Structural and Functional Insights into Methuselah Genes of Plutella xylostella (Lepidoptera: Plutellidae): Evolutionary Adaptations and Their Responses to Chlorantraniliprole
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Insects
2.2. Leaf-Dip Bioassays
2.3. Insecticides
2.4. Documentation of Mth Genes in the P. xylostella Genome
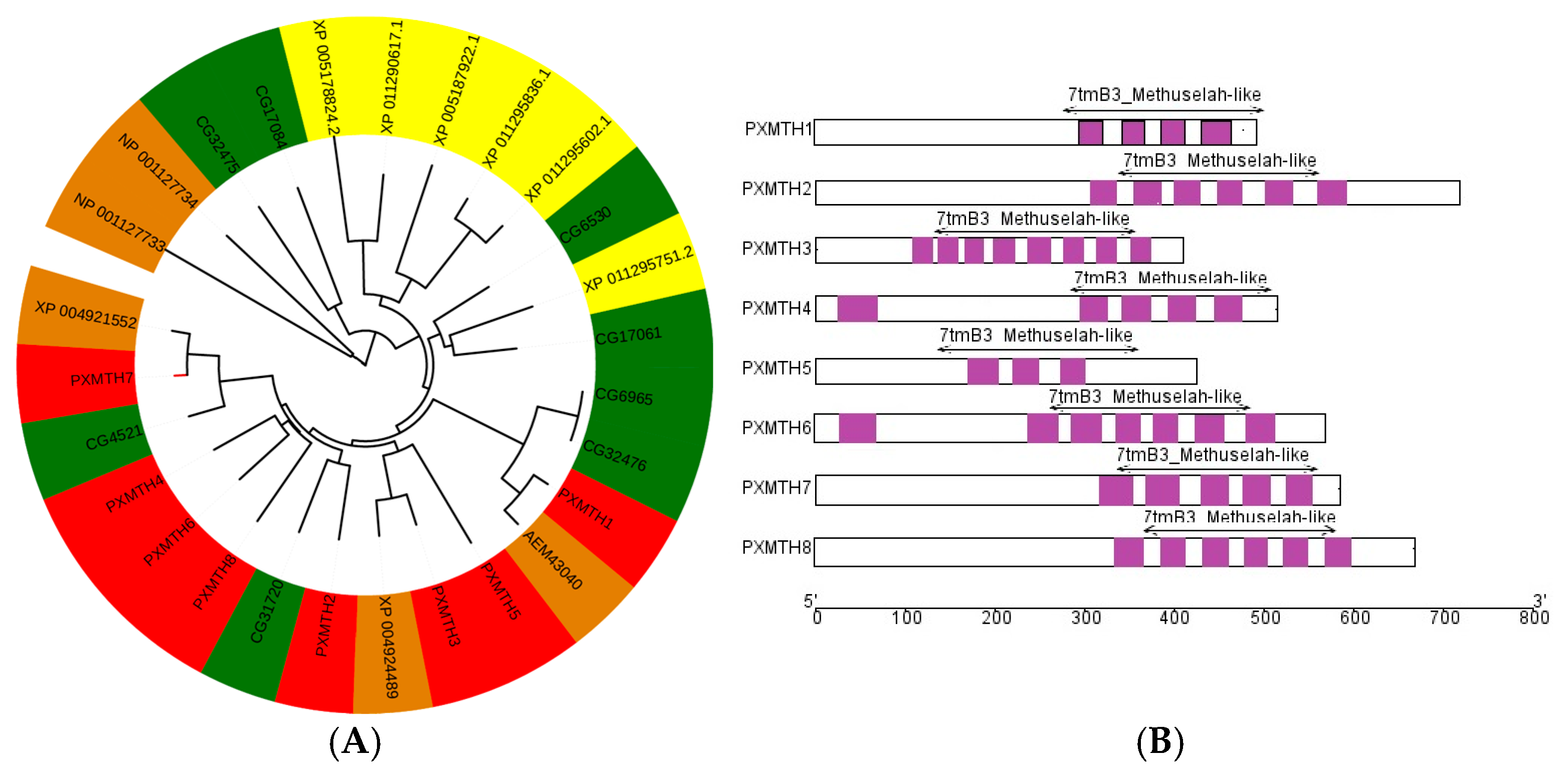
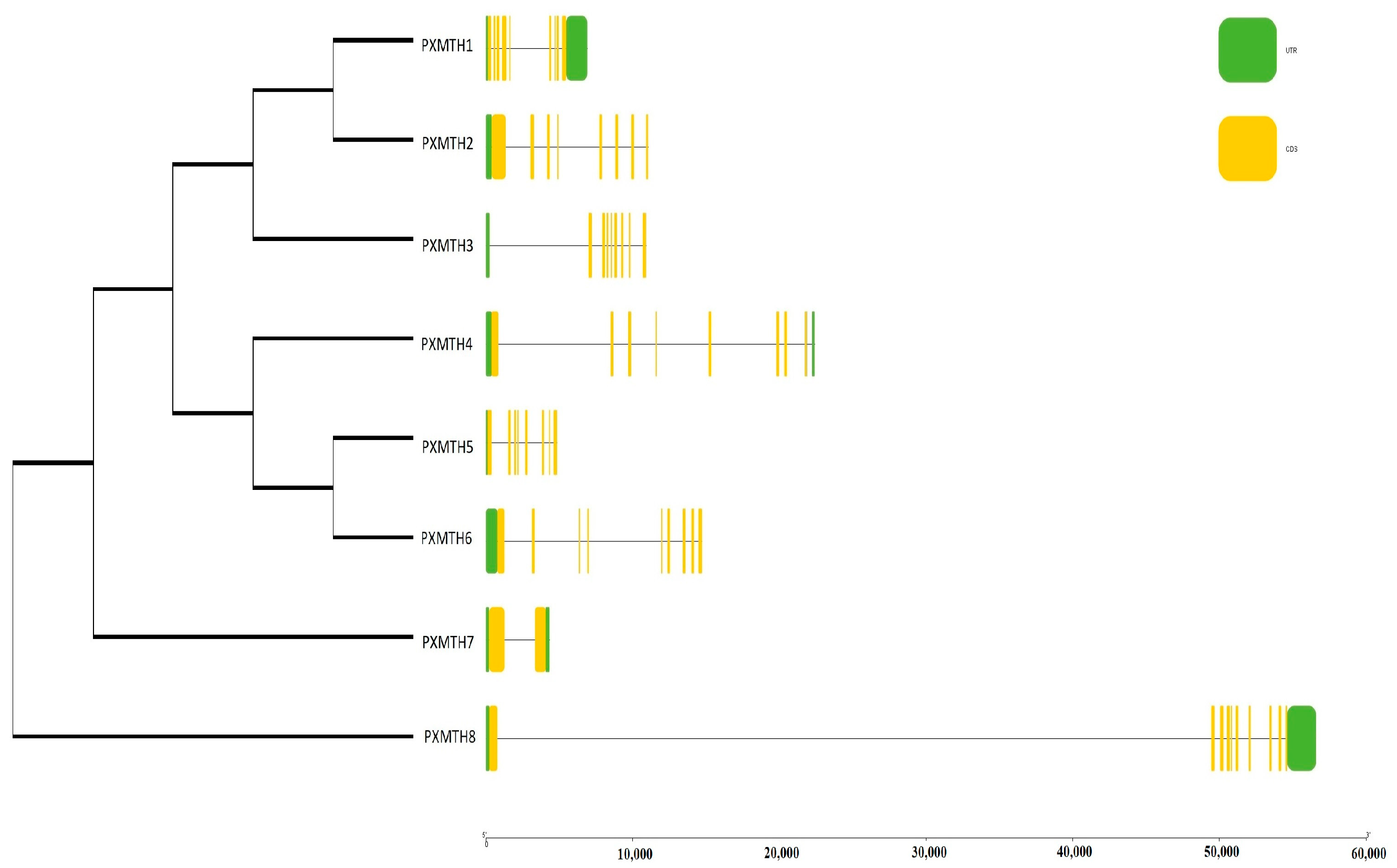
2.5. Phylogenetic Analysis and Chromosomal Mapping
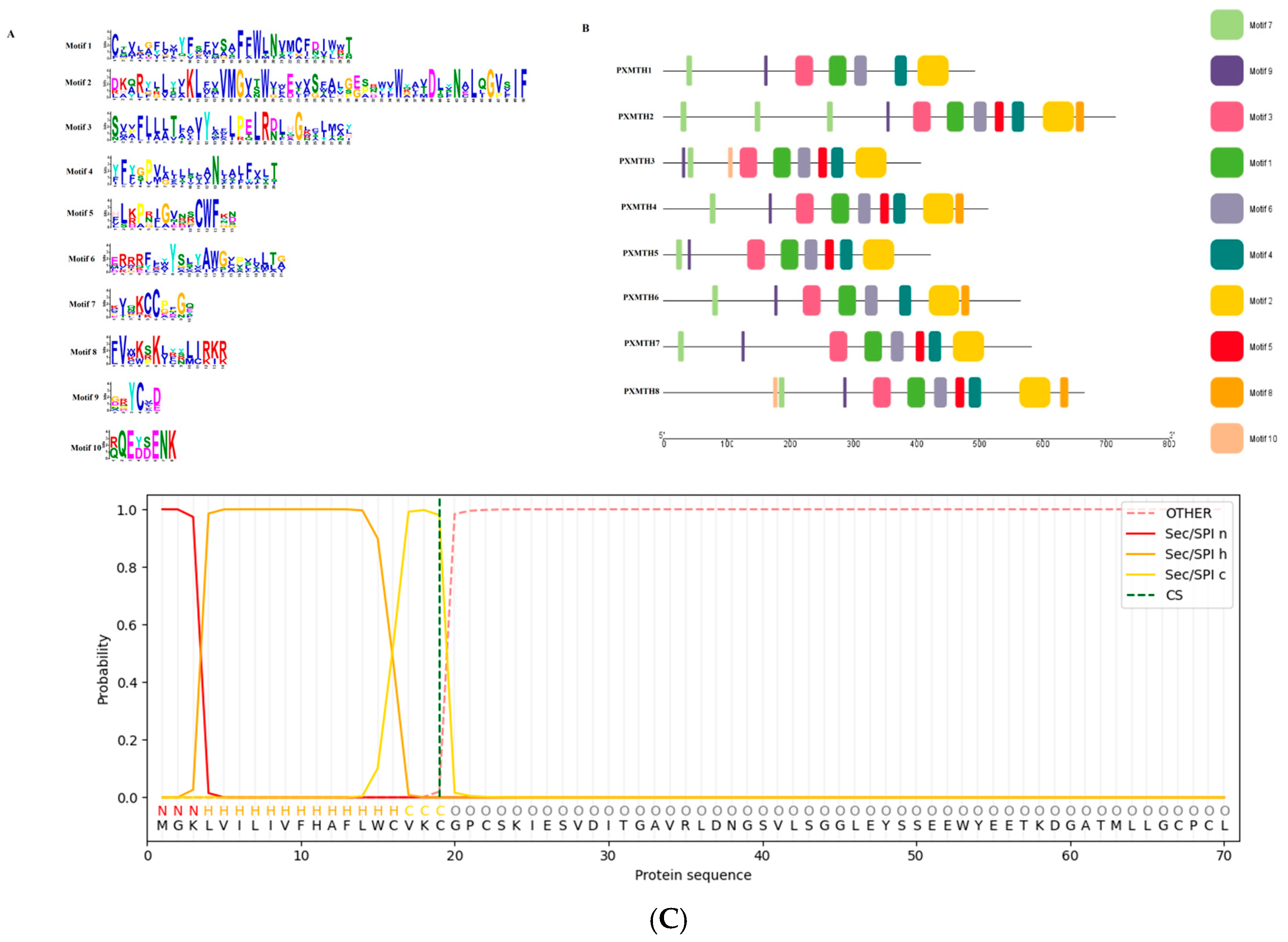
2.6. Gene Structure and Motif Analysis
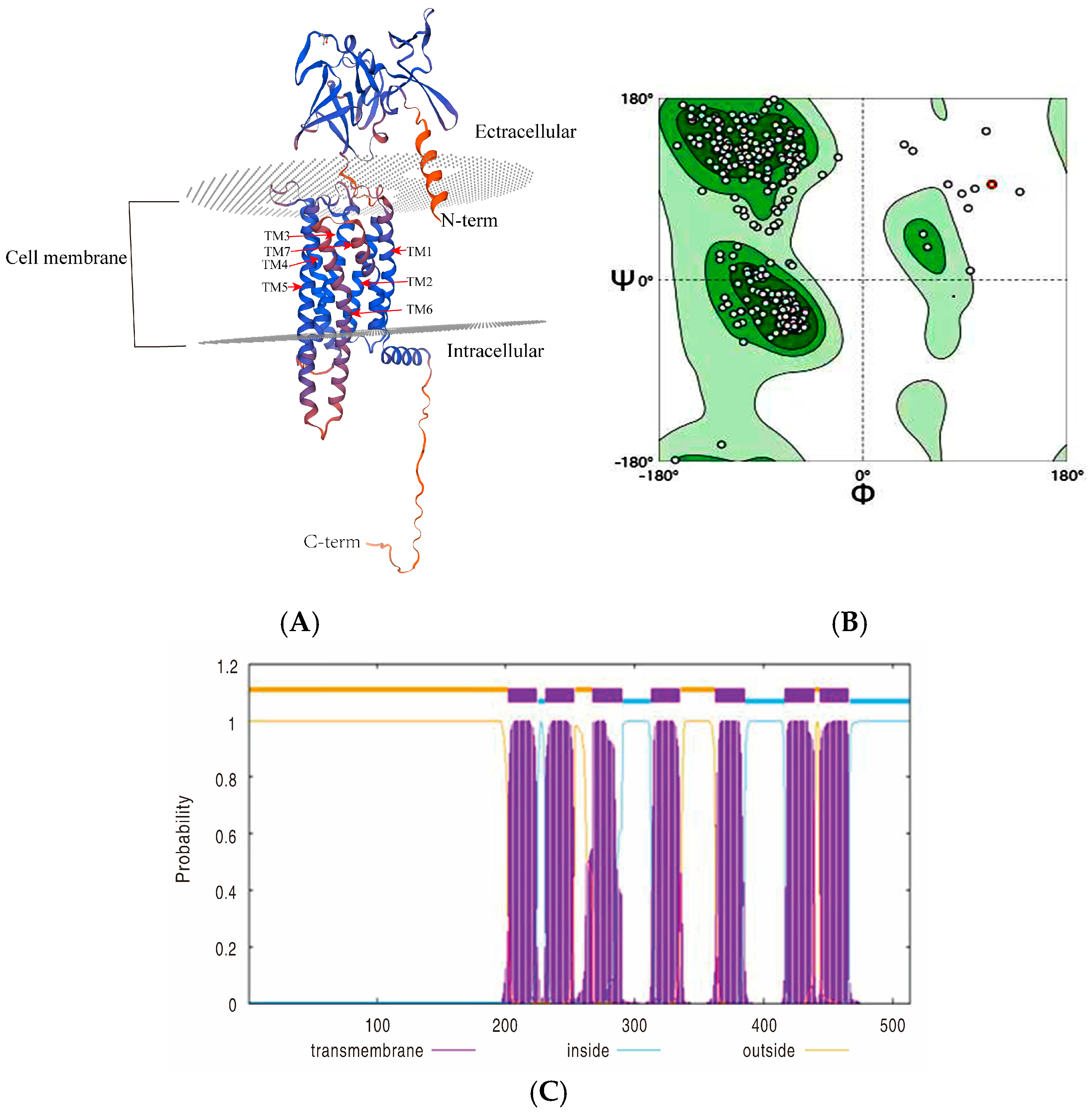
2.7. Protein Structure Analysis
2.7.1. Transmembrane Domain Prediction
2.7.2. Ramachandran Plot Analysis
2.8. Multiple Sequence Alignment and Phylogenetic Analysis
2.9. Signal Peptide Prediction
2.10. Expression Profiling of Mth Genes Under CAP Exposure
2.11. Functional Verification of Pxmth2 by RNA Interference (RNAi)
2.12. Generation of UAS-Pxmth2 Transgenic Drosophila
2.13. Statistical Analysis
3. Results
3.1. Identification and Distribution of Mth-Encoding Genes in the P. xylostella Genome
3.2. Characterization of Pxmth in P. xylostella
3.3. Phylogenetic and Domain Analysis
3.4. Gene Structure and Conserved Motif Analysis
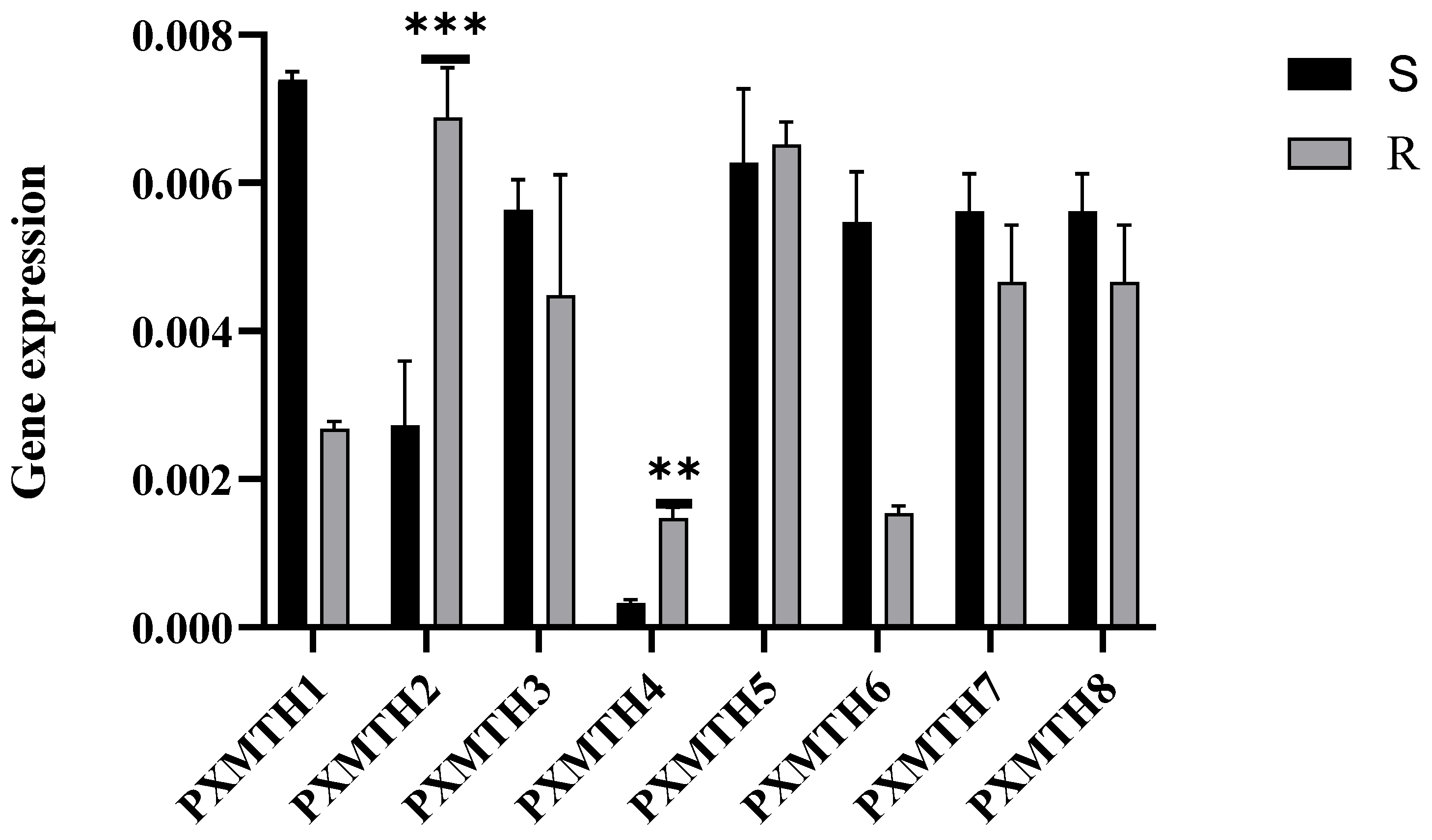
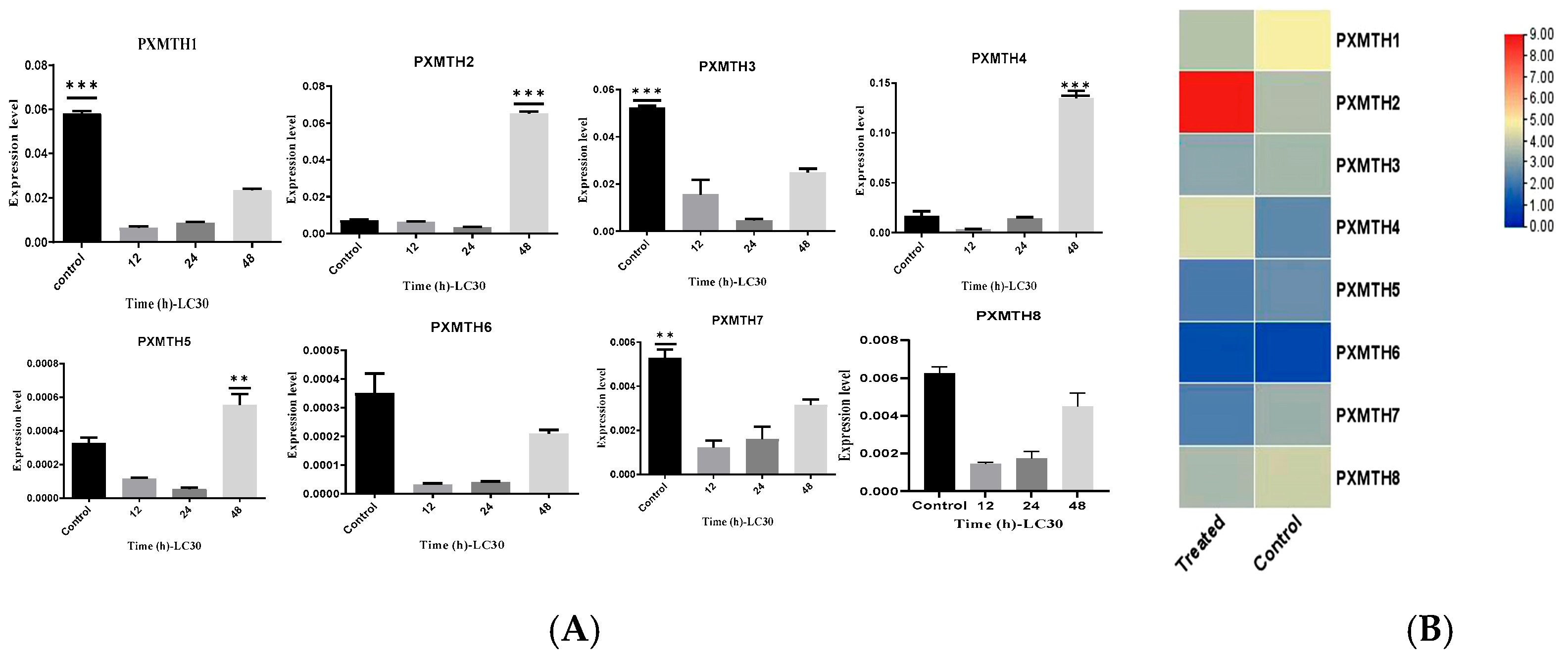
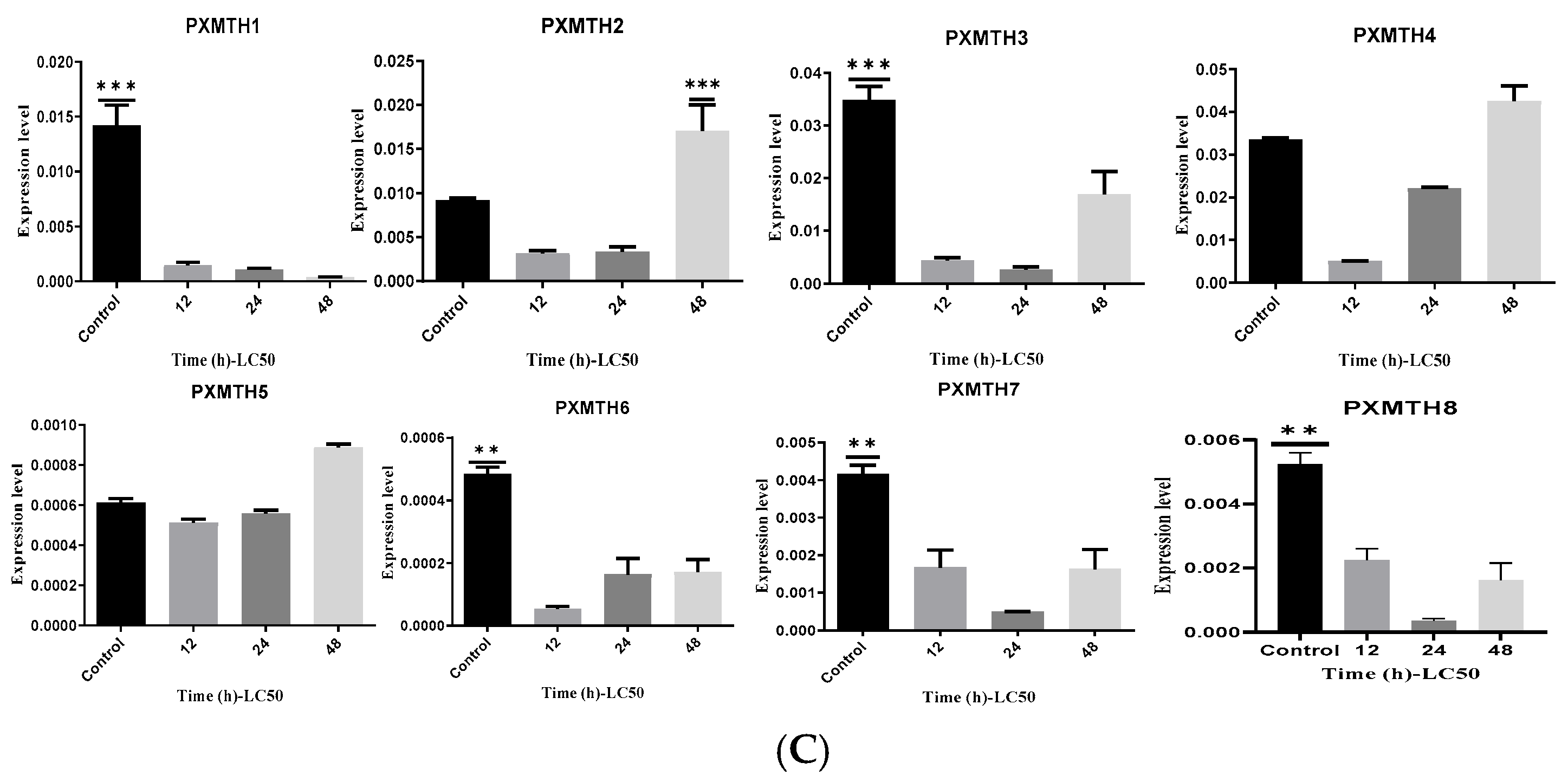
3.5. Expression Profiles of Mth Receptors in Response to Chlorantraniliprole
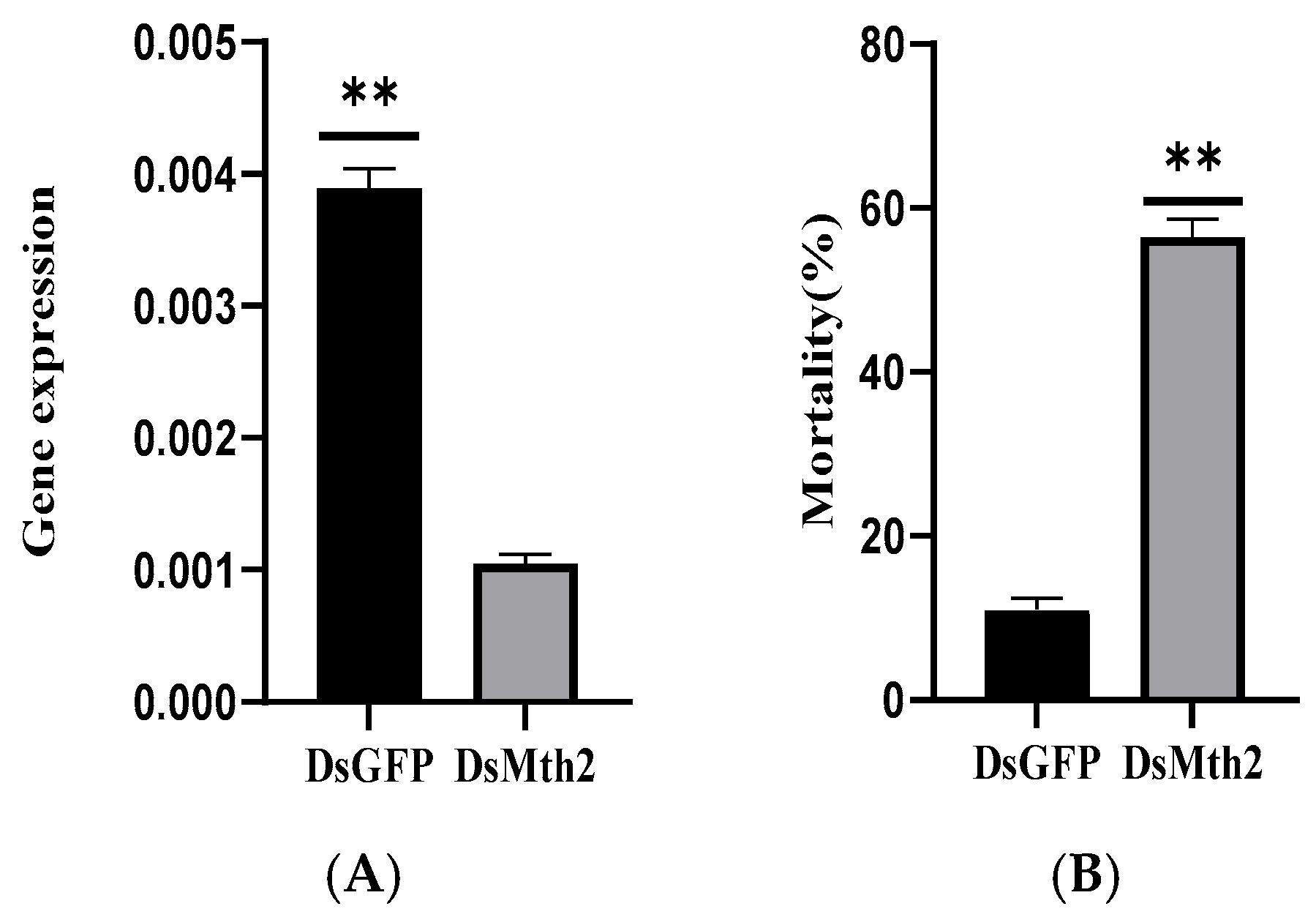
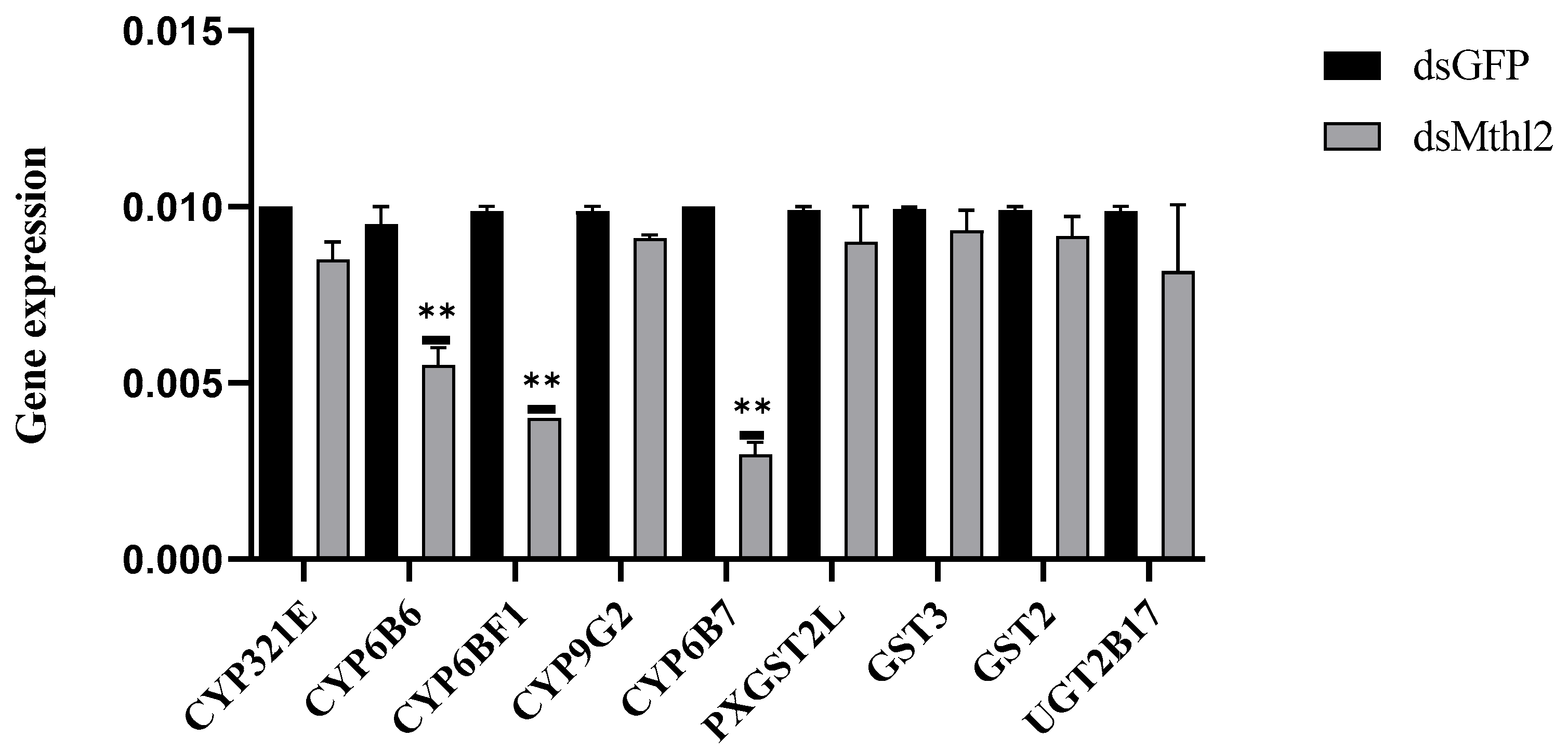
3.6. Investigation of RNAi of Pxmth2 in Resistance to CAP
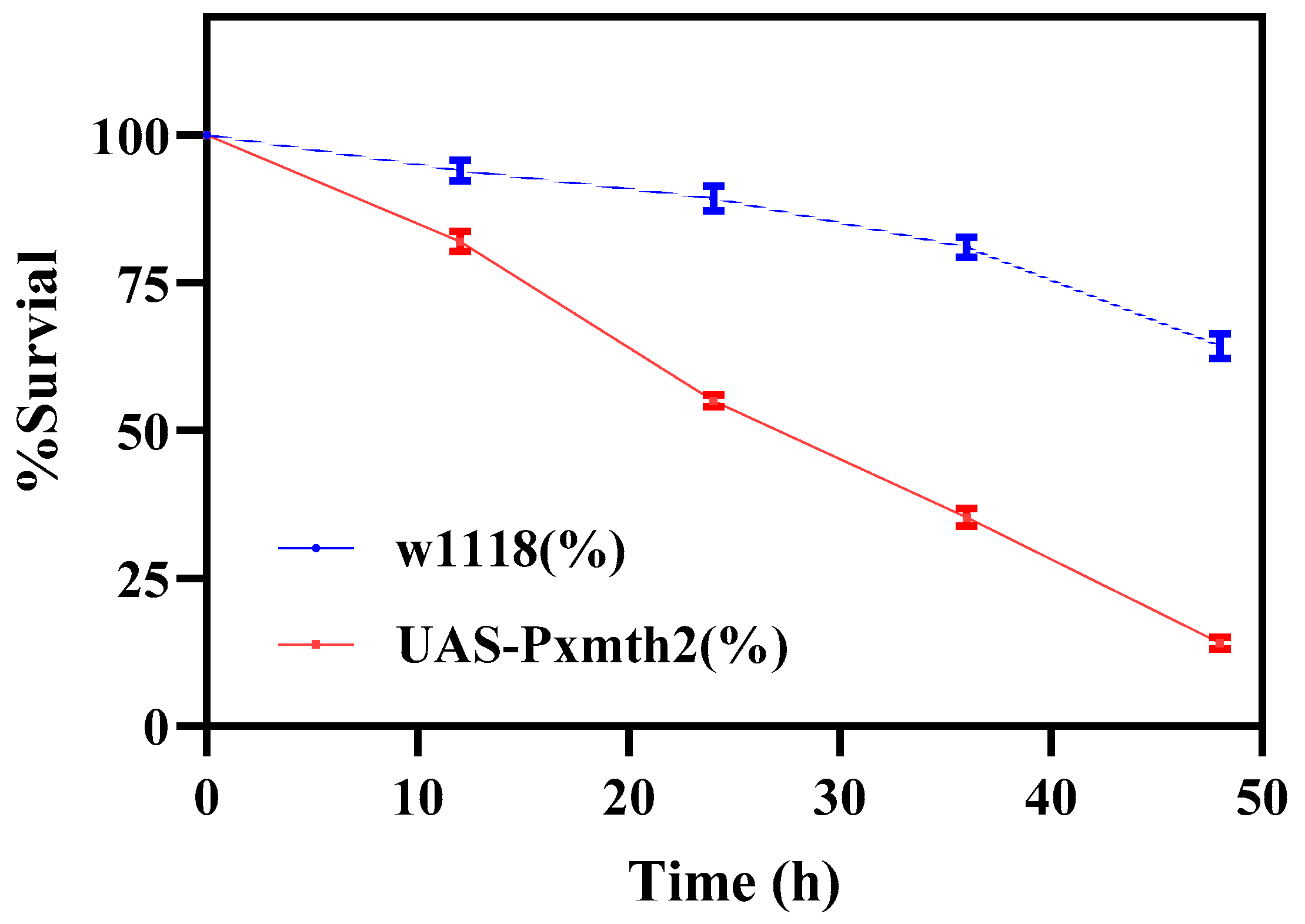
3.7. Pxmth2 Overexpression Confers CAP Resistance in Drosophila
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Garland, S.L. Are GPCRs still a source of new targets? J. Biomol. Screen. 2013, 18, 947–966. [Google Scholar] [CrossRef]
- Filmore, D. It’s a GPCR world. Mod. Drug Discov. 2004, 7, 24–28. [Google Scholar]
- Klabunde, T.; Hessler, G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem 2002, 3, 928–944. [Google Scholar] [CrossRef]
