Disentangling Gut Bacterial Community Patterns in Cryptocercus punctulatus and Comparing Their Metagenomes with Other Xylophagous Dyctioptera Insects
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Cryptocercus punctulatus Metagenome Sequencing and Data Processing
2.3. Downloaded Data for Comparative Analysis
2.4. Bioinformatics and Statistical Analyses
3. Results
3.1. Bacterial Gut Patterns in Cryptocercus punctulatus
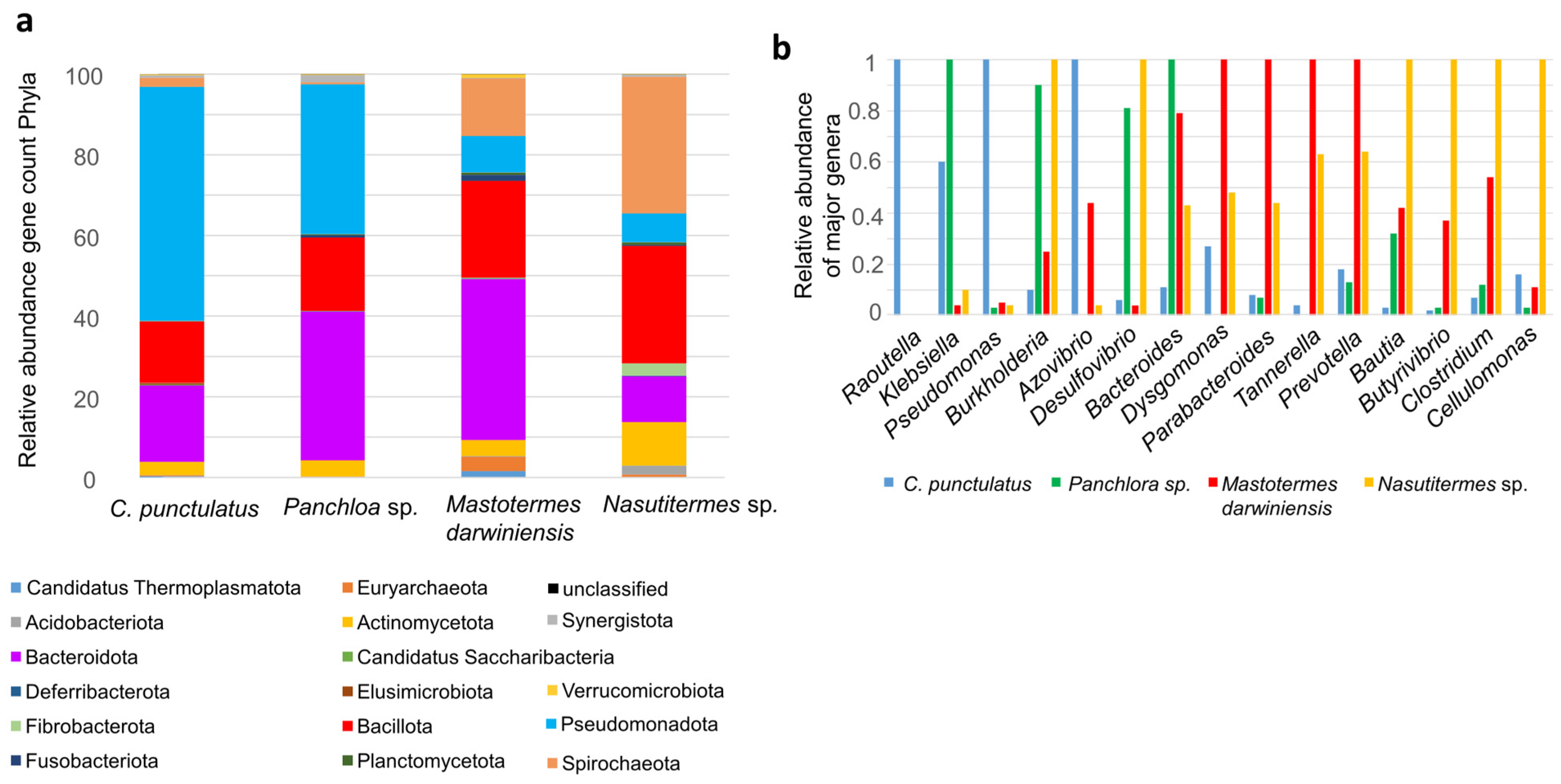
3.2. Shotgun Taxa of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
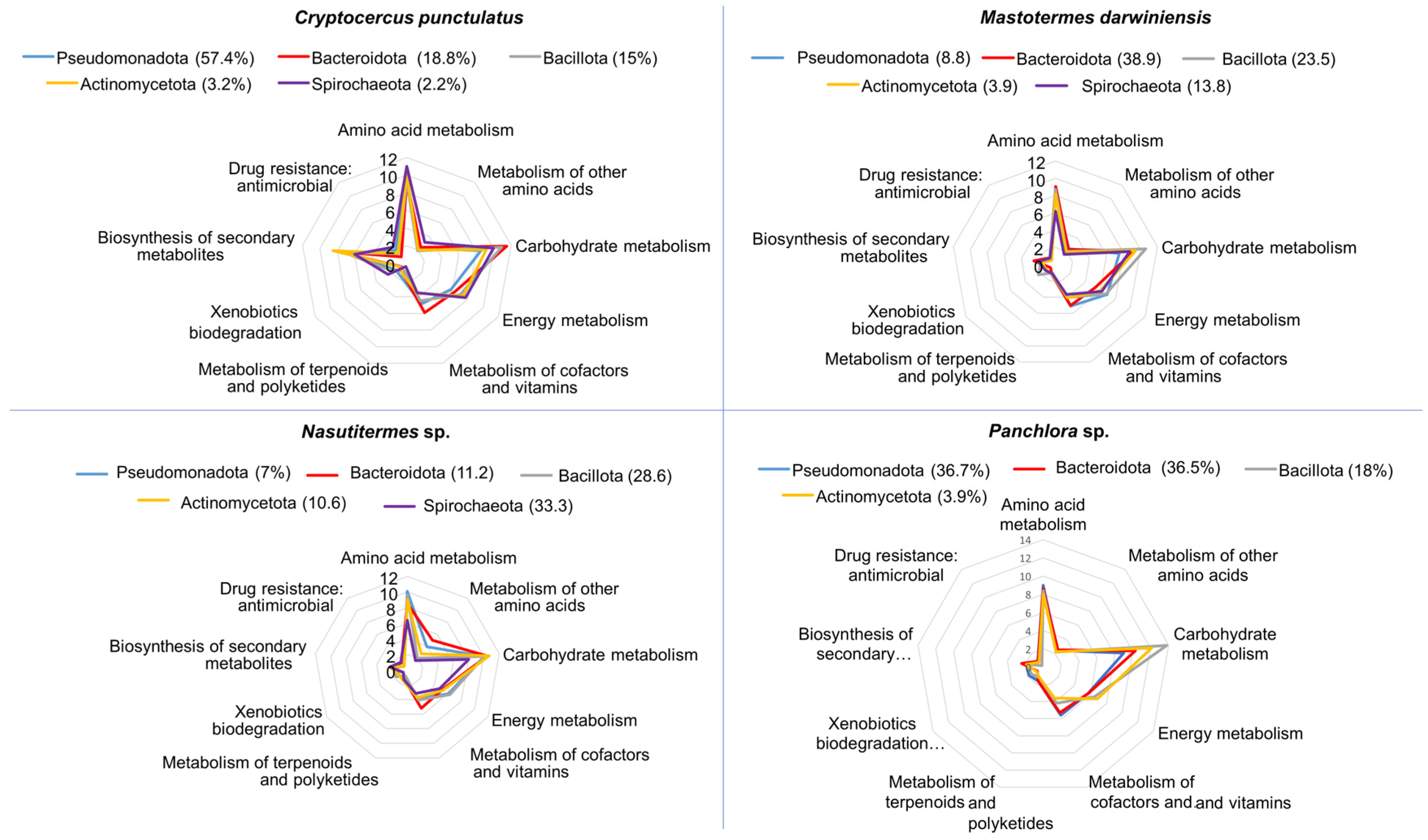
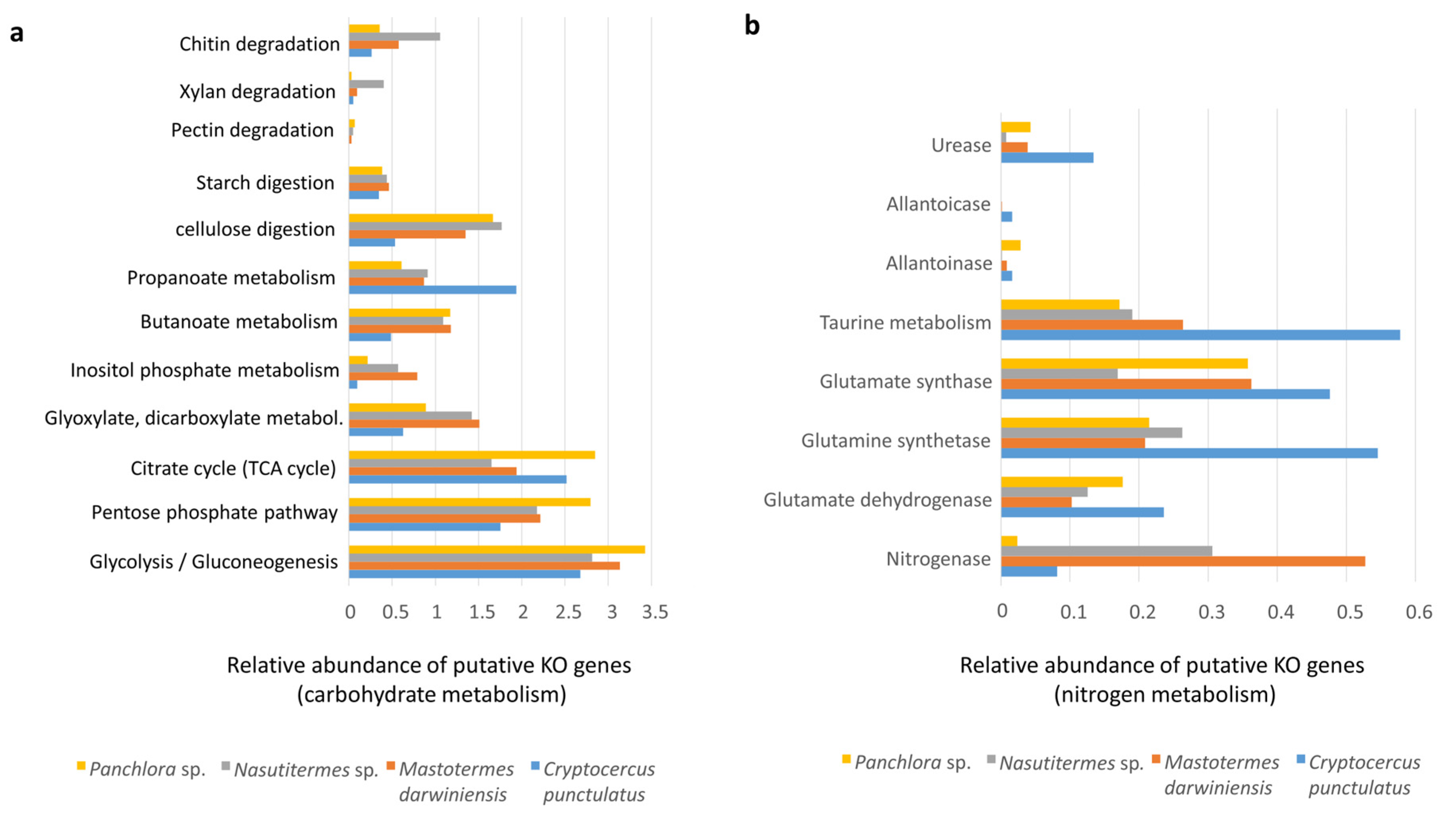
3.3. Functional Nutritional Shotgun Metagenome of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
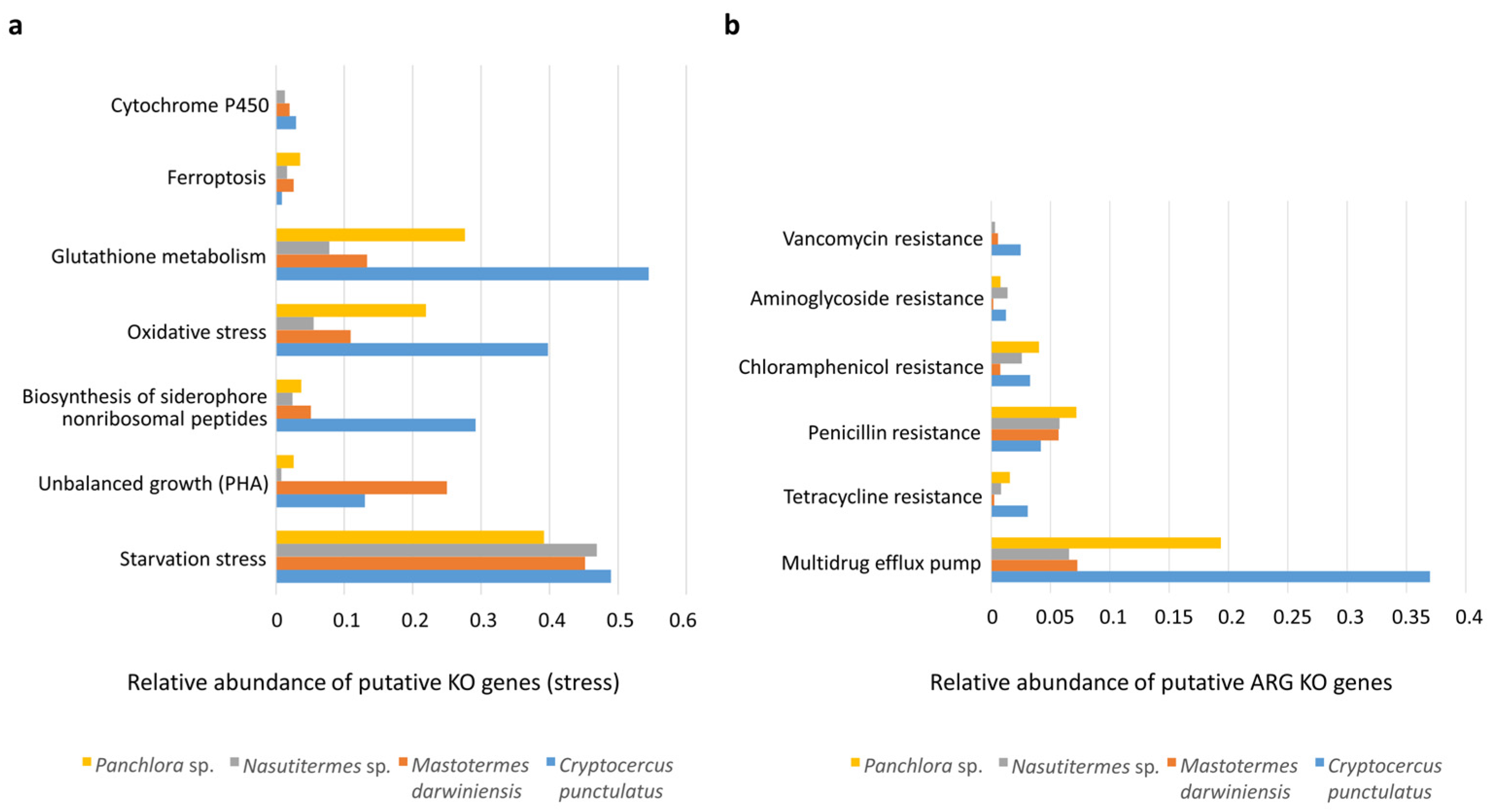
3.4. Functional Stress and Antibiotic Shotgun Metagenome of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
4. Discussion
4.1. Bacterial Gut Patterns from Cryptocercus punctulatus
4.2. Shotgun Taxa Metagenome of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
4.3. Functional Nutritional Shotgun Metagenome of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
4.4. Functional Stress and Antibiotic Shotgun Metagenome of Cryptocercus punctulatus and Other Xylophagous Dyctioptera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.D.; Geib, S.M.; Carlson, J.E.; Tien, M.; McKenna, D.; Hoover, K. Functional genomics and microbiome profiling of the Asian long horned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genom. 2014, 15, 1096. [Google Scholar] [CrossRef]
- Berlanga, M.; Guerrero, R. The holobiont concept: The case of xylophagous termites and cockroaches. Symbiosis 2016, 68, 49–60. [Google Scholar] [CrossRef]
- Scharf, M.E. Challenges and physiological implications of wood feeding in termites. Curr. Opin. Insect Sci. 2020, 41, 79–85. [Google Scholar] [CrossRef]
- Rowell, R.M.; Pettersen, R.; Tshabalada, M.A. Cell wall chemistry. In Handbook of Wood Chemistry and Wood Composites, 2nd ed.; Rowell, R.M., Ed.; CRC Press: Boca Raton, FL, USA, 2013; Volume 3, pp. 34–72. [Google Scholar]
- Christopher, L.P.; Yao, B.; Ji, Y. Lignin biodegradation with laccase-mediator systems. Front. Energy Res. 2014, 2, 12. [Google Scholar] [CrossRef]
- Montella, S.; Ventorino, V.; Lombard, V.; Henrissat, B.; Pepe, O.; Faraco, V. Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci. Rep. 2017, 7, 42623. [Google Scholar] [CrossRef]
