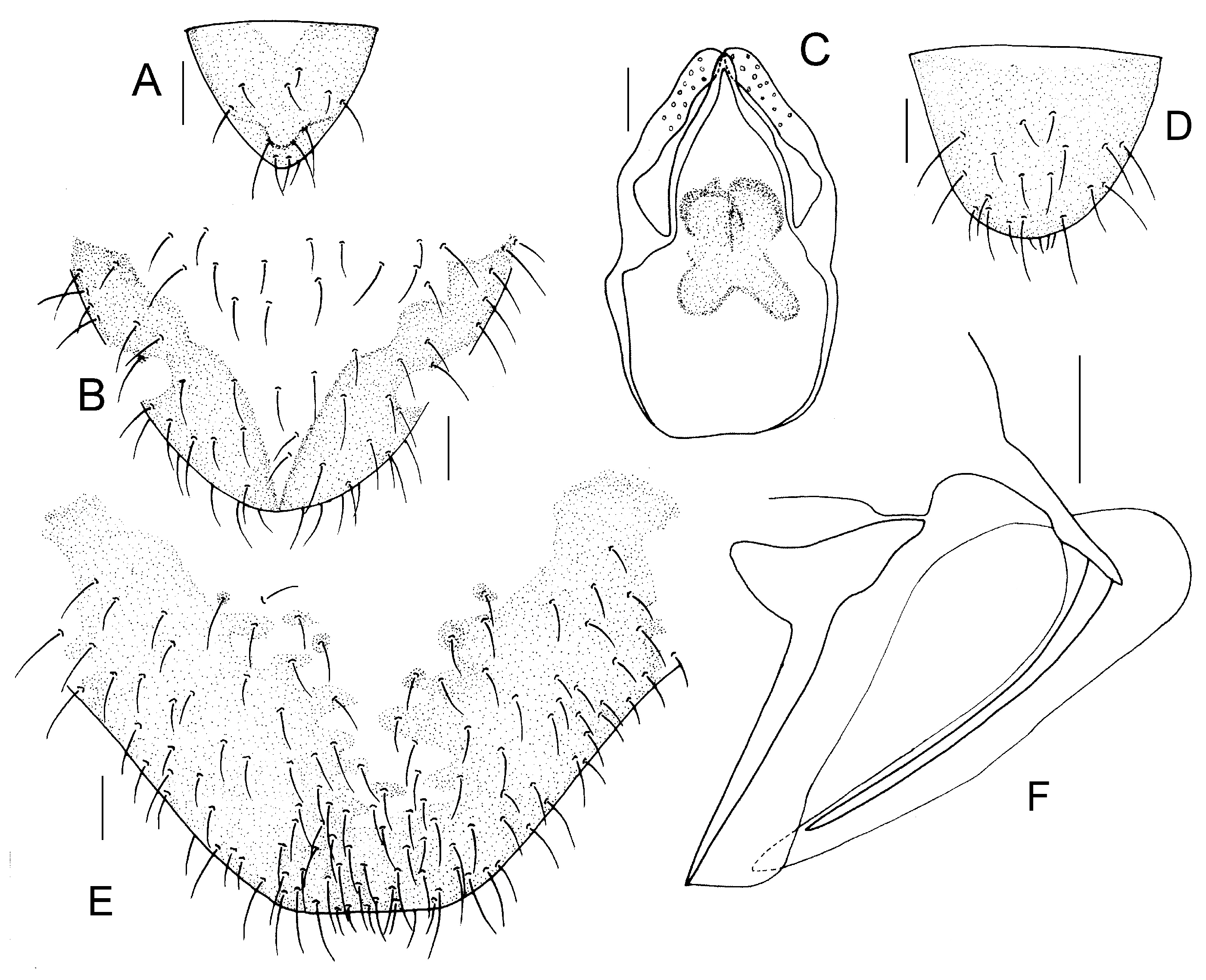
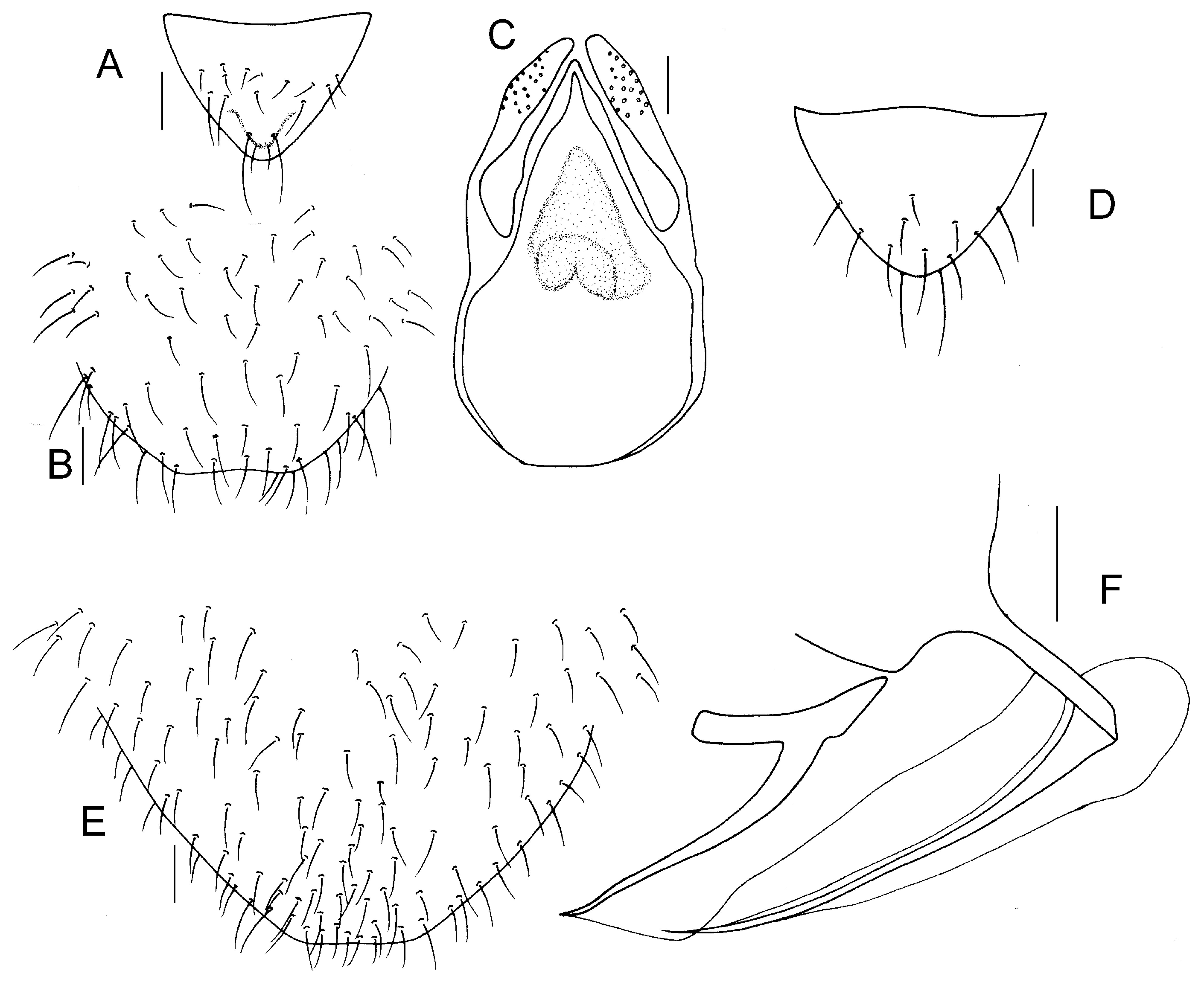
Genital segments strongly sclerotized. Epiproct (
Figure 7A) subtriangular. Paraproct with 44 trichobothria. Hypandrium (
Figure 7B) with strongly sclerotized area. Endophallus (
Figure 7C) more sclerotized than hypandrium, external parameres robust, with some punctures on broad end toward apex, and exceeding apex of aedeagal arch.