- Liu, N.; Li, T.; Wang, Y.; Liu, S. G-protein coupled receptors (GPCRs) in insects—A potential target for new insecticide development. Molecules 2021, 26, 2993. [Google Scholar] [CrossRef]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.-Y. Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef]
- Falkenburger, B.H.; Dickson, E.J.; Hille, B. Quantitative properties and receptor reserve of the DAG and PKC branch of Gq-coupled receptor signaling. J. Gen. Physiol. 2013, 141, 537–555. [Google Scholar] [CrossRef]
- Kim, C.; Cheng, C.Y.; Saldanha, S.A.; Taylor, S.S. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell 2007, 130, 1032–1043. [Google Scholar] [CrossRef]
- Chen-Goodspeed, M.; Lukan, A.N.; Dessauer, C.W. Modeling of Gαs and Gαi regulation of human type V and VI adenylyl cyclase. J. Biol. Chem. 2005, 280, 1808–1816. [Google Scholar] [CrossRef]
- Bradley, J.; Li, J.; Davidson, N.; Lester, H.A.; Zinn, K. Heteromeric olfactory cyclic nucleotide-gated channels: A subunit that confers increased sensitivity to cAMP. Proc. Natl. Acad. Sci. USA 1994, 91, 8890–8894. [Google Scholar] [CrossRef]
- Li, T.; Cao, C.; Yang, T.; Zhang, L.; He, L.; Xi, Z.; Bian, G.; Liu, N. A G-protein-coupled receptor regulation pathway in cytochrome P450-mediated permethrin-resistance in mosquitoes, Culex quinquefasciatus. Sci. Rep. 2015, 5, 17772. [Google Scholar] [CrossRef]
- Li, T.; Liu, N. The function of G-protein-coupled receptor-regulatory cascade in southern house mosquitoes (Diptera: Culicidae). J. Med. Entomol. 2018, 55, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Audsley, N.; Down, R.E. G protein coupled receptors as targets for next generation pesticides. Insect Biochem. Mol. Biol. 2015, 67, 27–37. [Google Scholar] [CrossRef]
- Patel, M.V.; Hallal, D.A.; Jones, J.W.; Bronner, D.N.; Zein, R.; Caravas, J.; Husain, Z.; Friedrich, M.; Vanberkum, M.F. Dramatic expansion and developmental expression diversification of the methuselah gene family during recent Drosophila evolution. J. Exp. Zool. B Mol. Dev. Evol. 2012, 318, 368–387. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ranjan, R.; Dawson-Scully, K.; Bronk, P.; Marin, L.; Seroude, L.; Lin, Y.J.; Nie, Z.; Atwood, H.L.; Benzer, S.; et al. Presynaptic regulation of neu rotransmission in Drosophila by the g protein-coupled receptor methuselah. Neuron 2002, 36, 105–119. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Seroude, L.; Benzer, S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 1998, 282, 943–946. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Zhang, Z.; Hua, Q.; Wang, M.; Shi, C.; Xue, L.; Zhang, R.; Xie, X. Methuselah regulates longevity via dTOR: A pathway revealed by small-molecule ligands. J. Mol. Cell Biol. 2015, 7, 280–282. [Google Scholar] [CrossRef]
- Li, C.; Wu, W.; Sang, M.; Liu, X.; Hu, X.; Yun, X.; Li, B. Comparative RNA-sequencing analysis of mthl1 functions and signal transductions in Tribolium castaneum. Gene 2014, 547, 310–318. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zhang, J.; Yang, C.J.; Lian, H.Y.; Yu, H.; Huang, X.M.; Cai, P. Coupling mechanism of electromagnetic field and thermal stress on Drosophila melanogaster. PLoS ONE 2016, 11, e0162675. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Huang, F.; Ghimire, M.N.; Leonard, B.R.; Siegfried, B.D.; Rangasamy, M.; Yang, Y.; Wu, Y.; Gahan, L.J.; Heckel, D.G.; et al. Efficacy of genetically modified Bt toxins against insects with different genetic mechanisms of resistance. Nat. Biotechnol. 2011, 29, 1128–1131. [Google Scholar] [CrossRef]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef]
- Xu, P.; Lu, B.; Xiao, H.; Fu, X.; Murphy, R.W.; Wu, K. The evolution and expression of the moth visual opsin family. PLoS ONE 2013, 8, e78140. [Google Scholar] [CrossRef]
- Lee, D.W.; Shrestha, S.; Kim, A.Y.; Park, S.J.; Yang, C.Y.; Kim, Y.; Koh, Y.H. RNA interference of pheromone biosynthesis activating neuropeptide receptor suppresses mating behavior by inhibiting sex pheromone production in Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2011, 41, 236–243. [Google Scholar] [CrossRef]