- Brune, A.; Dietrich, C. The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 2015, 69, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.; Tokuda, G.; Watanabe, H.; Rose, H.; Slaytor, M.; Maekawa, K.; Bandi, C.; Noda, H. Evidence from multiple gene sequences indicates that termites evolved from wood-feeding cockroaches. Curr. Biol. 2000, 10, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Klass, K.; Nalepa, C.; Lo, N. Wood-feeding cockroaches as models for termite evolution (Insecta: Dictyoptera): Cryptocercus vs. Parasphaeria boleiriana. Mol. Phyl. Evol. 2008, 46, 809–817. [Google Scholar] [CrossRef]
- Lampert, N.; Mikaelyan, A.; Brune, A. Diet is not the primary driver of bacterial community structure in the gut of litter-feeding cockroaches. BMC Microbiol. 2019, 30, 238. [Google Scholar] [CrossRef]
- Jahnes, B.C.; Sabree, Z.L. Nutritional symbiosis and ecology of host-gut microbe systems in the Blattodea. Curr. Opin. Insect Sci. 2020, 39, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, M.; Llorens, C.; Comas, J.; Guerrero, R. Gut bacterial community of the xylophagous cockroaches Cryptocercus punctulatus and Parasphaeria boleiriana. PLoS ONE 2016, 11, e0152400. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Aylward, F.O.; Carlos, C.; del Río, T.G.; Chovatia, M.; Fern, A.; Lo, C.-C.; Malfatti, S.A.; Tringe, S.G.; Currie, C.R.; et al. Major changes in microbial diversity and community composition across gut sections of a juvenile Panchlora cockroach. PLoS ONE 2017, 12, e0177189. [Google Scholar] [CrossRef]
- Berlanga, M. Functional symbiosis and communication in microbial ecosystems. The case of wood-eating termites and cockroaches. Int. Microbiol. 2015, 18, 159–169. [Google Scholar] [CrossRef]
- Jahnes, B.C.; Poudel, K.; Staats, A.M.; Sabree, Z.L. Microbial colonization promotes model cockroach gut tissue growth and development. J. Insect Physiol. 2021, 133, 104274. [Google Scholar] [CrossRef]
- Hiltemann, S.; Rasche, H.; Gladman, S.; Hotz, H.-R.; Larivière, D.; Blankenberg, D.; Jagtap, P.D.; Wollmann, T.; Bretaudeau, A.; Goué, N.; et al. Galaxy Training: A powerful framework for teaching! PLoS Comput. Biol. 2023, 19, e1010752. [Google Scholar] [CrossRef]