Adult female: Body (
Figure 6F) length 3.71 mm, length from postclypeus to wing tip 5.85 mm. IO: 0.58 mm; d: 0.25 mm; IO/d = 2.32; f1: 1.20 mm; f2: 1.05 mm; f3: 0.83 mm; FWL: 4.55 mm; FWW: 1.48 mm; HWL: 3.31 mm; HWW: 1.01 mm; t1: 0.47 mm; t2: 0.14 mm.
Color (in alcohol): Frontal area vertex (
Figure 6G) yellowish white, with a brown stripe crossing ocellus area and connecting compound eyes. Vertex (
Figure 6H) yellowish white. Antenna dark brown. Mouthparts yellowish, with dark brown postclypeus. Thorax brown. Leg yellowish white, hindleg with brown tibia. Abdomen dorsally purplish red, ventrally yellowish white, genital segments brown.
Forewing vertex (
Figure 6I) yellowish white, transparent. Anterior area of pterostigma yellowish, posterior area of pterostigma with dark brown markings. Hindwing with a brown stripe between Sc and R.
Genital segments sclerotized. Epiproct vertex (
Figure 7D) yellowish white, subtriangular. Paraproct with 25 trichobothria. Subgenital plate vertex (
Figure 7E) yellowish white, with distinctly sclerotized area. Gonapophyses vertex (
Figure 7F) yellowish white, strongly sclerotized.
Specimens examined. Holotype of
Stenopsocus dictyodromus, ♀, China, Guangdong, Shixing, Chebaling National Nature Reserve (600 m), 1991.IV.23, Fasheng Li (CAU) (
Figure S17);
Holotype of
Stenopsocus longitudinalis, ♀, China, Guangxi, Tianlin, Langping (1500 m), 1982.V.30, Fasheng Li (CAU) (
Figure S18);
Holotype of
Stenopsocus trisetus, ♀, China, Guangxi, Pingxiang, Daqingshan, 1963.V.15, Chikun Yang (CAU) (
Figure S19).
CHINA: 2♂♂2♀♀, Guangxi, Wuming, Mt. Damingshan (1230 m), 2013.V.20, Xingyue Liu (CAU); 1♀, Chongqing, Yintiaoling, 2022.VII.12, Leran Cao (CAU); 2♂♂8♀♀, Guangxi, Jinxiu, Yinshan Park (1150 m), 2016.V.22, Feiyang Liang (CAU); 2♀♀, Yunnan, Bingchuan, Mt. Jizushan (2228 m), 2016.VII.4, Liang Wang & Qicheng Yang (CAU); 1♀, Yunnan, Baoshan, Nankang (2048 m), 2015.VII.19, Yunlan Jiang (CAU); 1♀, Yunnan, Jinping, Jinhe Hotel (1792 m), 2016.VII.11, Ya’nan, Lv (CAU); ♂2♀♀, Yunnan, Malipo, Xiajinchang (1430 m), 2016.VII.27, Yunlan Jiang (CAU); 1♀, Fujian, Dehua, Leifengcun (850 m), 2010.VII.13, Xinyu Luo (CAU); 1♀, Chongqing, Yintiaoling Nature Reserve, Linkouzi Station (1248 m), 16.VIII.2022, Leran Cao (CAU).
Distribution. China (Guangdong, Guangxi, Yunnan, Chongqing, Fujian).
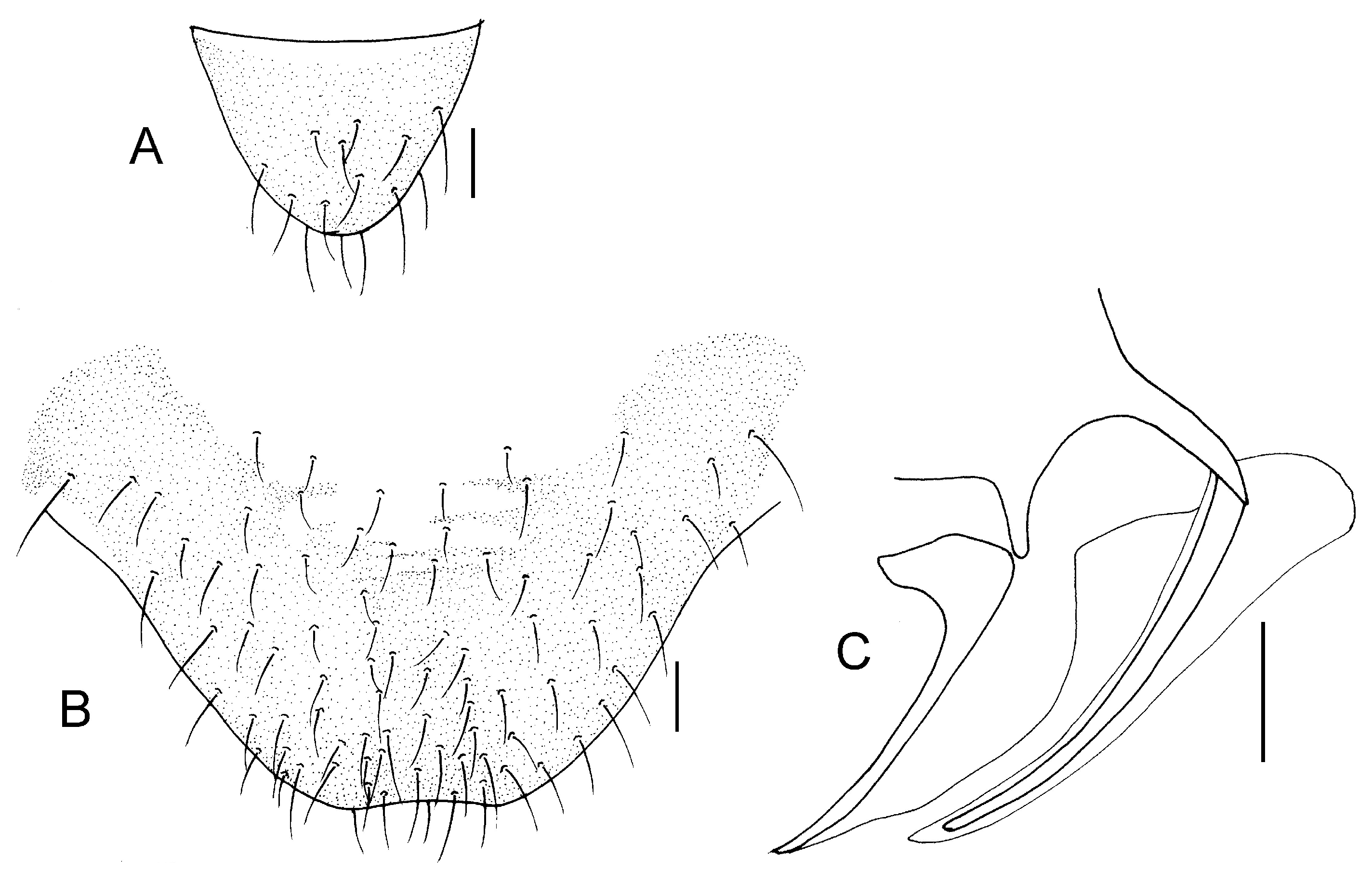
Remarks. The forewing markings display slight variations among different female individuals of this species. The brown marking within the forewing pterostigma exhibits continuous size variation, ranging from occupying less than half of the pterostigmal area to covering more than half of it (
Figure 8). We consider
S. beroni,
N. longitudinalis, and
N. trisetus to be synonymous with
N. dictyodromus based on the similarities in the forewing marking patterns and sclerotized terminalia. This species appears to be related to
N. gracillimus due to their similar head and forewing marking patterns; however, it can be distinguished from the latter by the sclerotized terminalia.