- Yin, F.; Lin, Q.; Wang, X.; Li, Z.; Feng, X.; Shabbir, M.Z. The glutathione S-transferase (PxGST2L) may contribute to the detoxification metabolism of chlorantraniliprole in Plutella xylostella (L.). Ecotoxicology 2021, 30, 1007–1016. [Google Scholar] [CrossRef]
- Zolfaghari, M.; Yin, F.; Jurat-Fuentes, J.L.; Xiao, Y.; Peng, Z.; Wang, J.; Yang, X.; Li, Z.-Y. Effects of Bacillus thuringiensis Treatment on Expression of Detoxification Genes in Chlorantraniliprole-Resistant Plutella xylostella. Insects 2024, 15, 595. [Google Scholar] [CrossRef]
- Zolfaghari, M.; Ghadamyari, M.; Sajedi, R.H. Resistance Mechanisms of a Field Population of Diamondback Moth, Plutella xylostella (Lepidoptera: Plutellidae), to Current Organophosphate Pesticides. J. Crop Prot. 2019, 8, 403–416. [Google Scholar]
- Tabashnik, T.; Cushing, N. Leaf residue vs. topical bioassays for assessing insecticide resistance in the diamond-back moth, Plutella xylostella L. FAO Plant Prot. Bull. 1987, 35, 11–14. [Google Scholar]
- Kahsay, R.Y.; Gao, G.; Liao, L. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 2005, 21, 1853–1858. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.-Y.; Zhu, Q.-H.; Chen, X.; Luo, J.-C. GSDS: A gene structure display server. Yi Chuan Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Duckert, P.; Brunak, S.; Blom, N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004, 17, 107–112. [Google Scholar] [CrossRef]
- Rinkevich, F.D.; Scott, J.G. Reduction of dADAR activity affects the sensitivity of Drosophila melanogaster to spinosad and imidacloprid. Pestic. Biochem. Physiol. 2012, 104, 163–169. [Google Scholar] [CrossRef]
- Cao, C.; Sun, L.; Du, H.; Moural, T.W.; Bai, H.; Liu, P.; Zhu, F. Physiological functions of a methuselah-like G protein coupled receptor in Lymantria dispar Linnaeus. Pestic. Biochem. Physiol. 2019, 160, 1–10. [Google Scholar] [CrossRef]