- Huntemann, M.; Ivanova, N.N.; Mavromatis, K.; Tripp, H.J.; Paez-Espino, D.; Tennessen, K.; Palaniappan, K.; Szeto, E.; Pillay, M.; Chen, I.-M.A.; et al. The standard operation procedure of the DOE-JGI metagenome annotation pipeline (MAP v.4). Stand. Genom. Sci. 2016, 11, 17. [Google Scholar] [CrossRef]
- Utami, Y.D.; Kuwahara, H.; Igai, K.; Murakami, T.; Sugaya, K.; Morikawa, T.; Nagura, Y.; Yuki, M.; Deevong, P.; Inoue, T.; et al. Genome analyses of uncultured TG2/ZB3 bacteria in “margulisbacteria” specifically attached to ectosymbiotic spirochetes of protists in the termite gut. ISME J. 2019, 13, 455–467. [Google Scholar] [CrossRef]
- Tai, V.; James, E.R.; Nalepa, C.A.; Scheffranhn, R.H.; Perlman, S.J.; Keeling, P.J. The role of host phylogeny varies in shaping microbial diversity in the hindguts of lower termites. Appl. Environ. Microbiol. 2015, 81, 1059–1070. [Google Scholar] [CrossRef]
- Dietrich, C.; Brune, A. The cockroach origin of the termite gut microbiota: Patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 2014, 80, 2261–2269. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.G.; Saccenti, E. The power of microbiome studies: Some consideration on which alpha and beta metrics to use and how to report results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef]
- Van Hul, M.; Cani, P.; Petitfils, C.; De Vos, W.; Tilg, H.; El-Omar, E. What defines a healthy gut microbiome? Gut 2024, 73, 333378. [Google Scholar] [CrossRef]
- Tai, V.; Carpenter, L.J.; Weber, P.K.; Nalepa, C.A.; Perlman, S.J.; Keeling, P.J. Genome evolution and nitrogen fixation in bacterial ectosymbionts of a protist inhabiting wood-feeding cockroaches. Appl. Environ. Microbiol. 2016, 82, 4682–4695. [Google Scholar] [CrossRef]
- Hansen, A.K.; Pers, D.; Russell, J.A. Symbiotic solutions to nitrogen limitation and amino acid imbalance in insect diets. Adv. Insect Physiol. 2020, 58, 161–205. [Google Scholar] [CrossRef]
- Esteves, F.; Rueff, J.; Kranendonk, M. The central role of cytochrome P450 in xenobiotic metabolism—A brief review on a fascinating enzyme family. J. Xenobiotics 2021, 11, 94–114. [Google Scholar] [CrossRef]
