A Female-Biased Chemosensory Protein PxutCSP19 in the Antennae of Papilio xuthus Tuned to Host Volatiles and Insecticides
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Butterflies and Dissection of Body Parts
2.2. RNA Isolation and cDNA Synthesis
2.3. Gene Identification
2.4. Sequence and Structure Analysis
2.5. Expression Profiling Analysis of PxutCSP19 in P. xuthus
2.6. Protein Expression and Purification
2.7. Host Volatiles and Insecticides
2.8. Fluorescence Competitive Binding Assays
2.9. Construction of Two Truncated PxutCSP19s
2.10. Statistical Analyses
3. Results
3.1. The Identification of CSP19 Orthologs in Papilio Butterflies
3.2. Tissue- and Sex-Specific Expression of PxutCSP19 in P. xuthus
3.3. Bacterial Expression and Purification of PxutCSP19
3.4. Binding Properties of PxutCSP19 to Host Volatiles
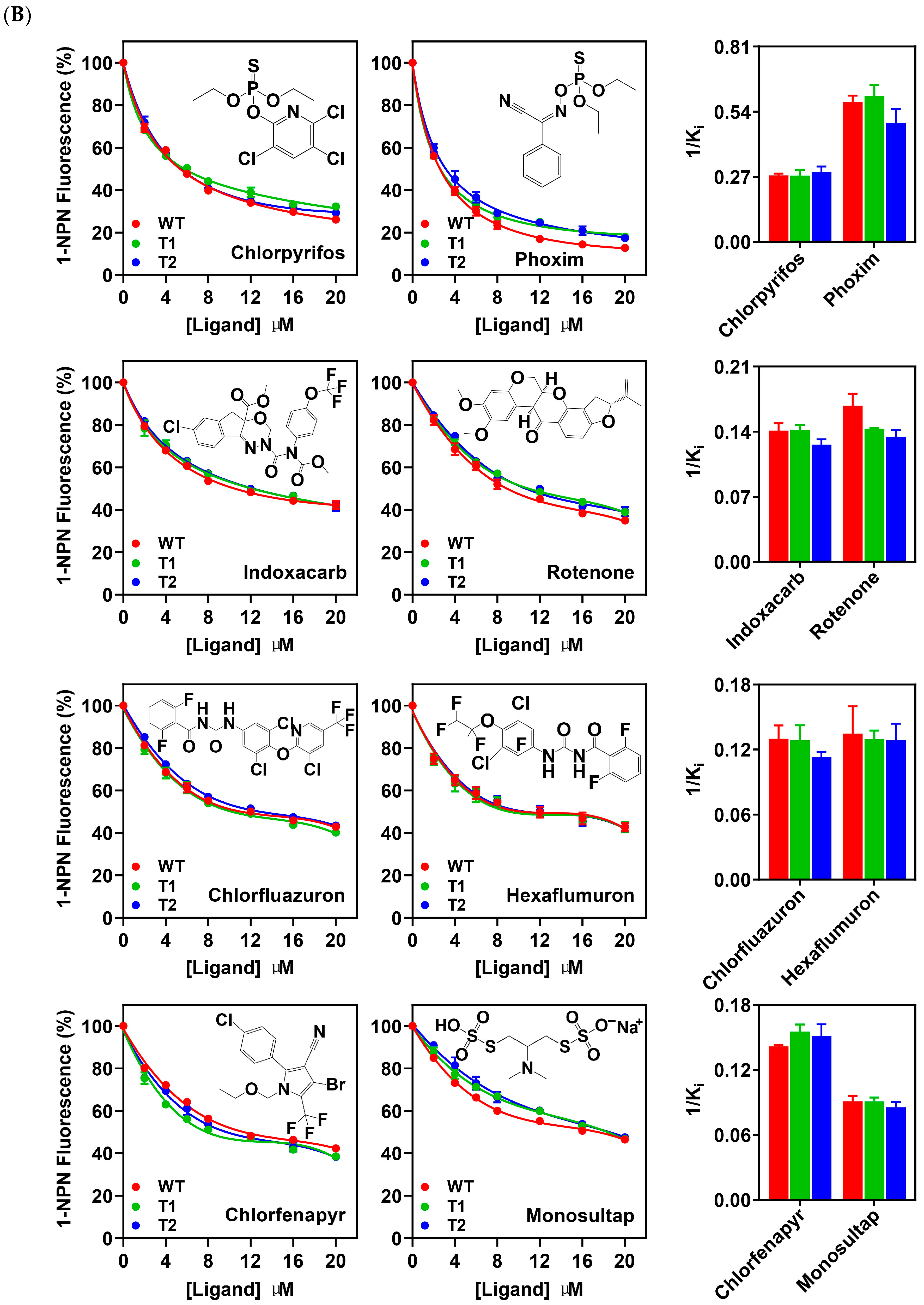
3.5. Binding Properties of PxutCSP19 to Insecticides
3.6. Effects of an Extended N-Terminus on the Binding Specificity of PxutCSP19 to Ligands
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mitter, C.; Davis, D.R.; Cummings, M.P. Phylogeny and evolution of Lepidoptera. Annu. Rev. Entomol. 2017, 62, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Van Nieukerken, E.; Kaila, L.; Kitching, I.; Kristensen, N.; Lees, D.; Minet, J.; Mitter, C.; Mutanen, M.; Regier, J.; Simonsen, T.; et al. Order Lepidoptera Linnaeus, 1758. In Animal Biodiversity: An Outline of Higher-Level Classification and Survey of Taxonomic Richness; Zhang, Z.Q., Ed.; Zootaxa; Magnolia Press: Waco, TX, USA, 2011; Volume 3148, pp. 212–221. [Google Scholar]
- Heppner, J.B. Butterflies and Moths (Lepidoptera). In Encyclopedia of Entomology; Capinera, J.L., Ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 626–672. [Google Scholar]
- Robinson, G.; Ackery, P.; Kitching, I.; Beccaloni, G.; Hernandez, L. Hostplants of the Moth and Butterfly Caterpillars of the Oriental Region; United Selangor Press: Kuala Lumpur, Malaysia, 2001. [Google Scholar]
- Yukawa, J.; Nishida, R.; Fukuda, H.; Inoue, R. Aristolochiaceae- and Asteraceae-feeding by larvae of Papilio xuthus L. (Lepidoptera: Papilionidae) in Japan: A review. Entomol. Sci. 2019, 22, 355–364. [Google Scholar] [CrossRef]
- Häuser, C.; De Jong, R.; Lamas, G.; Robbins, R.; Smith, C.; Vane-Wright, R. Papilionidae–Revised GloBIS/GART Species Checklist (2nd Draft). 2005. Available online: https://www.insectsonline.de/frames/papilio.htm (accessed on 5 April 2024).