- Sun, L.; Liu, P.; Zhang, C.; Du, H.; Wang, Z.; Moural, T.W.; Zhu, F.; Cao, C. Ocular albinism type 1 (OA1) regulates deltamethrin tolerance in Lymantria dispar and Drosophila melanogaster. Front. Physiol. 2019, 10, 766. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, P.; Zhang, J.; Zheng, C.; Zhang, J.; Chen, N. Pheromone-binding proteins in the Asian gypsy moth females, Lymantria dispar, recognizing the sex pheromone and plant volatiles. Arch. Insect Biochem. Physiol. 2018, 99, e21477. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.F.; Chen, P.; Liu, F.T.; Zheng, S.C.; Ye, H.; Mo, M.H. RNA interference in moths: Mechanisms, applications, and progress. Genes 2016, 7, 88. [Google Scholar] [CrossRef]
- Kim, Y.H.; Issa, M.S.; Cooper, A.M.; Zhu, K.Y. RNA interference: Applications and advances in insect toxicology and insect pest management. Pestic. Biochem. Physiol. 2015, 120, 109–117. [Google Scholar] [CrossRef]
- Lin, W.; Yu, Y.; Zhou, P.; Zhang, J.; Dou, L.; Hao, Q.; Chen, H.; Zhu, S. Identification and knockdown of the olfactory receptor (OrCo) in gypsy moth, Lymantria dispar. Int. J. Biol. Sci. 2015, 11, 772–780. [Google Scholar] [CrossRef]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Yun, X.; Wang, Y.; Sang, M.; Liu, X.; Hu, X.; Li, B. Methuselah-like genes affect development, stress resistance, lifespan and reproduction in Tribolium castaneum. Insect Mol. Biol. 2014, 23, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, L.E.; Ghildyal, P.; Fischer, K.E.; Hu, H.; Ja, W.W.; Eaton, B.A.; Wu, Y.; Austad, S.N.; Ranjan, R. Modulation of methuselah expression targeted to Drosophila insulin-producing cells extends life and enhances oxidative stress resistance. Aging Cell 2013, 12, 121–129. [Google Scholar] [CrossRef]
- Friedrich, M.; Jones, J.W. Gene ages, nomenclatures, and functional diversification of the Methuselah/Methuselah—Like GPCR family in Drosophila and Tribolium. J. Exp. Zool. Part B 2016, 326, 453–463. [Google Scholar] [CrossRef]
- Li, C.; Chen, M.; Sang, M.; Liu, X.; Wu, W.; Li, B. Comparative genomic analysis and evolution of family-B G protein-coupled receptors from six model insect species. Gene 2013, 519, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, S.; Deng, M.-G.; Nawaz, M.; Lin, Q.-S. Genome-Wide Identification of G-Protein Coupled Receptors (GPCRs) and Their Expression Profile in Response to β-Cypermethrin Stress in Zeugodacus cucurbitae. Pestic. Biochem. Physiol. 