- Xu, J.; Strange, J.P.; Welker, D.L.; James, R.R. Detoxification and stress response genes expressed in a western North American bumble bee, Bombus huntii (Hymenoptera: Apidae). BMC Genom. 2013, 14, 5075–5082. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Li, J.; Liu, M.; Liu, Z. Midgut transcriptome of the cockroach Periplaneta americana and its microbiota: Digestion, detoxification and oxidative stress response. PLoS ONE 2016, 11, e0155254. [Google Scholar] [CrossRef]
- Conrad, M.; Kagan, V.E.; Bayir, H.; Pagnussat, G.C.; Head, B.; Traber, M.G.; Stockwell, B.R. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Develop. 2018, 32, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Salem, H.; Bauer, E.; Strauss, A.S.; Vogel, H.; Marz, M.; Kaltenpoth, M. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. Royal Soc. B Biol. Sci. 2014, 281, 20141838. [Google Scholar] [CrossRef]
- Gupta, A.; Nair, S. Dynamics of insect-microbiome interactions influence host and microbial symbiont. Front. Microbiol. 2020, 11, 1357. [Google Scholar] [CrossRef]
- Bukin, Y.; Galachyants, Y.; Morozov, I.; Bukin, S.; Zakharenko, A.; Zemskaya, T. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci. Data 2019, 6, 190007. [Google Scholar] [CrossRef] [PubMed]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Deissova, T.; Zapletalová, M.; Kunovský, L.; Kroupa, R.; Grolich, T.; Kala, Z.; Linhartová, P.B.; Lochman, J. 16S rRNA gene primer choice impacts off-target amplification in human gastrointestinal tract biopsies and microbiome profiling. Sci. Rep. 2023, 13, 12577. [Google Scholar] [CrossRef]
- Addison, B.; Winange, M.C.D.; Pu, Y.; Ragauskas, A.J.; Harman-Ware, A.E. Solid-satate NMR at natural isotopic abundance for bioenergy application. Biotechnol. Biofuels Bioprod. 2025, 18, 46. [Google Scholar] [CrossRef]
- Nalepa, C.A. Origin of termite eusociality: Trophallaxis integrates the social, nutricional, and microbial environments. Ecol. Èntomol. 2015, 40, 323–335. [Google Scholar] [CrossRef]
- Nalepa, C.A. Early development of nymphs and establishment of hindgut symbiosis in Cryptocercus punctulatus (Dictyoptera: Cryptocercidae). Anna. Entomol. Soc. Am. 1990, 83, 786–789. [Google Scholar] [CrossRef]
- Waltmann, A.; Willcox, A.C.; Balasubramanian, S.; Mayorí, K.B.; Guerrero, S.M.; Sanchez, R.S.S.; Roach, J.; Pino, C.C.; Gilman, R.H.; Bern, C.; et al. Hindgut microbiota in laboratory-reared and wild Triatoma infestans. PLoS Neglected Trop. Dis. 2019, 13, e0007383. [Google Scholar] [CrossRef]