Stenopsocus eucallus Li & Yang, 1988 [
45]: 174. Type locality: China (Guizhou: Fanjingshan).
Stenopsocus metastictus Li, 2002 [
2]: 629. Type locality: China (Guangxi: Jinxiu).
syn. nov. Diagnosis. This species is characterized by a dark brown head with yellowish-white vertex, forewing R with a dark brown spot, posterior margin of pterostigma with brown stripe, hindwing with a brown spot between anterior margin and M, and strongly sclerotized genitalia.
Redescription. Adult male: Body (
Figure 9A) length 2.46 mm, length from postclypeus to wing tip 4.82 mm. IO: 0.38 mm; d: 0.29 mm; IO/d = 1.31; f1: 1.01 mm; f2: 0.87 mm; f3: 0.71 mm; FWL: 3.49 mm; FWW: 1.18 mm; HWL: 2.58 mm; HWW: 0.81 mm; t1: 0.44 mm; t2: 0.12 mm.
Color (in alcohol): Head (
Figure 9B,C) dark brown, vertex with a subtrapezoidal yellowish area. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown. Thorax dark brown. Leg yellowish. Abdomen yellowish white, genital segments brown.
Forewing (
Figure 9D) transparent. R dark brown. Pterostigma yellowish, posterior margin with narrow brown marking which does not extend along r-rs. Hindwing with dark brown markings present between Sc and R.
Genital segments strongly sclerotized. Epiproct (
Figure 10A) subtriangular. Paraproct with 32 trichobothria. Subgenital plate (
Figure 10B) sclerotized strongly. Endophallus (
Figure 10A) strongly sclerotized, external parameres robust, with some punctures on broad end toward apex, and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 9E) length 3.20 mm, length from postclypeus to wing tip 5.48 mm. IO: 0.67 mm; d: 0.25 mm; IO/d = 2.28; f1: 1.18 mm; f2: 1.10 mm; f3: 0.85 mm; FWL: 4.29 mm; FWW: 1.41 mm; HWL: 3.24 mm; HWW: 0.95 mm; t1: 0.41 mm; t2: 0.13 mm.
Color similar to male, slightly darker. Forewing (
Figure 9H) markings much larger than those in male, R with a brown marking. Hindwing (
Figure 9I) with dark brown markings present between Sc and R. Abdomen with 1–3 segments purplish red, 4–6 segments laterally purplish.
Genital segments strongly sclerotized. Epiproct (
Figure 10D) subtrapezoidal. Paraproct with 20 trichobothria. Subgenital plate (
Figure 10E) with a broad sclerotized area. External valve short and robust, with a flat tip, almost perpendicular to dorsal valve (
Figure 10F).
Specimens examined. Holotype of
Stenopsocus eucallus, ♀, Guizhou, Jiangkou, Mt. Fanjingshan (850 m), 1986.VIII.14, Fasheng Li (CAU) (
Figure S20);
Holotype of
Stenopsocus metastictus, ♀, Guangxi, Jinxiu, Mt. Dayaoshan (800 m), 1982.VI.12, Fasheng Li (CAU) (
Figure S21).
CHINA: 2♀♀, Guangxi, Jinxiu, Yinshan Park (1150 m), 2016.VII.22, Xingyue Liu (CAU); 2♂♂1♀, Yunnan, Pingbian, Daweishan Park, 2016.VII.16, Ya’nan Lv (CAU); 1♂, Yunnan, Baoshan, Xiaoheishan (2116 m), 2015.VII.20, Yunlan Jiang (CAU); 1♂, Yunnan, Baoshan, Mt. Gaoligongshan (2148 m), 2015.VII.20, Yunlan Jiang (CAU); 1♀, Sichuan, Mt. Emei, Linggongli, 2015.VIII.25, Tingting Zhang (CAU); 1♀, Xizang, Motuo (1117 m), 2014.VII.25, Yan Li (CAU).
Distribution. China (Yunnan, Guangxi, Guizhou, Sichuan, Xizang).
Remarks. Combined with the molecular delimitation results, this species shows stable coloration in the body and wings. However, there are differences in the markings of the forewings between male and female individuals. The R vein of male has no obvious markings, while that of female has a brown spot; the posterior marginal spot of the pterostigma in male is significantly narrower. We consider S. metastictus to be synonymous with N. eucallus based on the similarities in the forewing marking patterns and yellowish vertex. This species appears to be closely related to N. formosanus by the forewing marking patterns, but can be distinguished from the latter species by the hindwing with a larger brown spot.
(5) Neostenopsocus externus (Banks, 1937)
Stenopsocus externus Banks, 1937 [
38]: 259. Type locality: China (Taiwan: Taihoku).
Stenopsocus hemiostictus Li, 2002 [
2]: 659. Type locality: China (Yunnan: Kunming).
syn. nov. Stenopsocus phaneostriatus Li, 2002 [
2]: 663. Type locality: China (Yunnan: Kunming).
syn. nov. Diagnosis. See Liang et al., 2015 [
20].
Specimens examined. Holotype of
Stenopsocus externus, ♀, China, Taiwan, Taipei, 1934.V.2, Judson Linsley Gressitt (MCZ) (
Figure S22);
Paratype of
Stenopsocus externus, ♀, China, Taiwan, Arisan, 1934.V.29, Judson Linsley Gressitt (MCZ) (
Figure S22);
Holotype of
Stenopsocus hemiostictus, ♀, China, Yunnan, Kunming, Xishan (2100 m), 1981.V.16, Fasheng Li (CAU) (
Figure S23);
Holotype of
Stenopsocus phaneostriatus, ♀, China, Yunnan, Kunming, Xishan (1900 m), 1981.V.18, Fasheng Li (CAU) (
Figure S24).
CHINA: 1♀, Taiwan, Kaohsiung, Shoushan Park, 2013.V.31, Xinyu Luo; 1♀, Taiwan, Yilan, Wushibi, 2013.VI.8, Xinyu Luo (CAU); 1♀, Zhejiang, Li’an, Mt. Tianmu (300–1000 m), 2016.VII.27, Feiyang Liang (CAU); 1♂2♀♀, Guangxi, Wuming, Daminshan, 2016.V.25, Feiyang Liang (CAU); 2♀♀, Fujian, Xiamen, Mt. Dongpingshan, 2021.VII.2, Yuchen Zheng (CAU); 1♂, Guizhou, Mt. Leigongshan, Xiaodanjiang (664 m), 2014.VII.23, Lu Yue (CAU); 1♀, Guizhou, Mt. Leigongshan, Xiaodanjiang (664 m), 2014.VII.23, Yuting Dai (CAU); 1♀, Guangxi, Pingxiang, Lanhuagu, 2014.V.8, Xiumei Lu (CAU); 1♂, Guangxi, Nanning, Guangxi Academy of Forestry Sciences, 2013.V.14, Xingyue Liu (CAU); 1♀, Guangxi, Fangchenggang, Wangle Nature Reserve, 2013.V.18, Xingyue Liu (CAU); 2♂♂, Guangxi, Fangchenggang, Shiwandashan Forest Park, 2013.V.17, Guoquan Wang (CAU); 1♂, Guangxi, Fangchenggang, Jinhuacha Nature Reserve, 2014.VI.7, Xingyue Liu (CAU); 1♂, Yunnan, Menglun, Xishuangbanna Tropical Botanical Garde, 2015.IV.10-13, Feiyang Liang (CAU); 1♀, Guangxi, Baise, Songshuping, 2013.VIII.1, Feiyang Liang (CAU); 1♂4♀♀, Henan, Xinxian, Jiulongtan (200 m), 2014.VI.18, Xingyue Liu (CAU); 1♀, Xizang, Linzhi, Lulang, 2012.VII.18, Chenliang Zhang & Jianyun Wang (CAU); 1♂1♀, Hubei, Yinshan, Mt. Wujiashan (715 m), 2014.VI.30, Wencheng Chang (CAU); 1♂1♀, Hunan, Xiangtan, Hunan University of Science and Technology, 2019.V.30, Feiyang Liang (HNUST). LAOS: 1♂, Khammouan, Phou Hi Poun NBCA, near Tha Long (550 m), 2016.IV.1, Xingyue Liu (CAU). VIETNAM: ♀, Kon Tum, Chu Mom Ray National Park, 2012.VIII.1, Feiyang Liang (CAU).