- Crava, C.M.; Bobkov, Y.V.; Sollai, G.; Anfora, G.; Crnjar, R.; Cattaneo, A.M. Chemosensory receptors in the larval maxilla of Papilio hospiton. Front. Ecol. Evol. 2022, 9, 795994. [Google Scholar] [CrossRef]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yin, M.Z.; Ma, S.; Gu, N.; Qian, L.F.; Zhang, Y.N.; Li, X.M. Ligand-binding properties of chemosensory protein 1 in Callosobruchus chinensis to mung bean volatiles. Pestic. Biochem. Physiol. 2023, 192, 105394. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.H.; Wang, S.Y.; Zhang, X.Y.; Ji, P.; Liu, J.T.; Wang, G.R.; Wu, K.M.; Guo, Y.Y.; Zhou, J.J.; Zhang, Y.J. Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition. PLoS ONE 2012, 7, e42871. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.-N.; Yao, Y.-J.; Liang, Y.-L.; Wang, Z.-Q.; Li, Y.-H.; Liu, N.-Y. Functional characterization of four antenna-biased chemosensory proteins in Dioryctria abietella reveals a broadly tuned olfactory DabiCSP1 and its key residues in ligand-binding. Pestic. Biochem. Physiol. 2023, 197, 105678. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.G.; Li, D.Z.; Wang, M.Q. Chemosensory proteins used as target for screening behaviourally active compounds in the rice pest Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Insect Mol. Biol. 2019, 28, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Khuhro, S.A.; Yan, Q.; Liao, H.; Zhu, G.H.; Sun, J.B.; Dong, S.L. Expression profile and functional characterization suggesting the involvement of three chemosensory proteins in perception of host plant volatiles in Chilo suppressalis (Lepidoptera: Pyralidae). J. Insect Sci. 2018, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Waris, M.I.; Younas, A.; Ul Qamar, M.T.; Hao, L.; Ameen, A.; Ali, S.; Abdelnabby, H.E.; Zeng, F.-F.; Wang, M.-Q. Silencing of chemosensory protein gene NlugCSP8 by RNAi induces declining behavioral responses of Nilaparvata lugens. Front. Physiol. 2018, 9, 379. [Google Scholar] [CrossRef]
- Waris, M.I.; Younas, A.; Adeel, M.M.; Duan, S.-G.; Quershi, S.R.; Kaleem Ullah, R.M.; Wang, M.-Q. The role of chemosensory protein 10 in the detection of behaviorally active compounds in brown planthopper, Nilaparvata lugens. Insect Sci. 2020, 27, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Pan, Y.F.; Ma, Y.F.; Wang, J.; He, M.; He, P. Binding affinity characterization of an antennae-enriched chemosensory protein from the white-backed planthopper, Sogatella furcifera (Horvath), with host plant volatiles. Pestic. Biochem. Physiol. 2018, 152, 1–7. [Google Scholar] [CrossRef]