- Baltar, J.; Pavan, M.; Correa-Antonio, J.; Couto-Lima, D.; Maciel-De-Freitas, R.; David, M. Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil. Viruses 2023, 15, 1309. [Google Scholar] [CrossRef] [PubMed]
- Tinker, K.; Ottesen, E. Differences in Gut Microbiome Composition Between Sympatric Wild and Allopatric Laboratory Populations of Omnivorous Cockroaches. Front. Microbiol. 2021, 12, 703785. [Google Scholar] [CrossRef]
- Kundu, P.; Manna, B.; Majumder, S.; Ghosh, A. Species-wide metabolic interaction network for understanding natural lignocellulose digestion in termite gut microbiota. Sci. Rep. 2019, 9, 16329. [Google Scholar] [CrossRef]
- Marynowska, M.; Goux, X.; Sillam-Dussès, D.; Rouland-Lefèvre, C.; Halder, R.; Wilmes, P.; Gawron, P.; Roisin, Y.; Delfosse, P.; Calusinska, M. Compositional and functional characterization of biomass-degrading microbial communities in guts and plant fiber- and soil-feeding higher termites. Microbiome 2020, 8, 96. [Google Scholar] [CrossRef]
- Vera-Ponce de Leon, A.; Jahnes, B.C.; Duan, J.; Camuy-Vélez, L.A.; Sabree, Z.L. Cultivable, host-specific Bacteroidetes symbionts exhibit diverse polysaccharolytic strategies. Appl. Environ. Microbiol. 2020, 86, e00091-20. [Google Scholar] [CrossRef]
- Saati-Santamaria, Z.; Rivas, R.; Kolarik, M.; Garcia-Fraile, P. A new perspective of Pseudomonas—Host interactions: Distribution and potential ecological functions of the genus Pseudomonas within the Bark Beetle holobiont. Biology 2021, 10, 164. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, F.; Lu, X. Diversity and functional roles of the gut microbiota in Lepidopteran insects. Microorganisms 2022, 10, 1234. [Google Scholar] [CrossRef]
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Brune, A. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. USA 2018, 155, E11996–E12004. [Google Scholar] [CrossRef]
- Gazal, V.; Bailez, O.; Viana-Bailez, A.M.; Aguilar Menenzes, E.; Barsanulfo Menendez, E. Decayed wood affecting the attraction of the pest arboretum termite Nasutitermes corniger (Isoptera: Termitidae) to resource foods. Sociobiology 2012, 59, 287–295. [Google Scholar] [CrossRef]
- Hu, H.; Rodrigues da Costa, R.; Pilgaard, B.; Schiott, M.; Lange, L.; Poulsen, M. Fungiculture in termites is associated with mycolytic gut bacterial community. mSphere 2019, 4, e00165-19. [Google Scholar] [CrossRef]