Distribution. China (Taiwan, Guangxi, Hunan, Hubei, Henan, Fujian, Yunnan, Guizhou, Sichuan, Gansu, Hebei, Xizang, Shanghai); Laos (Khammouan); Vietnam (Kon Tum).
Remarks. Li [
2] and Liang et al. [
20] reported this species is widely distributed in southern China. In the present study, we matched the male and female of this species based on the DNA barcoding. This species is diagnosable by the brown pedicel and the distal half of R1 with a brown marking. Li [
2] described
S. hemiostictus and
S. phaneostriatus from Kunming and Yunnan, noting that their antennae have yellowish 1st antennomere, brown 2nd–10th antennomeres, and half of the posterior margin of the pterostigma on the forewings bears brown markings. After examining the relevant specimens, we found that these characteristics are consistent with the diagnostic features of
S. externus, and consider
S. hemiostictus and
S. phaneostriatus to be synonyms of
N. externus.
Stenopsocus foliaceus Li, 1997 [
40]: 434. Type locality: China (Hubei: Xingshan).
Diagnosis. This species is characterized by a dark brown head, forewing R with brown spot, pterostigma with a subtriangle brown spot, and a strongly sclerotized terminalia.
Redescription. Adult male: Body (
Figure 11A) length 2.13 mm, length from postclypeus to wing tip 3.92 mm. IO: 0.39 mm; d: 0.30 mm; IO/d = 1.30; f1: 0.82 mm; f2: 0.63 mm; f3: 0.51 mm; FWL: 2.91 mm; FWW: 1.06 mm; HWL: 2.31 mm; HWW: 0.72 mm; t1: 0.32 mm; t2: 0.11 mm.
Color (in alcohol): Head (
Figure 11C) dark brown. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown. Thorax dark brown. Leg brown to dark brown, with femur mostly yellowish white. Abdomen yellowish white, genital segments dark brown.
Forewing (
Figure 11D) transparent. R dark brown. Pterostigma yellowish, posterior margin with narrow brown marking which does not extend along r-rs. Hindwing (
Figure 11E) with pale brown markings present between Sc and R.
Genital segments strongly sclerotized. Epiproct (
Figure 12A) subtriangular. Paraproct with 29 trichobothria. Hypandrium (
Figure 12B) sclerotized strongly. Endophallus (
Figure 12C) strongly sclerotized, external parameres robust, with some punctures on broad end toward apex, and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 11F) length 2.90 mm, length from postclypeus to wing tip 4.64 mm. IO: 0.52 mm; d: 0.19 mm; IO/d = 2.74; f1: 0.91 mm; f2: 0.67 mm; f3: 0.52 mm; FWL: 3.62 mm; FWW: 1.22 mm; HWL: 2.63 mm; HWW: 0.82 mm; t1: 0.34 mm; t2: 0.13 mm.
Color similar to male, slightly darker. Forewing (
Figure 11I) markings much larger than those in male, R with a brown marking. Abdomen dorsally purplish red, ventrally yellowish white, genital segments dark brown.
Genital segments strongly sclerotized. Epiproct (
Figure 12D) subtrapezoidal. Paraproct with 33 trichobothria. Subgenital plate (
Figure 12E) with a broad sclerotized area. External valve short and robust, with a round apex (
Figure 12F).
Specimens examined. Holotype of
Stenopsocus foliaceus, ♀, China, Hubei, Xingshan, Longmenhe (1300 m), 1994.IX.13, Fasheng Li (CAU) (
Figure S25).
CHINA: 1♂, Sichuan, Mt. Emei, Leidongping (2300 m), 2012.IX.2, Ding Yang (CAU); 1♀, Sichuan, Mt. Emei, Leidongping (2300 m), 2012.IX.3, Xuankun Li (CAU); 4♀♀, Shaanxi, Lingwu, 2013.VII.13, Wanzhi Cai & Jianyun Wang (CAU).
Distribution. China (Hubei, Sichuan, Shaanxi).
Remarks. This species differs from N. eucallus by the dark brown vertex, and differs from N. melanocephalus by the forewing R with a large brown spot.
Stenopsocus hexagonus Li, 2002 [
2]: 633. Type locality: China (Xizang: Bomi).
Diagnosis. This species is characterized by a yellowish vertex, dark brown antenna, yellowish-white leg, and forewing with a Y-shape marking near the pterostigma.
Redescription. Adult male: Unknown.
Adult female: Body (
Figure 13A) length 2.78 mm, length from postclypeus to wing tip 5.05 mm. IO: 0.51 mm; d: 0.19 mm; IO/d = 2.73; f1: 1.03 mm; f2: 0.71 mm; f3: 0.69 mm; FWL: 4.37 mm; FWW: 1.44 mm; HWL: 3.12 mm; HWW: 0.93 mm; t1: 0.46 mm; t2: 0.13 mm.
Color (in alcohol): Head (
Figure 13B) brown, vertex (
Figure 13C) yellowish white. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax dark brown. Leg yellowish white. Abdomen dorsally pale purplish red, ventrally yellowish white, genital segments brown.
Forewing (
Figure 13D) transparent. Pterostigma yellowish, posterior margin with narrow brown stripe extending along r-rs. Hindwing (
Figure 13E) immaculate.
Genital segments strongly sclerotized. Epiproct (
Figure 14A) subtrapezoidal. Paraproct with 24 trichobothria. Subgenital plate (
Figure 14B) with a broad sclerotized area. External valve short and robust, almost perpendicular to dorsal valve (
Figure 14C).
Specimens examined. Holotype of
Stenopsocus hexagonus, ♀, China, Xizang, Bomi, Zhamu (2800 m), 1978.VII.22, Fasheng Li (CAU) (
Figure S26).
CHINA: 3♀♀, Xizang, Linzhi, 2012.IX.22-X.1, Malaise trap (CAU).
Distribution. China (Xizang).
Remarks. This species can be distinguished from N. polyceratus by a forewing R without markings, and the narrower makings near the pterostigma.
Stenopsocus kunmingiensis Li, 2002 [
2]: 681. Type locality: China (Yunnan: Kunming).
Stenopsocus pavonicus Li, 2002 [
2]: 697. Type locality: China (Yunnan: Mengzhe).
syn. nov. Diagnosis. This species is characterized by a yellowish head with a brown stripe crossing the ocellus area and connecting compound eyes, 1st–5th flagellomeres yellowish-brown with brown tips, posterior margin of pterostigma with very narrow brown stripe, and weakly sclerotized terminalia.
Redescription. Adult male: Body (
Figure 15A) length 2.44 mm, length from postclypeus to wing tip 4.86 mm. IO: 0.42 mm; d: 0.33 mm; IO/d = 1.27; f1: 0.97 mm; f2: 0.86 mm; f3: 0.70 mm; FWL: 3.86 mm; FWW: 1.31 mm; HWL: 2.94 mm; HWW: 0.94 mm; t1: 0.47 mm; t2: 0.14 mm.