- Li, J.-Q.; Zhu, R.; Yao, W.-C.; Yu, H.-P.; Huang, J.-R.; Wang, Z.; Sun, X.-Y.; Yuan, D.-H.; Sun, Y.-Y.; Emam, S.S.; et al. Chemosensory protein 2 of male Athetis lepigone is involved in the perception of sex pheromones and maize volatiles. J. Agric. Food Chem. 2023, 71, 6277–6287. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Li, F.; Yan, X.; Hao, C. Expression profiles and binding properties of the chemosensory protein PxylCSP11 from the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J. Insect Sci. 2020, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Ye, Z.-F.; Yang, K.; Dong, S.-L. Antenna-predominant and male-biased CSP19 of Sesamia inferens is able to bind the female sex pheromones and host plant volatiles. Gene 2014, 536, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Jacquin-Joly, E.; Vogt, R.G.; François, M.-C.; Nagnan-Le Meillour, P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 2001, 26, 833–844. [Google Scholar] [CrossRef]
- Hua, J.-F.; Zhang, S.; Cui, J.-J.; Wang, D.-J.; Wang, C.-Y.; Luo, J.-Y.; Lv, L.-M. Identification and binding characterization of three odorant binding proteins and one chemosensory protein from Apolygus lucorum (Meyer-Dur). J. Chem. Ecol. 2012, 38, 1163–1170. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Tahir Ul Qamar, M.; Shaaban, M.; Prager, S.M.; Wang, M.-Q. Functional analysis of the chemosensory protein MsepCSP8 from the oriental armyworm Mythimna separata. Front. Physiol. 2018, 9, 872. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Chang, X.-Q.; Shaaban, M.; Abdelnabby, H.; Ul Qamar, M.T.; Wang, M.-Q. A chemosensory protein MsepCSP5 involved in chemoreception of oriental armyworm Mythimna separata. Int. J. Biol. Sci. 2018, 14, 1935–1949. [Google Scholar] [CrossRef]
- Hua, J.; Fu, Y.; Zhou, Q.; Huang, Y.; Li, H.; Chen, T.; Ma, D.; Li, Z. Three chemosensory proteins from the sweet potato weevil, Cylas formicarius, are involved in the perception of host plant volatiles. Pest Manag. Sci. 2021, 77, 4497–4509. [Google Scholar] [CrossRef]
- Li, C.; Sun, K.; Li, D.; Liu, D. Functional characterization of chemosensory protein AmalCSP5 From apple buprestid beetle, Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 2020, 114, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; He, X.; Lehane, S.; Lehane, M.; Hertz-Fowler, C.; Berriman, M.; Field, L.M.; Zhou, J.-J. Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour. Insect Mol. Biol. 2012, 21, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wan, W.; Xie, M.; Mao, J.; Dong, Z.; Lu, S.; He, J.; Xie, F.; Liu, G.; Dai, X.; et al. Chromosome-level reference genome assembly and gene editing of the dead-leaf butterfly Kallima inachus. Mol. Ecol. Res. 2020, 20, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- The Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 2012, 487, 94–98. [Google Scholar] [CrossRef]