2024, 202, 105919. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Song, X.; Chen, X.; Liu, X.; Sang, M.; Wu, W.; Yun, X.; Hu, X.; Li, B. Identification and comparative analysis of G protein-coupled receptors in Pediculus humanus humanus. Genomics 2014, 104, 58–67. [Google Scholar] [CrossRef]
- Pandey, A.; Khatoon, R.; Saini, S.; Vimal, D.; Patel, D.K.; Narayan, G.; Chowdhuri, D.K. Efficacy of methuselah gene mutation toward tolerance of dichlorvos exposure in Drosophila melanogaster. Free Radic. Biol. Med. 2015, 83, 54–65. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef]
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef]
- Liu, N.; Zhu, F.; Xu, Q.; Pridgeon, J.W.; Gao, X. Behavioral change, physiological modification, and metabolic detoxification: Mechanisms of insecticide resistance. Acta Entomol. Sin. 2006, 49, 671–679. [Google Scholar]
- Ranson, H.; Claudianos, C.; Ortelli, F.; Abgrall, C.; Hemingway, J.; Sharakhova, M.V.; Unger, M.F.; Collins, F.H.; Feyereisen, R. Evolution of supergene families associated with insecticide resistance. Science 2002, 298, 179–181. [Google Scholar] [CrossRef]
- Zhu, F.; Gujar, H.; Gordon, J.R.; Haynes, K.F.; Potter, M.F.; Palli, S.R. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci. Rep. 2013, 3, 1456. [Google Scholar] [CrossRef]
- Zhu, F.; Moural, T.W.; Shah, K.; Palli, S.R. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genom. 2013, 14, 174. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect CYP genes and P450 enzymes. In Insect Molecular Biology and Biochemistry; Gilbert, L.I., Ed.; Elsevier Academic Press: Cambridge, MA, USA, 2012; pp. 236–316. [Google Scholar]
- Liu, N.; Zhu, F. House fly cytochrome P450s: Their role in insecticide resistance and strategies in the isolation and characterization. In Recent Advances in Entomological Research; Liu, T., Kang, L., Eds.; Higher Education Press: Beijing, China; Springer: Berlin/Heidelberg, Germany, 2011; pp. 246–257. [Google Scholar]