Color (in alcohol): Head (
Figure 15B,C) yellowish white. Frontal area with a brown stripe crossing ocellus area and connecting compound eyes. Antenna with yellowish-brown scape and pedicel, and blackish-brown flagella. Mouthparts yellowish, apex of maxillary palpus brown, remaining segments of maxillary palpus whitish. Prothorax yellowish, mid- and metathorax laterally yellowish and dorsally brown. Leg yellowish, with apex of tibiae, 1st tarsomeres, and entire 2nd tarsomere pale brown. Abdomen yellowish white, only trichobothrial area brown.
Forewing (
Figure 15D) transparent. Pterostigma pale yellowish, posterior margin with narrow brown marking. Hindwing (
Figure 15E) immaculate.
Genital segments weakly sclerotized. Epiproct (
Figure 16A) subtriangular. Paraproct with 31 trichobothria. Hypandrium (
Figure 16B) without distinctly sclerotized area. Endophallus (
Figure 16C) weakly sclerotized; external parameres robust, with some punctures on broad end toward apex and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 15F) length 3.66 mm, length from postclypeus to wing tip 5.79 mm. IO: 0.57 mm; d: 0.23 mm; IO/d = 2.48; f1: 1.23 mm; f2: 1.03 mm; f3: 0.77 mm; FWL: 4.43 mm; FWW: 1.41 mm; HWL: 3.15 mm; HWW: 0.95 mm; t1: 0.36 mm; t2: 0.11 mm.
Color similar to male, slightly darker. Forewing (
Figure 15I) markings much larger than those in male, R with a brown marking. Abdomen with 1–3 segments purplish red, 4–6 segments laterally purplish.
Genital segments strongly sclerotized. Epiproct (
Figure 16D) subtrapezoidal. Paraproct with 20 trichobothria. Subgenital plate (
Figure 16E) with a broad sclerotized area. External valve short and robust, with a flat tip, almost perpendicular to dorsal valve (
Figure 16F).
Specimens examined. Holotype of Stenopsocus kunmingiensis, ♂, China, Yunnan, Kunming (1900 m), 1981.V.18, Fasheng Li (CAU).
CHINA: 3♂♂2♀♀, Xiajinchang (1430 m), Malipo, Yunnan, 2016.VII.27, Yunlan Jiang (CAU).
Distribution. China (Yunnan).
Remarks. This species is one with a relatively light body color in the genus Neostenopsocus; most of its body is pale yellow with few brown markings. In particular, the head only bears a brown band between the compound eyes, while all other areas are yellow.
Stenopsocus maculosus Li & Yang, 1988 [
45]: 73. Type locality: China (Guizhou: Fanjingshan).
Stenopsocus brevicapitus Li, 1997 [
40]: 429. Type locality: China (Sichuan: Wushan).
syn. nov. Stenopsocus dactylinus Li, 1997 [
40]: 431. Type locality: China (Sichuan: Wushan).
syn. nov. Stenopsocus daozheniensis Li, 2005 [
42]: 92. Type locality: China (Guizhou: Dashahe).
syn. nov. Stenopsocus isotomus Li, 2002 [
2]: 673. Type locality: China (Sichuan: Emeishan).
syn. nov. Stenopsocus perspicuus Li, 1997 [
40]: 427. Type locality: China (Sichuan: Wushan).
syn. nov. Stenopsocus shennongjiaensis Li, 2002 [
2]: 624. Type locality: China (Hubei: Shennongjia).
syn. nov. Stenopsocus wuxiaensis Li, 1997 [
40]:432. Type locality: China (Sichuan: Wushan).
syn. nov. Stenopsocus xilingxianicus Li, 1997 [
40]: 430. Type locality: China (Hubei: Xingshan).
syn. nov. Diagnosis. This species is characterized by a large body (female), mostly black–brown head, and a head equal in length (from vertex to margin of labrum in frontal view) and width (distance of the outer margins of the two compound eyes).
Redescription. Adult male: Body (
Figure 17A) length 2.76 mm, length from postclypeus to wing tip 5.46 mm. IO: 0.36 mm; d: 0.27 mm; IO/d = 1.33; f1: 1.07 mm; f2: 0.93 mm; f3: 0.76 mm; FWL: 4.60 mm; FWW: 1.75 mm; HWL: 3.36 mm; HWW: 1.13 mm; t1: 0.50 mm; t2: 0.18 mm.
Color (in alcohol): Head (
Figure 17B,C) dark brown. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus. Thorax dark brown. Leg yellowish, with dark brown tibia. Abdomen yellowish white, genital segments pale brown with brown trichobothrial area.
Forewing (
Figure 17D) transparent. R dark brown. Pterostigma without distinct markings. Hindwing (
Figure 17E) immaculate.
Genital segments weakly sclerotized. Epiproct (
Figure 18A) subtriangular. Paraproct with 29 trichobothria. Hypandrium (
Figure 18B) without distinct sclerotized area. Endophallus (
Figure 18C) weakly sclerotized; external parameres robust, with some punctures on broad end toward apex and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 17F) length 3.70 mm, length from postclypeus to wing tip 6.90 mm. IO: 0.36 mm; d: 0.16 mm; IO/d = 2.25; f1: 1.14 mm; f2: 0.81 mm; f3: 0.76 mm; FWL: 5.12 mm; FWW: 1.76 mm; HWL: 3.45 mm; HWW: 1.06 mm; t1: 0.52 mm; t2: 0.18 mm.
Color similar to male, slightly darker. Forewing (
Figure 17I) with a brown stripe along posterior margin of Pterostigma.
Genital segments sclerotized. Epiproct (
Figure 18D) subtrapezoidal. Paraproct with 36 trichobothria. Subgenital plate (
Figure 18E) with a V-shape sclerotized area. External valve (
Figure 18F) short and robust, with a round tip.
Specimens examined. Holotype of
Stenopsocus maculosus, ♀, China, Guizhou, Jiangkou, Fanjingshan Nature Reserve, 1983.VII.13, Fasheng Li (CAU) (
Figure S27);
Holotype of
Stenopsocus brevicapitus, ♀, China, Sichuan, Wushan, Liziping (1850 m), 1994.IX.21, Fasheng Li (CAU) (
Figure S28);
Holotype of
Stenopsocus dactylinus, ♀, China, Sichuan, Wushan, Liziping (1850 m), 1994.IX.21, Fasheng Li (CAU) (
Figure S29);
Holotype of
Stenopsocus daozheniensis, ♀, Guizhou, Daozhen, Dashahe Nature Reserve (1400 m), 2004.V.25, Maofa Yang (CAU) (
Figure S30);
Holotype of
Stenopsocus isotomus, ♂, China, Sichuan, Mt. Emei, Jiulaodong (2070 m), 1978.IX.18, Fasheng Li (CAU) (
Figure S31);
Holotype of
Stenopsocus perspicuus, ♀, China, Sichuan, Wushan, Liziping (1850 m), 1994.IX.21, Fasheng Li (CAU) (
Figure S32);
Holotype of
Stenopsocus shennongjiaensis, ♂, China, Hubei, Shennongjia Nature Reserve, Dayanwu, Hubei, 1984.VI.19, Chikun Yang (CAU) (
Figure S33);
Holotype of
Stenopsocus wuxiaensis, ♀, China, Sichuan, Wushan, Liziping (1850 m), 1994.IX.21, Fasheng Li (CAU) (
Figure S34);
Holotype of
Stenopsocus xilingxianicus, ♀, China, Hubei, Xingshan, Longmen River (1300 m), 1994.IX.13, Fasheng Li (CAU) (
Figure S35).