- Kanost, M.R.; Arrese, E.L.; Cao, X.; Chen, Y.R.; Chellapilla, S.; Goldsmith, M.R.; Grosse-Wilde, E.; Heckel, D.G.; Herndon, N.; Jiang, H.; et al. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem. Mol. Biol. 2016, 76, 118–147. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.L.; Clarke, D.F.; East, P.D.; Elfekih, S.; Gordon, K.H.J.; Jermiin, L.S.; McGaughran, A.; Oakeshott, J.G.; Papanikolaou, A.; Perera, O.P.; et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-C.; Nuo, S.-M.; Wang, Z.-Q.; Yang, A.-J.; Liu, N.-Y. Identification and expression profiling of chemosensory membrane protein genes in Achelura yunnanensis (Lepidoptera: Zygaenidae). Comp. Biochem. Physiol. D Genom. Proteom. 2021, 40, 100876. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechselberger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 10606–10613. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.-N.; Nuo, S.-M.; Xiao, H.-Y.; Zhao, Y.-J.; Zhu, J.-Y.; Liu, N.-Y. The ionotropic receptor gene family in Lepidoptera and Trichoptera: Annotation, evolutionary and functional perspectives. Genomics 2021, 113, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.N.; Yang, A.J.; Wu, C.; Xiao, H.Y.; Guo, Y.R.; Liu, N.Y. Genome-wide analysis of odorant-binding proteins in Papilio xuthus with focus on the perception of two PxutGOBPs to host odorants and insecticides. J. Agric. Food. Chem. 2022, 70, 10747–10761. [Google Scholar] [CrossRef] [PubMed]
- Simon, P. Q-Gene: Processing quantitative real-time RT–PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Janovjak, H.; Miserez, A.R.; Dobbie, Z. Processing of gene expression data generated by quantitative real-time RT–PCR. BioTechniques 2002, 32, 1372–1374, 1376, 1378–1379. [Google Scholar] [PubMed]
- Zhang, W.; Tan, S.; Xi, W.; Yang, J.; Liao, Q.; Lan, J.; Lv, Y.; Tang, J. Comparison of volatile components in fresh and dried Zanthoxylum bungeanum Maxim. Food Sci. Biotechnol. 2019, 28, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Dönmez, Ş. Determination of volatile components of Citrus flowers and leaves growing in Hatay, Türkiye. BioResources 2024, 19, 2935–2947. [Google Scholar] [CrossRef]
- González-Mas, M.C.; Rambla, J.L.; López-Gresa, M.P.; Blázquez, M.A.; Granell, A. Volatile compounds in Citrus essential oils: A comprehensive review. Front. Plant Sci. 2019, 10, 12. [Google Scholar] [CrossRef]
- Azam, M.; Song, M.; Fan, F.; Zhang, B.; Xu, Y.; Xu, C.; Chen, K. Comparative analysis of flower volatiles from nine Citrus at three blooming stages. Int. J. Biol. Sci. 2013, 14, 22346–22367. [Google Scholar] [CrossRef]
- Yao, Y.J.; Yin, N.N.; Pu, L.M.; Yang, A.J.; Liu, N.Y. Three chemosensory proteins enriched in antennae and tarsi of Rhaphuma horsfieldi differentially contribute to the binding of insecticides. Pestic. Biochem. Physiol. 2024, 199, 105797. [Google Scholar] [CrossRef]