- Zhu, F.; Parthasarathy, R.; Bai, H.; Woithe, K.; Kaussmann, M.; Nauen, R.; Harrison, D.A.; Palli, S.R. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2010, 107, 8557–8562. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Liu, N. Differential expression of CYP6A5 and CYP6A5v2 in pyrethroid resistant house flies, Musca domestica. Arch. Insect Biochem. Physiol. 2008, 67, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Liu, N.; Wen, Z. Insect cytochromes P450: Diversity, insecticide resistance and tolerance to plant toxins. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1998, 121, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Z.; Zou, C.; Cao, C. Transcription profiling of 12 Asian gypsy moth (Lymantria dispar) cytochrome P450 genes in response to insecticides. Arch. Insect Biochem. Physiol. 2014, 85, 181–194. [Google Scholar] [CrossRef]
- Sun, Y.; Zou, P.; Yu, X.Y.; Chen, C.; Yu, J.; Shi, L.N.; Hong, S.C.; Zhou, D.; Chang, X.L.; Wang, W.J.; et al. Functional characterization of an arrestin gene on insecticide resistance of Culex pipiens pallens. Parasites Vectors 2012, 5, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, L.; Zhang, L.; Liu, N. Role of G-protein-coupled receptor-related genes in insecticide resistance of the mosquito, Culex quinquefasciatus. Sci. Rep. 2014, 4, 6474. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Rename | Chr | Start | End | Strand | A.A | Exon | Intron | MW (kDs) | PI | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LOC105382322 | PXMTH1 | 25 | 4,719,631 | 4,726,549 | F | 493 | 9 | 8 | 56.67 | 6.23 | 0.326 |
| LOC119693055 | PXMTH2 | 8 | 3,392,336 | 3,414,747 | R | 514 | 10 | 9 | 57.1 | 8.05 | 0.215 |
| LOC105397179 | PXMTH3 | 25 | 4,705,051 | 4,71,5964 | F | 407 | 9 | 8 | 45.86 | 7.81 | 0.434 |
| LOC105392921 | PXMTH4 | 4 | 7,187,785 | 7,198,855 | F | 715 | 9 | 8 | 79.82 | 7.65 | 0.002 |
| LOC105382325 | PXMTH5 | 25 | 4,680,720 | 4,685,582 | F | 423 | 8 | 7 | 47.14 | 8.13 | 0.365 |
| LOC105383750 | PXMTH6 | 8 | 2,418,343 | 2,433,086 | F | 565 | 9 | 8 | 64.67 | 7.99 | 0.081 |
| LOC119693713 | PXMTH7 | 10 | 148,466 | 152,815 | F | 583 | 2 | 1 | 59.84 | 8.76 | 0.408 |