- Tong, R.L.; Aguilera-Olivares, D.; Chouvenc, T.; Su, N.-Y. Nitrogen content of the exuviae of Coptotermes gestroi (Wasmann) (Blattodea: Rhinotermitidae). Heliyon 2021, 7, e06697. [Google Scholar] [CrossRef]
- Stentz, R.; Osborne, S.; Horn, N.; Li, A.W.H.; Hautefort, I.; Bongaerts, R.; Rouyer, M.; Bailey, P.; Shears, S.B.; Hemmings, A.M.; et al. A bacterial homolog of eukaryotic inositol phosphate signaling enzyme mediates cross-kingdom dialog in the mammalian gut. Cell Rep. 2014, 6, 646–656. [Google Scholar] [CrossRef]
- Yang, P.; Liu, W.; Chen, Y.; Gong, A.-D. Engineering the glyoxylate cycle for chemical bioproduction. Front. Bioeng. Biotechnol. 2022, 10, 1066651. [Google Scholar] [CrossRef]
- Fisher, S.H. Regulation of nitrogen metabolism in Bacillus subtilis: Vive la difference. Mol. Microbiol. 1999, 32, 223–232. [Google Scholar] [CrossRef]
- Reitzer, L. Nitrogen assimilation and global regulation in Escherichia coli. Ann. Rev. Microbiol. 2003, 57, 155–176. [Google Scholar] [CrossRef]
- Rohwerder, T. New structural insights into bacterial sulfoacetaldehyde and taurine metabolism. Biochem. J. 2020, 477, 1367–1371. [Google Scholar] [CrossRef]
- Ge, S.-X.; Li, T.-F.; Ren, L.-L.; Zong, S.-X. Host-plant adaptation in xylophagous insect-microbiome systems: Contributions of longicorns and gut symbionts revealed by parallel metatranscriptome. iScience 2023, 26, 106680. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.; Moeller, A.H. The effects of temperature on animal gut microbiomes. Front. Microbiol. 2020, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, M.; Palau, M.; Guerrero, R. Gut microbiota dynamics and functionality in Reticulitermes grassei after a 7-day dietary shift and ciprofloxacin treatment. PLoS ONE 2018, 13, e0209789. [Google Scholar] [CrossRef]
- Zhao, M.; Lin, X.; Guo, X. The role of insect symbiotic bacteria in metabolizing phytochemicals and agrochemicals. Insects 2022, 13, 583. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.S.; Aylward, F.O.; Adams, S.M.; Erbilgin, N.; Aukema, B.H.; Currie, C.R.; Suen, G.; Raffa, K.F. Mountain pine beetles colonizing historical and naïve host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microbiol. 2013, 79, 3468–3475. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, J.; Zhu, F.; Shen, Z.; Jiang, H.; Li, Z.; Liu, X.; Xu, H. Function of cytochrome P450s and gut microbiome in biopesticide adaptation of Grapholita molesta on different host diets. Int. J. Mol. Sci. 2023, 24, 15435. [Google Scholar] [CrossRef]