CHINA: 1♂1♀, Sichuan, Mt. Emei, Leidongping to Jinding, 2013.VI.29, Xingyue Liu & Feiyang Liang (CAU); 1♂, Sichuan, Mt. Emei, Linggongli, 2011.VII.5, Xingyue Liu (CAU); 1♀, Sichuan, Mt. Emei, Leidongping, 2015.VIII.23, Tingting Zhang (CAU); 1♀, Hubei, Shennongjia Nature Reserve, Dalongtan, 2012.VII.29, Tingting Zhang (CAU); 1♀, Shaanxi, Lingwu, 2011.VII.25, Wanzhi Cai & Jianyun Wang (CAU); 1♀, Gansu, Wenxian, Qiujiaba Station, 2012.VII.29, Sipei Liu (CAU).
Distribution. China (Guizhou, Sichuan, Hubei, Shaanxi, Gansu).
Remarks. This species can be distinguished from other
Neostenopsocus based on the whole black–brown vertex, and the length of its head nearly equal to its width in the frontal view. Li & Yang [
45] described
N. maculosus from Mt. Fanjingshan, Guizhou. Subsequently, Li [
2,
40] described six additional species from three adjacent collection localities:
S. brevicapitus,
S. dactylinus,
S. perspicuus, and
S. wuxiaensis from Liziping;
S. shennongjiaensis from Shennongjia; and
S. xilingxianicus from Xingshan. After examining the type specimens of these six species, we found that the differences among these specimens primarily lie in the depth of body color: the body color of
S. wuxiaensis and
S. shennongjiaensis is relatively dark, while the body color of the specimens of other species is lighter. Besides this, there are no other obvious differences. Therefore, we consider these species to be the same species. These lighter-colored specimens may be individuals that have just emerged and have not yet fully developed their coloration. These specimens can be distinguished from the type specimen of
N. maculosus by the forewing markings, specifically the markings along the posterior margin of the pterostigma, which are significantly narrower than those in
N. maculosus. However, we collected several specimens from Shennongjia that exhibit broad markings consistent with those of
N. maculosus. Therefore, these six species are considered synonyms of
N. maculosus, as are
N. daozheniensis and
N. isotomus. Notably, the shape of head in
N. maculasus is similar to the genera
Malostenopsocus and
Stenopsocus, with the length of head nearly equal to its width, which was identified by Liang & Liu [
5] as a synapomorphy for these two genera.
Stenopsocus maximalis Li, 1997 [
40]: 433. Type locality: China (Hubei: Xingshan).
Diagnosis. This species is characterized by a dark brown labrum and big markings behind pterostigma.
Redescription. Adult male: Unknown.
Adult female: Body (
Figure 19A) length 2.98 mm, length from postclypeus to wing tip 4.91 mm. IO: 0.47 mm; d: 0.17 mm; IO/d = 2.61; f1: 1.12 mm; f2: 0.70 mm; f3: 0.64 mm; FWL: 3.87 mm; FWW: 1.36 mm; HWL: 2.74 mm; HWW: 0.94 mm; t1: 0.37 mm; t2: 0.10 mm.
Color (in alcohol): Head (
Figure 19B,C) dark brown. Antenna with 1–6 antennomeres blackish brown, 7–13 antennomeres white. Postclypeus, labrum, maxilla stipes, and base of mandible darkish brown. Other part of mouthparts yellowish. Thorax dark brown. Leg yellowish white, hindleg with brown tibia. Abdomen dorsally pale purplish, ventrally yellowish white, genital segments dark brown.
Forewing (
Figure 19D) transparent, wing margin mostly pale brown; costal vein from base to pterostigma and R brown, a big brown marking around crossvein r-rs. Hindwing (
Figure 19E) transparent and glabrous.
Genitalia sclerotized. Epiproct (
Figure 20A) subtrapezoid, with round apex. Paraproct with 22 trichobothrias. Subgenital plate (
Figure 20B) with distinctly “U” shape sclerotized region, middle sclerotized region broad. Gonapophyses (
Figure 20C) sclerotized; external valve broad, perpendicular to dorsal valve; dorsal and ventral valve blade shaped.
Specimens examined. Holotype of
Stenopsocus maximalis, ♀, China, Hubei, Xingshan, Longmen River (1300 m), 1994.IX.13, Fasheng Li (CAU) (
Figure S36).
CHINA: 4♀♀, Yunnan, Yuanjiang Nature Reserve, Yangchajie, 2015.IV.20, Feiyang Liang (CAU); 1♀, Xizang, Motuo, 2013.IX.12, Jianyuan Wang & Yun Ji (CAU); 2♀♀, Sichuan, Mt. Emei, 2015.VIII.25, Tingting Zhang (CAU); 1♀, Yunnan, Nujiang, Pianma, 2013.VII. 6, Ziqiang Sun (CAU); 3♀♀, Yunnan, Boshan, Mt. Baihualing, 1900 m, 2015.VII.26, Lu Yue (CAU); 1♀, Yunnan, Mt. Gaoligong, Xiaoheishan (2116 m), 2015.VII.20, Yunlan Jiang (CAU); 1♀, Yunnan, Nanjiang, Mt. Wuliang (2221 m), 2016.VII.16, Qicheng Yang (CAU). Nepal: 3♀♀, Pokhara, Mt. Annapurna (2000 m), 2014.VII. 2, Feiyang Liang (CAU).
Distribution. China (Hubei, Chongqing, Yunan, Xizang), Nepal (Pokhara).
Remarks. This species appears to be closely related to N. capacimacularus and N. makii, having similar body color and markings of forewing, but it can be distinguished from the latter two species by the black–brown vertex and labrum.
Stenopsocus melanocephalus Li, 1997 [
40]: 437. Type locality: China (Sichuan: Wushan).
Diagnosis. This species is characterized by a whole brown or dark brown antenna, forewing R without marking, pterostigma with brown marking extending along r-rs, and hindleg with dark brown tibia.
Redescription. Adult male: Body (
Figure 21A) length 2.79 mm, length from postclypeus to wing tip 5.41 mm. IO: 0.37 mm; d: 0.29 mm; IO/d = 1.28; f1: 1.13 mm; f2: 1.01 mm; f3: 0.79 mm; FWL: 4.63 mm; FWW: 1.66 mm; HWL: 3.47 mm; HWW: 1.13 mm; t1: 0.46 mm; t2: 0.16 mm.
Color (in alcohol): Head (
Figure 21B,C) dark brown. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax dark brown. Leg yellowish, with dark brown tibiae. Abdomen dorsally purplish brown, ventrally yellowish white, genital segments brown.
Forewing (
Figure 21D) transparent. R dark brown. Pterostigma yellowish, posterior margin with narrow brown marking, which does not extend along r-rs. Hindwing (
Figure 21E) with pale brown markings present between Sc and R.
Genital segments strongly sclerotized. Epiproct (
Figure 22A) subtriangular. Paraproct with 26 trichobothria. Hypandrium (
Figure 22B) with V-shaped sclerotized area. Endophallus (
Figure 22C) strongly sclerotized; external parameres robust, with some punctures on broad end toward apex and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 21F) length 3.92 mm, length from postclypeus to wing tip 6.33 mm. IO: 0.44 mm; d: 0.24 mm; IO/d = 1.82; f1: 1.38 mm; f2: 1.11 mm; f3: 0.84 mm; FWL: 5.37 mm; FWW: 1.13 mm; HWL: 3.60 mm; HWW: 1.13 mm; t1: 0.51 mm; t2: 0.18 mm.
Color similar to male, slightly darker. Forewing (
Figure 21I) markings much larger than those in male, R with a brown marking. Abdomen purplish red, with 4–8 segments ventrally yellowish white.