| LOC105380334 | PXMTH8 | 8 | 8,697,740 | 8,754,344 | R | 666 | 10 | 9 | 74.42 | 8.39 | −0.019 |
| Insecticide | Population | N a | LC50 (mg/L) | 95% CI of LC50 b | X2 (df) c | RR d |
|---|---|---|---|---|---|---|
| Chlorantraniliprole | Resistant | 150 | 63.2 | 46.66-70.2 | 0.90 (3) | 42.1 |
| Control | 150 | 1.5 | 0.8-2.3 | 0.90 (3) | 1 |
| Insecticide | Population | N a | LC50 (mg/L) | 95% CI of LC50 b | X2 (df) c | RR d |
|---|---|---|---|---|---|---|
| chlorantraniliprole | Transgenic Drosophila | 420 | 0.7 | 0.6–1 | 7 (4) | 2.9 |
| Control (WT) | 420 | 0.24 | 0.21–0.28 | 1.57 (4) | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zolfaghari, M.; Yin, F.; Shabbir, S.; Chen, Q.; Xiao, Y.; Peng, Z.; Li, Z.-Y.; Zalucki, M.P. Structural and Functional Insights into Methuselah Genes of Plutella xylostella (Lepidoptera: Plutellidae): Evolutionary Adaptations and Their Responses to Chlorantraniliprole. Insects 2025, 16, 1092. https://doi.org/10.3390/insects16111092
Zolfaghari M, Yin F, Shabbir S, Chen Q, Xiao Y, Peng Z, Li Z-Y, Zalucki MP. Structural and Functional Insights into Methuselah Genes of Plutella xylostella (Lepidoptera: Plutellidae): Evolutionary Adaptations and Their Responses to Chlorantraniliprole. Insects. 2025; 16(11):1092. https://doi.org/10.3390/insects16111092
Chicago/Turabian StyleZolfaghari, Maryam, Fei Yin, Samina Shabbir, Qichun Chen, Yong Xiao, Zhengke Peng, Zhen-Yu Li, and Myron P. Zalucki. 2025. "Structural and Functional Insights into Methuselah Genes of Plutella xylostella (Lepidoptera: Plutellidae): Evolutionary Adaptations and Their Responses to Chlorantraniliprole" Insects 16, no. 11: 1092. https://doi.org/10.3390/insects16111092
APA StyleZolfaghari, M., Yin, F., Shabbir, S., Chen, Q., Xiao, Y., Peng, Z., Li, Z.-Y., & Zalucki, M. P. (2025). Structural and Functional Insights into Methuselah Genes of Plutella xylostella (Lepidoptera: Plutellidae): Evolutionary Adaptations and Their Responses to Chlorantraniliprole. Insects, 16(11), 1092. https://doi.org/10.3390/insects16111092