- Calvello, M.; Guerra, N.; Brandazza, A.; D’Ambrosio, C.; Scaloni, A.; Dani, F.R.; Turillazzi, S.; Pelosi, P. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell. Mol. Life Sci. 2003, 60, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Terrado, M.; Okon, M.; McIntosh, L.P.; Plettner, E. Ligand- and pH-induced structural transition of gypsy moth Lymantria dispar pheromone-binding protein 1 (LdisPBP1). Biochemistry 2020, 59, 3411–3426. [Google Scholar] [CrossRef]
- Xu, W.; Xu, X.; Leal, W.S.; Ames, J.B. Extrusion of the C-terminal helix in navel orangeworm moth pheromone-binding protein (AtraPBP1) controls pheromone binding. Biochem. Biophys. Res. Commun. 2011, 404, 335–338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gong, L.; Luo, Q.; Rizwan-ul-Haq, M.; Hu, M.Y. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. Bull. Entomol. Res. 2012, 102, 600–609. [Google Scholar] [CrossRef]
- Xiao, H.Y.; Li, G.C.; Wang, Z.Q.; Guo, Y.R.; Liu, N.Y. Combined transcriptomic, proteomic and genomic analysis identifies reproductive-related proteins and potential modulators of female behaviors in Spodoptera litura. Genomics 2021, 113, 1876–1894. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Cui, S.; Tian, Z.; Zhang, Y.; Chen, G.; Gao, X.; Tian, Z.; Chen, H.; Guo, J.; Zhou, Z. OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: Chrysomelidae). Front. Physiol. 2019, 10, 1290. [Google Scholar] [CrossRef]
- Inoue, T.A.; Suetake, M.; Nishidzu, N.; Yokohari, F.; Niihara, K.; Fukuda, T. Behavioral and electrophysiological study on eight Japanese Papilio species with five hostplant volatiles and linalool. J. Chem. Ecol. 2023, 49, 397–407. [Google Scholar] [CrossRef]
- Baur, R.; Feeny, P. Comparative electrophysiological analysis of plant odor perception in females of three Papilio species. Chemoecology 1994, 5, 26–36. [Google Scholar] [CrossRef]
- Luo, Y.-J.; Wang, Y.; Zhou, Q.; He, J.; Li, X. Olfactory and behavioral responses of Papilio polytes (Lepidoptera: Papilioidae) adults to volatiles from the branches and leaves of citrus. Acta Entomol. Sin. 2023, 66, 1612–1625. [Google Scholar]
- Zhang, L.; Su, Q.-F.; Wang, L.-S.; Lv, M.-W.; Hou, Y.-X.; Li, S.-S. Linalool: A ubiquitous floral volatile mediating the communication between plants and insects. J. Syst. Evol. 2023, 61, 538–549. [Google Scholar] [CrossRef]
- Liu, X.; Liao, W.; Wu, Z.; Pei, Y.; Wei, Z.; Lu, M. Binding properties of odorant-binding protein 7 to host volatiles in larvae of Spodoptera frugiperda. J. Agric. Food Chem. 2023, 71, 20671–20679. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q.; Jiang, H.B.; Fan, J.Y.; Liu, T.Y.; Meng, L.W.; Liu, Y.; Yu, H.Z.; Dou, W.; Wang, J.J. An odorant-binding protein of Asian citrus psyllid, Diaphorina citri, participates in the response of host plant volatiles. Pest Manag. Sci. 2021, 77, 3068–3079. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, Z.; Si, P.; Liu, Y.; Zhou, Q.; Wang, G. Characterization of a specific odorant receptor for linalool in the Chinese citrus fly Bactrocera minax (Diptera: Tephritidae). Insect Biochem. Mol. Biol. 2020, 122, 103389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X.; Wang, G.; Liu, F.; Liu, Y. An odorant receptor of the green mirid bug, Apolygus lucorum, tuned to linalool. Insect Biochem. Mol. Biol. 2022, 144, 103764. [Google Scholar] [CrossRef]