Genital segments strongly sclerotized. Epiproct (
Figure 22D) subtrapezoidal. Paraproct with 28 trichobothria. Subgenital plate (
Figure 22E) with a broad sclerotized area. External valve short and robust, with a flat tip, almost perpendicular to dorsal valve (
Figure 22F).
Specimens examined. Holotype of
Stenopsocus melanocephalus, ♀, Sichuan, Mt. Wushan, Liziping (1850 m), 1994.IX.21, Fasheng Li (CAU) (
Figure S37).
CHINA: 1♂, Sichuan, Mt. Emei, Leidongping, 2012.IX.2, Ding Yang (CAU); 1♀, Sichuan, Mt. Emei, Leidongping, 2012.IX.3, Xuankun Li (CAU); 4♀, Shaanxi, Lingwu, 2013.VII.13, Jianyun Wang & Wanzhi Cai (CAU); 1♀, Hunan, Sangzhi, Mt. Tianpingshan (1300 m), 2012.VII.23, Mingchao Huang (CAU); 4♂♂, Yunnan, Pingbian, Daweishan Nature Reserve, 2016.VII.17, Ya’nan Lv (CAU); 2♀♀, Yunnan, Pingbian, Daweishan Nature Reserve, 2016.VII.17, Ya’nan Lv (CAU); 2♂♂2♀♀, Guangxi, Jinxiu, Yinshan Park (1500 m), 2016.V.22, Feiyang Liang (CAU).
Distribution. China (Yunnan, Sichuan, Guangxi, Shaanxi).
Remarks. This species appears to be closely related to N. anthracinus and N. tibialis, having similar forewing marking patterns, but it can be distinguished from the latter two species by the whole blackish-brown vertex.
Stenopsocus nepalensis New, 1971 [
46]: 207. Type locality: Nepal (Bagmati).
Diagnosis. The females are characterized by a yellowish-white vertex, forewing R with a brown spot, posterior margin of pterostigma with dark brown stripe, hindwing with a narrow brown strip between Sc and R, and strongly sclerotized terminalia. The males are similar to the females, but lack the forewing marking on the R.
Redescription. Adult male: Body (
Figure 23A) length 2.66 mm, length from postclypeus to wing tip 4.63 mm. IO: 0.34 mm; d: 0.30 mm; IO/d = 1.13; f1: 0.93 mm; f2: 0.86 mm; f3: 0.66 mm; FWL: 3.53 mm; FWW: 1.24 mm; HWL: 2.71 mm; HWW: 0.83 mm; t1: 0.42 mm; t2: 0.14 mm.
Color (in alcohol): Head (
Figure 23B) dark brown, vertex (
Figure 23C) with a subtrapezoidal yellowish area. Antenna blackish brown. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax brown. Leg yellowish, with yellow-brown tibia. Abdomen yellowish white, genital segments brown.
Forewing (
Figure 23D) transparent. R dark brown. Pterostigma yellowish, posterior margin with narrow brown marking, which does not extend along r-rs. Hindwing (
Figure 25E) with dark brown markings present between Sc and R.
Genital segments strongly sclerotized. Epiproct (
Figure 24A) subtriangular, weakly sclerotized. Paraproct with 32 trichobothria. Hypandrium (
Figure 24B) sclerotized strongly. Endophallus (
Figure 24C) strongly sclerotized, external parameres robust, with some punctures on broad end toward apex and not exceeding apex of aedeagal arch; aedeagal arch narrow.
Adult female: Body (
Figure 23F) length 3.46 mm, length from postclypeus to wing tip 5.27 mm. IO: 0.53 mm; d: 0.21 mm; IO/d = 2.52; f1: 1.30 mm; f2: 1.03 mm; f3: 0.75 mm; FWL: 4.29 mm; FWW: 1.41 mm; HWL: 3.24 mm; HWW: 0.95 mm; t1: 0.41 mm; t2: 0.13 mm.
Color similar to male, slightly darker. Forewing (
Figure 23I) markings much larger than those in male, R with a brown marking. Abdomen with 1–3 segments purplish red, 4–6 segments laterally purplish.
Genital segments strongly sclerotized. Epiproct (
Figure 24D) subtrapezoidal. Paraproct with 20 trichobothria. Subgenital plate (
Figure 24E) with a broad sclerotized area. External valve short and robust, with a flat tip, almost perpendicular to dorsal valve (
Figure 24F).
Specimens examined. CHINA: ♀, Xizang, Motuo, 2014.VII.12, Jianyun Wang & Yun Ji (CAU); 1♂1♀, Yunnan, Mt. Huanglianshan Nature Reserve, Yakou station, 2016.VII.9, Ya’nan Lv (CAU); 1♀, Guizhou, Mt. Fanjingshan, Tongkuangchang (850 m), 1988.VIII.14, Fasheng Li (CAU); 1♂, Guangxi, Jinxiu, Yinshan Park (1150 m), 2016.V.22, Feiyang Liang (CAU); 1♂3♀♀, Yunnan, Pingbian, Mt. Daweishan (1962 m), Ya’nan Lv (CAU); 1♀, Sichuan, Mianyang, Laohegou (1400–1800 m), 2016.V.14-15, Hui Dong (CAU); 1♀, Yunnan, Mt. Gaoligong Nature Reserve, Nankang Station (2048 m), 2015.VII.7, Lu Yue (CAU). NEPAL: 3♀♀, Pokhara, Mt. Annapura (2000 m), 2014.VII.17, Feiyang Liang (CAU).
Distribution. China (Yunnan, Guizhou, Sichuan, Guangxi, Xizang), Nepal (Bagmati, Pokhara).
Remarks. This species is newly recorded herein from China. It differs diagnostically from N. eucallus by the forewing R vein without markings in females.
Stenopsocus polyceratus Li, 2002 [
2]: 613. Type locality: China (Ningxia: Liupanshan).
Diagnosis. This species differs from the other species of Neostenopsocus by the forewing with a y-shaped brown marking between the pterostigma and Rs.
Redescription. Adult male: Unknown.
Adult female. Body (
Figure 25A) length 2.78 mm, length from postclypeus to wing tip 5.14 mm. IO: 0.49 mm; d: 0.20 mm; IO/d = 2.45; f1: 0.98 mm; f2: 0.73 mm; f3: 0.58 mm; FWL: 4.13 mm; FWW: 1.54 mm; HWL: 2.87 mm; HWW: 1.00 mm; t1: 0.44 mm; t2: 0.12 mm.
Color (in alcohol): Head (
Figure 25B) dark brown, vertex (
Figure 25C) with a yellowish oval area, frontal area yellowish brown, antenna with 1–9 antennomeres dark brown, 9–13 antennomeres whitish. Mouthparts mostly yellowish with blackish-brown postclypeus, apex of maxillary palpus pale brown, remaining segments of maxillary palpus whitish. Thorax brown or dark brown. Fore- and midleg mostly yellowish; hind leg mostly brown, with trochanter, middle of tibia, and entire 2nd tarsomere whitish. Abdomen purplish brown, with 4–8 segments ventrally whitish; genital segments dark brown.
Forewing (
Figure 25D) transparent. Mid of R with a brown spot, anterior margin of pterostigma yellowish brown. Pterostigma yellowish, a y-shaped dark brown marking between posterior margin of pterostigma and Rs. Hindwing (
Figure 25E) with a dark brown marking present between Sc and R.
Genital segments strongly sclerotized. Epiproct (
Figure 26A) subtriangular. Paraproct with 22 trichobothria. Subgenital plate (
Figure 26B) with a broad sclerotized area. External valve (
Figure 26C) short, thumb shaped.