- Brune, A.; Emerson, D.; Breznak, J.A. The termite gut microflora as an oxygen sink: Microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 1995, 61, 2681–2687. [Google Scholar] [CrossRef]
- Berlanga, M.; Paster, B.J.; Guerrero, R. The taxophysiological paradox: Changes in the intestinal microbiota of the xylophagous cockroach Cryptocercus punctulatus depending on the physiological state of the host. Int. Microbiol. 2009, 12, 227–236. [Google Scholar] [CrossRef]
- Pester, M.; Brune, A. Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 2007, 1, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.-A.T.; Chen, Q.-L.; He, J.-Z.; Hu, H.-W. Microbial regulation of natural antibiotic resistance: Understanding the protist-bacteria interactions for evolution of soil resistome. Sci. Total Environ. 2020, 705, 135882. [Google Scholar] [CrossRef]
- Levy, S.B.; Marshall, B.M. Honeybees and tetracycline resistance. mBio 2013, 4, e00045-13. [Google Scholar] [CrossRef]
- Liu, C.C.; Yao, H.Y.; Wang, C.W. Black soldier fly larvae can effectively degrade oxytetracycline bacterial residue by means of the gut bacterial community. Front. Microbiol. 2021, 12, 663972. [Google Scholar] [CrossRef]
- Maestre-Carballa, L.; Navarro-López, V.; Martinez-Garcia, M. A Resistome roadmap: From the human body to pristine environments. Front. Microbiol. 2022, 13, 858831. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Greening, C. Termite-engineered microbial communities of termite nest structures: A new dimension to the extended phenotype. FEMS Microbiol. Rev. 2022, 46, fuac034. [Google Scholar] [CrossRef] [PubMed]
- Llop, P.; Latorre, A.; Moya, A. Experimental epidemiology of antibiotic resistance: Looking for an appropriate animal model system. Microbiol. Spectrum. 2018, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bogri, A.; Jensen, E.E.B.; Borchert, A.V.; Brinch, C.; Otani, S.; Aarestrup, F.M. Transmission of antimicrobial resistance in the gut microbiome of gregarious cockroaches: The importance of interaction between antibiotic exposed and non-exposed populations. mSystems 2024, 9, e0101823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Wang, T.; Xu, N.; Lu, T.; Hong, W.; Penuelas, J.; Gillings, M.; Wang, M.; Gao, W.; et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef] [PubMed]