- Cattaneo, A.M.; Witzgall, P.; Kwadha, C.A.; Becher, P.G.; Walker, W.B. Heterologous expression and functional characterization of Drosophila suzukii OR69a transcript variants unveiled response to kairomones and to a candidate pheromone. J. Pest Sci. 2022, 96, 1149–1171. [Google Scholar] [CrossRef]
- Huff, R.M.; Pitts, R.J. Functional conservation of Anopheline linalool receptors through 100 million years of evolution. Chem. Senses 2022, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ni, S.; Wang, Y.; Li, R.; Sun, H.; Ye, X.; Tian, Z.; Zhang, Y.; Liu, J. The chemosensory protein 1 contributes to indoxacarb resistance in Plutella xylostella (L.). Pest Manag. Sci. 2023, 79, 2456–2468. [Google Scholar] [CrossRef]
- Peng, X.; Qu, M.J.; Wang, S.J.; Huang, Y.X.; Chen, C.; Chen, M.H. Chemosensory proteins participate in insecticide susceptibility in Rhopalosiphum padi, a serious pest on wheat crops. Insect Mol. Biol. 2021, 30, 138–151. [Google Scholar] [CrossRef]
- Wu, C.; Yin, N.; Guo, Y.; Wang, Z.; Liu, N. Two antenna-enriched odorant binding proteins in Dioryctria abietella tuned to general odorants and insecticides. Insects 2022, 13, 1145. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Mao, Y.; Zhang, L. Binding properties of four antennae-expressed chemosensory proteins (CSPs) with insecticides indicates the adaption of Spodoptera litura to environment. Pestic. Biochem. Physiol. 2018, 146, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Zhang, X.-C.; Zhu, R.; Yao, W.-C.; Xu, J.-W.; Wang, M.; Ren, J.-Y.; Xu, C.-Z.; Huang, Z.-R.; Zhang, X.-W.; et al. Computational and experimental approaches to decipher the binding mechanism of general odorant-binding protein 2 from Athetis lepigone to chlorpyrifos and phoxim. J. Agric. Food Chem. 2021, 69, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Mohamed, A.; Cattaneo, A.M.; Huang, X.; Keyhani, N.O.; Gu, M.; Zang, L.; Zhang, W. Odorant-binding proteins and chemosensory proteins in Spodoptera frugiperda: From genome-wide identification and developmental stage-related expression analysis to the perception of host plant odors, sex pheromones, and insecticides. Int. J. Mol. Sci. 2023, 24, 5595. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, R.; Du, Y.; Gao, B.; Gui, F.; Lu, K. Olfactory perception of herbicide butachlor by GOBP2 elicits ecdysone biosynthesis and detoxification enzyme responsible for chlorpyrifos tolerance in Spodoptera litura. Environ. Pollut. 2021, 285, 11740. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Jiang, Y.; Zhang, L.; Cai, Y. Effects of insecticides chlorpyrifos, emamectin benzoate and fipronil on Spodoptera litura might be mediated by OBPs and CSPs. Bull. Entomol. Res. 2018, 108, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.S.; Buckmeier, B.G.; Griffith, W.; Olafson, P.U.; Perez de Leon, A.A.; Renthal, R. Odorant-binding protein from the stable fly (Stomoxys calcitrans) has a high-histidine N-terminal extension that binds transition metals. Insect Biochem. Mol. Biol. 2022, 141, 103707. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tian, X.; Gui, L.; Wang, F.; Zhang, G. Key amino acid residues involved in binding interactions between Bactrocera minax odorant-binding protein 3 (BminOBP3) and undecanol. Insects 2023, 14, 745. [Google Scholar] [CrossRef]
- Lagarde, A.; Spinelli, S.; Qiao, H.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of a novel type of odorant-binding protein from Anopheles gambiae, belonging to the C-plus class. Biochem. J. 2011, 437, 423–430. [Google Scholar] [CrossRef]
- Wogulis, M.; Morgan, T.; Ishida, Y.; Leal, W.S.; Wilson, D.K. The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism. Biochem. Biophys. Res. Commun. 2006, 339, 157–164. [Google Scholar] [CrossRef]
- Lartigue, A.; Gruez, A.; Briand, L.; Blon, F.; Bezirard, V.; Walsh, M.; Pernollet, J.C.; Tegoni, M.; Cambillau, C. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J. Biol. Chem. 2004, 279, 4459–4464. [Google Scholar] [CrossRef] [PubMed]
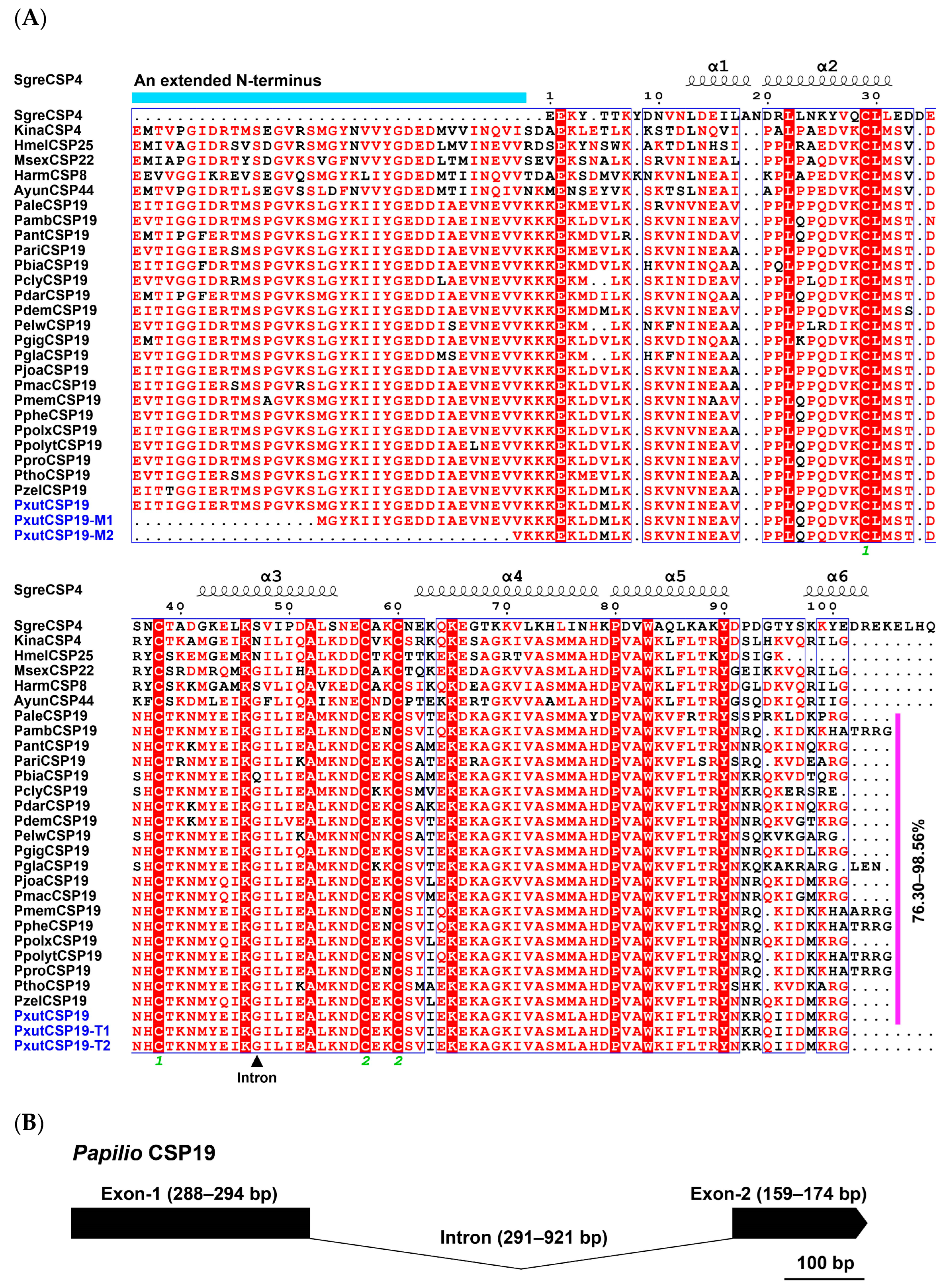

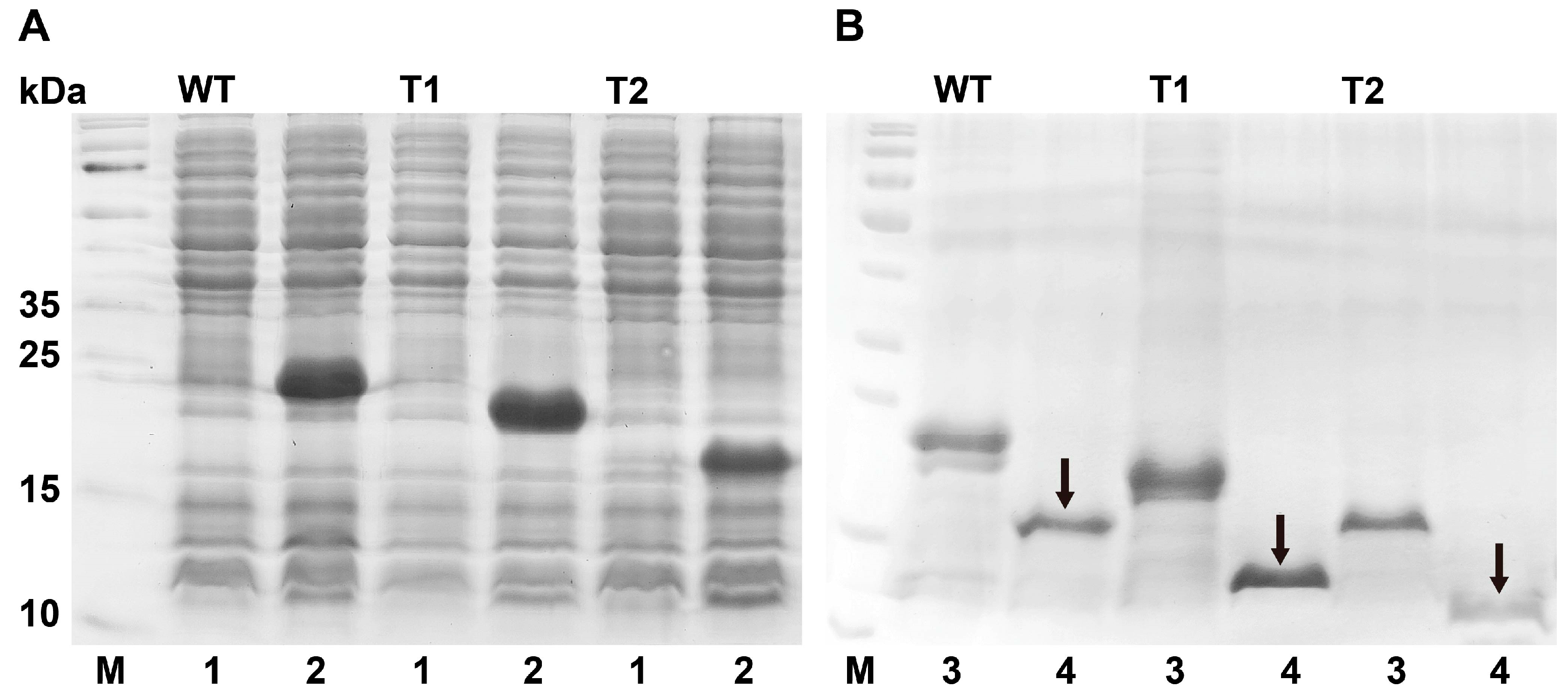
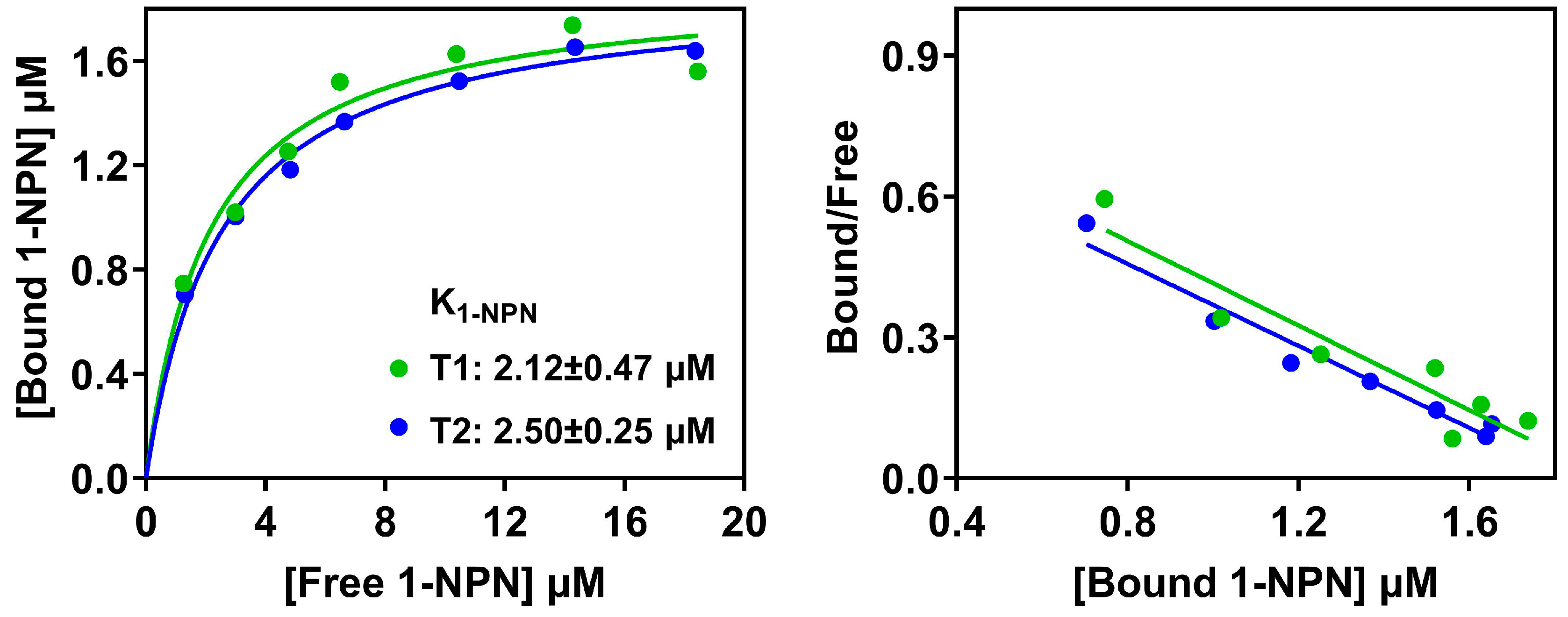
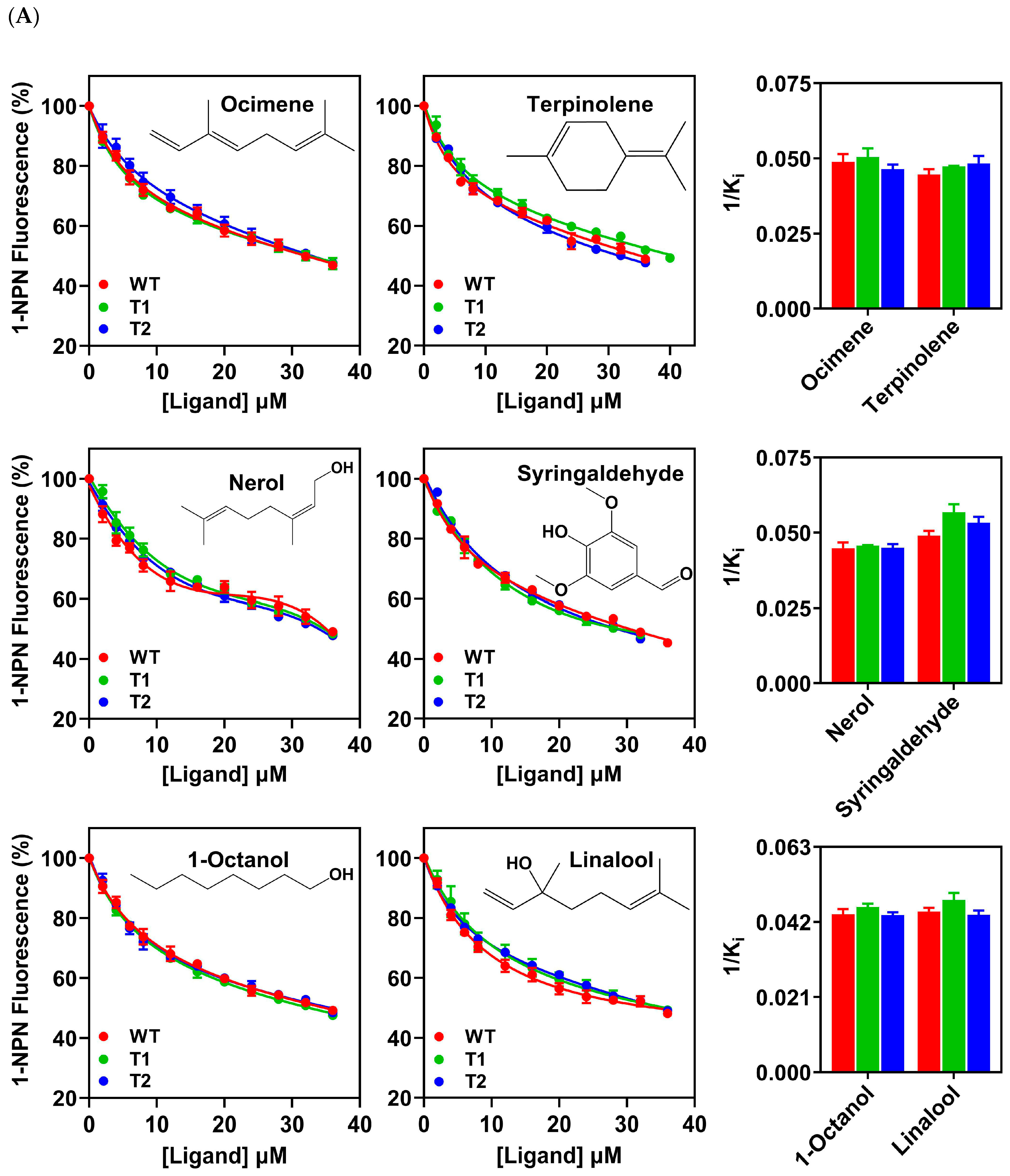
| Species | ORF(bp)/ Full-Length | Signal Peptide (aa) | Exon Length (bp) | Intron | Orientation | Chromosome | Note | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon-1 | Exon-2 | Number | Length (bp) | Phase | ||||||
| P. polytes | 471/Yes | 17 | 294 | 165 | 1 | 498 | 1 | + | DF823923.1 | |
| P. xuthus | 462/Yes | 17 | 294 | 165 | 1 | 578 | 1 | + | Scaffold 50 | |
| P. machaon | 462/Yes | 17 | 294 | 165 | 1 | 388 | 1 | – | Scaffold 4 | |
| P. memnon | 471/Yes | 17 | 294 | 174 | 1 | 518 | 1 | + | Scaffold 26 | |
| P. dardanus | 462/Yes | 17 | 294 | 165 | 1 | 291 | 1 | + | Scaffold 115 | |
| P. bianor | 462/Yes | 17 | 294 | 165 | 1 | 671 | 1 | – | TM66B3WUVR 8 | |
| P. aristodemus | 456/Yes | 16 | 291 | 162 | 1 | 326 | 1 | – | JZZY7UMRXG 8 | |
| P. glaucus | 462/Yes | 16 | 288 | 171 | 1 | 792 | 1 | – | scaffold_2208 | |
| P. clytia | 453/Yes | 17 | 288 | 162 | / | / | / | + | cov_5.555223 + cov_5.608469 | |
| P. antimachus | 462/Yes | 17 | 294 | 165 | 1 | 502 | 1 | – | cov_36.791104 | |
| P. slateri | 378/No | 17 | / | / | / | / | / | + | cov_5.088006 | Missing ~25 amino acids in the middle of this gene |
| P. alexanor | 462/Yes | 17 | 294 | 165 | 1 | 629 | 1 | – | cov_28.334434 | |