Specimens examined. Holotype of
Stenopsocus polyceratus, ♀, Ningxia, Mt. Liupanshan (2100 m), 1992.VII.30, Fasheng Li (CAU) (
Figure S38).
CHINA: 13♀♀, Shaanxi, Lingwu, 2013.VII.13, Wanzhi Cai & Jianyun Wang (CAU); 3♀♀, Ningxia, Zhongwei, Shapotou (1200 m), 1992.VIII.1, Fasheng Li (CAU); 4♀♀, Ningxia, Mt. Liupanshan, Liangdian Gorge, 2012.VIII.2, Yang Zhao (CAU); 2♀♀, Ningxia, Liupanshan Botanical Garden, 2012.VII.28, Yang Zhao (CAU); 4♀♀, Ningxia, Mt. Liupanshan, Xiaonanchuan, 2012.VII.27, Yang Zhao (CAU); 1♀, Sichuan, Mt Emei, Leidongping, 2011.VII.5, Xingyue Liu (CAU); 1♀, Sichuan, Mt Emei, Leidongping, 2011.VII.14, Yingying Wang (CAU).
Distribution. China (Gansu, Shaanxi, Ningxia, Sichuan).
Remarks. Li [
2] described
S. polyceratus from Gansu and
S. brachyodicrus from Ningxia. Based on examination of additional specimens from Shaanxi, Ningxia, and Sichuan, we synonymize
S. brachyodicrus under
S. polyceratus based on the similar color of head, antenna, and markings on forewing.
Stenopsocus zonatus Li, 1989 [
39]: 32. Type locality: China (Shaanxi: Foping).
Stenopsocus angustistriatus Li, 2002 [
2]: 665. Type locality: China (Guizhou: Pingtang).
syn. nov. Stenopsocus genostictus Li, 2002 [
2]: 690. Type locality: China (Yunnan: Ruili).
syn. nov. Stenopsocus macrocheirus Li, 2002 [
2]: 704. Type locality: China (Gansu: Wenxian).
syn. nov. Stenopsocus thermophiles Li, 2002 [
2]: 705. Type locality: China (Guizhou: Luodian).
syn. nov. Stenopsocus xiangxiensis Li, 1992 [
41]: 691. Type locality: China (Hunan: Zhangjiajie).
syn. nov. Diagnosis. This species is characterized by a yellowish vertex, dark brown antenna with yellowish-brown pedicel and scape, posterior margin of pterostigma with narrow brown stripe, and weakly sclerotized terminalia.
Redescription. Adult male: Body (
Figure 27A) length 2.67 mm, length from postclypeus to wing tip 4.72 mm. IO: 0.37 mm; d: 0.26 mm; IO/d = 1.43; f1: 1.23 mm; f2: 1.17 mm; f3: 0.65 mm; FWL: 3.88 mm; FWW: 1.34 mm; HWL: 2.63 mm; HWW: 0.85 mm; t1: 0.39 mm; t2: 0.12 mm.
Color (in alcohol): Head (
Figure 27B) yellowish brown, vertex (
Figure 27C) yellowish. Antenna dark brown. Mouthparts yellowish white, with brown postclypeus. Thorax dark brown. Leg with yellowish-white femur and yellowish-brown tibia. Abdomen yellowish white. Genital segments yellowish white with pale brown paraproct.
Forewing (
Figure 27D) transparent. Posterior margin of pterostigma with narrow brown marking. Hindwing (
Figure 27E) immaculate.
Genital segments weakly sclerotized. Epiproct (
Figure 28A) subtriangular. Paraproct sclerotized, with 44 trichobothria. Hypandrium (
Figure 28B) without distinctly sclerotized area. Endophallus (
Figure 28C) sclerotized, external parameres robust, with some punctures on broad end toward apex and exceeding apex of aedeagal arch.
Adult female: Body (
Figure 27E) length 3.32 mm, length from postclypeus to wing tip 5.86 mm. IO: 0.44 mm; d: 0.20 mm; IO/d = 2.20; f1: 1.10 mm; f2: 1.07 mm; f3: 0.78 mm; FWL: 4.59 mm; FWW: 1.50 mm; HWL: 3.26 mm; HWW: 1.00 mm; t1: 0.42 mm; t2: 0.13 mm.
Color (in alcohol): Head (
Figure 27F,G) brown. Frontal area with a pair of yellowish areas. Antenna dark brown. Mouthparts yellowish, with dark brown postclypeus. Prothorax yellowish brown, mid- and metathorax brown. Leg yellowish, hindleg with yellowish-brown tibia. Abdomen yellowish, genital segments yellowish with pale brown paraproct.
Forewing (
Figure 27H) transparent. Anterior area of pterostigma yellowish, posterior area of pterostigma with dark brown stripe. Hindwing (
Figure 27I) with a pale brown stripe between Sc and R.
Genital segments weakly sclerotized. Epiproct (
Figure 28D) subtriangular. Paraproct with 28 trichobothria. Subgenital plate (
Figure 28E) without distinctly sclerotized area. Gonapophyses (
Figure 28F) weakly sclerotized.
Specimens examined. Paratype of
S.
angustistriatus, ♀, China, Guizhou, Guiyang, Huaxi (1000), 1981.VI.9, Fasheng Li (CAU) (
Figure S39);
Holotype of
S.
genostictus, ♀, China, Yunnan, Ruili, Jiegao (750 m), 1981.V.5, Chikun Yang (CAU) (
Figure S40);
Holotype of
S.
macrocheirus, ♀, China, Gansu, Wenxian, 1980.VIII.7, Fasheng Li (CAU) (
Figure S41);
Paratype of
S.
thermophiles, ♀, China, Gansu, Kangxian, Lianghe (800 m), Chikun Yang (CAU) (
Figure S42);
Holotype of
S.
xiangxiensis,♀, China, Hunan, Zhangjiajie (600–1500 m), 1985.X.13, Fasheng Li (CAU) (
Figure S43).
CHINA: 2♂♂2♀♀, Shaanxi, Zhouzhi, Houzhenzi (1278 m), 2014.VII.17, Xiumei Lu (CAU); 2♀♀, Shaanxi, Lingwu, 2013.VII.13, Wanzhi Cai & Jianyun Wang (CAU); ♂, Shaanxi, Foping, Taiguping (1269 m), 2014.VIII.23, Xiumei Lu (CAU); 2♂♂2♀♀, Yunnan, Malipo, Xiajinchang (1430 m), 2016.VII.27, Yulan Jiang (CAU); 1♀, Guizhou, Luodian (850 m), 1981.VIII.8, Fasheng Li (CAU); 1♂1♀, Yunnan, Nanjian, Mt. Wuliangshan (2221 m), 2016.VII.17, Qicheng Yang (CAU); 1♀, Yunnan, Jinping, Maandi (1020 m), 2016.VII.13 Ya’nan Lv; VIETNAM: 1♀, Kon Tum, Chu Mom Ray National Park, 2012.VIII.1, Feiyang Liang (CAU).
Distribution. China (Guizhou, Yunnan, Guangdong, Guangxi, Shaanxi, Hunan), Vietnam (Kon Tum).
Remarks. There are intraspecific differences in the head markings of females, which are mainly displayed in the different brown areas in the frontal and genal regions. Combined with morphological characters and molecular species identification characters, N. angustistriatus, N. genostictus, N. macrocheirus, N. thermophilus, and N. xiangxisnsis are considered synonyms of N. zonatus. In addition, we also matched the male and female of this species based on the DNA barcoding. There are differences in marking patterns of forewing pterostigma between females and males of this species. The posterior margin of the pterostigma on the forewings of females bears a narrow brown band, whereas males have no obvious markings on their pterostigma. This species can be distinguished from N. dictyodromus by the yellowish scape and pedicel and the weakly sclerotized terminalia.