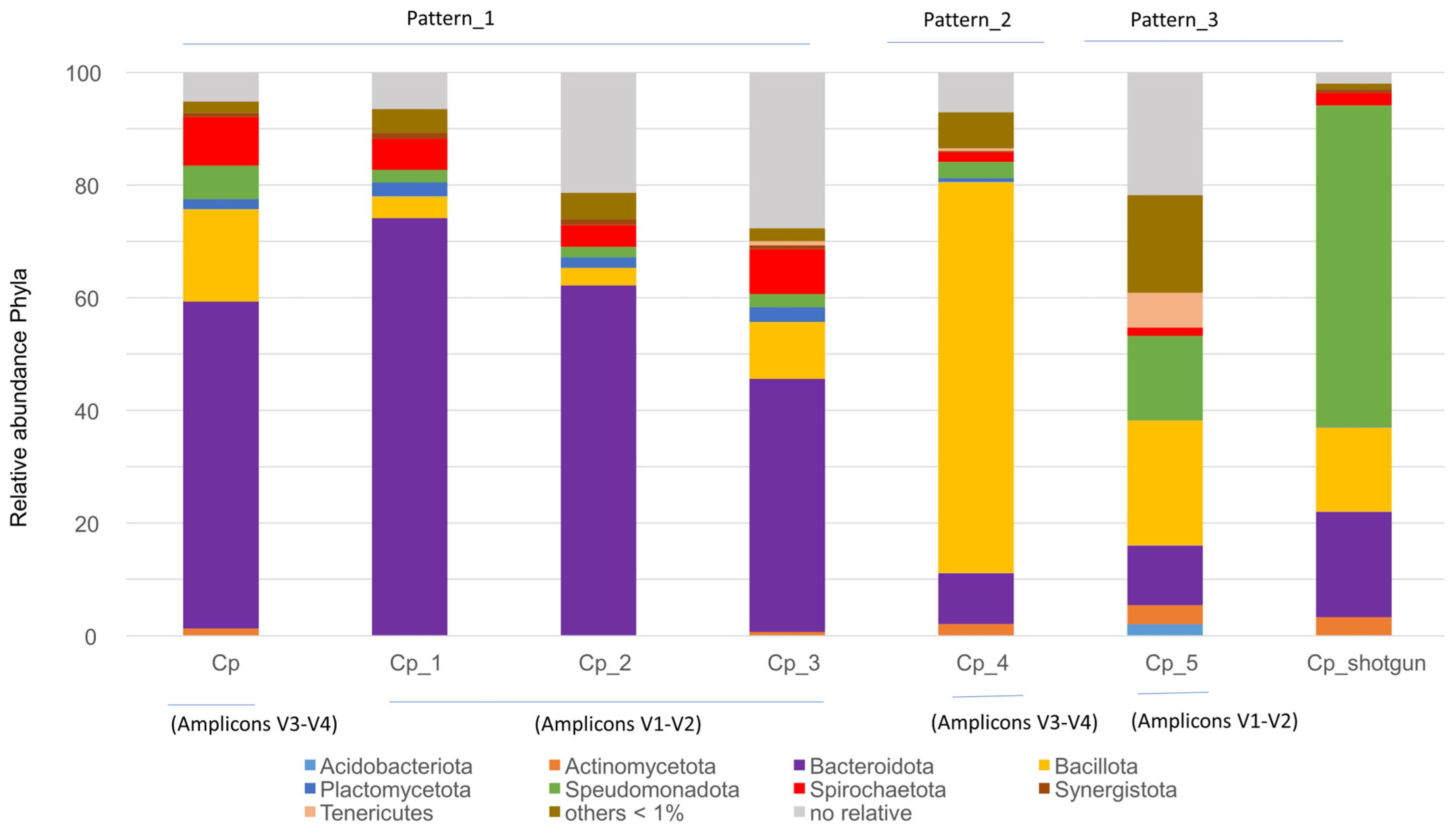
| Sample | Field Collected | 16S rRNA | Intestinal Tract | Valid Reads | Silva Clustering (OTU) |
|---|---|---|---|---|---|
| Cp | North Carolina (NC) or Virginia (VA), USA | V3–V4 | Entire gut | 148,434 | 40,364 |
| Cp_1 | NC (Mt. Collins), USA | V1–V2 | Hindguts | 12,352 | 10,513 |
| Cp_2 | VA (Mountain Lake), USA | V1–V2 | Hindguts | 10,297 | 9034 |
| Cp_3 | NC (South Mountains) USA | V1–V2 | Hindguts | 9322 | 4445 |
| Cp_4 | NC (Heywood Country) USA | V3–V4 | Hindguts | 36,158 | 10,388 |
| Cp_5 | VA (USA) and maintained in laboratory conditions | V1–V2 | Entire gut | 5221 | 2356 |
| Dictyoptera | Collected | JGI Database Gold | Annotated Gens | % Assembled (Genes KO) |
|---|---|---|---|---|
| Mastotermes darwiniensis | Townsville, Australia | Ga0068304 | 1,421,148 | 6.80 |
| Nasutitermes sp. | Murphy’s Creek, Queensland, Australia | Ga0072940 | 1,121,956 | 18.49 |
| Panchlora sp. | Gamboa Forest, Panama | Ga0026008 | 108,739 | 33.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berlanga, M.; Miñana-Galbis, D.; Guerrero, R. Disentangling Gut Bacterial Community Patterns in Cryptocercus punctulatus and Comparing Their Metagenomes with Other Xylophagous Dyctioptera Insects. Insects 2025, 16, 1128. https://doi.org/10.3390/insects16111128
Berlanga M, Miñana-Galbis D, Guerrero R. Disentangling Gut Bacterial Community Patterns in Cryptocercus punctulatus and Comparing Their Metagenomes with Other Xylophagous Dyctioptera Insects. Insects. 2025; 16(11):1128. https://doi.org/10.3390/insects16111128
Chicago/Turabian StyleBerlanga, Mercedes, David Miñana-Galbis, and Ricardo Guerrero. 2025. "Disentangling Gut Bacterial Community Patterns in Cryptocercus punctulatus and Comparing Their Metagenomes with Other Xylophagous Dyctioptera Insects" Insects 16, no. 11: 1128. https://doi.org/10.3390/insects16111128
APA StyleBerlanga, M., Miñana-Galbis, D., & Guerrero, R. (2025). Disentangling Gut Bacterial Community Patterns in Cryptocercus punctulatus and Comparing Their Metagenomes with Other Xylophagous Dyctioptera Insects. Insects, 16(11), 1128. https://doi.org/10.3390/insects16111128