| P. ambrax | 471/Yes | 17 | 294 | 174 | 1 | 446 | 1 | – | cov_5.029448 | |
| P. phestus | 471/Yes | 17 | 294 | 174 | 1 | 467 | 1 | – | cov_3.748094 | |
| P. polyxenes | 462/Yes | 17 | 294 | 174 | 1 | 498 | 1 | + | Scaffold3300 | |
| P. zelicaon | 459/Yes | 17 | 294 | 165 | / | / | / | – | cov_68.359788 + cov_43.479663 | |
| P. protenor | 471/Yes | 17 | 294 | 174 | 1 | 724 | 1 | + | ctg7 | |
| P. gigon | 462/Yes | 17 | 294 | 165 | 1 | 409 | 1 | – | 0KYF3WRZ5T 8 | |
| P. thoas | 456/Yes | 16 | 291 | 162 | / | / | / | / | cov_54.059850 + cov_21.006012 + cov_45.507022 | |
| P. joanae | 462/Yes | 17 | 294 | 165 | / | / | / | + | cov_10.450777 + cov_17.501926 | |
| P. demoleus | 462/Yes | 17 | 294 | 165 | 1 | 358 | 1 | + | ctg11 | |
| P. elwesi | 453/Yes | 17 | 288 | 162 | 1 | 921 | 1 | – | Chromosome 16 | |
| P. helenus | 402/No | 17 | / | / | / | / | / | – | MHAD71OSIT 15 | Missing ~23 amino acids in the middle of this gene |
| Compound | PxutCSP19 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wildtype (WT) | T1 | T2 | |||||||
| F.D. (%) | IC50 (μM) | Ki (μM) | F.D. (%) | IC50 (μM) | Ki (μM) | F.D. (%) | IC50 (μM) | Ki (μM) | |
| Host volatiles | |||||||||
| Alkenes | |||||||||
| α-Pinene | 34.65 | >20 | – | ||||||
| (1R)-(+)-α-Pinene | 38.38 | >20 | – | ||||||
| β-Pinene | 31.21 | >20 | – | ||||||
| Myrcene | 32.40 | >20 | – | ||||||
| Farnesene | 23.21 | >20 | – | ||||||
| Ocimene | 42.70 | 31.27 ± 1.73 | 20.61 ± 1.14 | 43.11 | 31.71 ± 1.86 | 19.93 ± 1.17 | 35.19 | 32.82 ± 1.09 | 21.61 ± 0.72 |
| 3-Carene | 35.12 | >20 | – | ||||||
| α-Caryophyllene | 7.48 | >20 | – | ||||||
| β-Caryophyllene | NB | – | |||||||
| Limonene | 20.36 | >20 | – | ||||||
| Sabinene | 33.51 | >20 | – | ||||||
| γ-Terpinene | 46.40 | >36 | – | ||||||
| α-Terpinene | 40.75 | >36 | – | ||||||
| α-Phellandrene | 34.09 | >20 | – | ||||||
| Camphene | 34.52 | >20 | – | ||||||
| Terpinolene | 44.05 | 34.11 ± 1.31 | 22.48 ± 0.87 | 40.12 | 33.64 ± 0.16 | 21.14 ± 0.10 | 38.49 | 31.57 ± 1.67 | 20.78 ± 1.10 |
| β-Elemene | NB | – | |||||||
| p-Isopropyl toluene | 32.32 | >20 | – | ||||||
| Alcohols | |||||||||
| 1-Hexanol | 38.95 | >20 | – | ||||||
| (E)-2-Hexen-1-ol | 35.63 | >20 | – | ||||||
| (Z)-3-Hexen-1-ol | 35.80 | >20 | – | ||||||
| 1-Octanol | 42.92 | 34.46 ± 1.11 | 22.71 ± 0.73 | 43.54 | 34.48 ± 0.70 | 21.67 ± 0.44 | 42.67 | 34.61 ± 0.64 | 22.79 ± 0.42 |
| Phenethyl alcohol | 39.66 | >20 | – | ||||||
| Nerolidol | 12.36 | >20 | – | ||||||
| Farnesol | 27.04 | >20 | – | ||||||
| Linalool | 45.65 | 33.84 ± 0.84 | 22.30 ± 0.55 | 39.75 | 33.14 ± 1.34 | 20.83 ± 0.85 | 40.26 | 34.58 ± 0.90 | 22.77 ± 0.59 |
| Geraniol | 39.03 | – | |||||||
| Nerol | 44.09 | 33.99 ± 1.46 | 22.40 ± 0.96 | 39.39 | 34.75 ± 0.03 | 21.84 ± 0.02 | 40.79 | 33.78 ± 0.85 | 22.24 ± 0.56 |
| 4-Terpineol | 45.13 | >36 | – | ||||||
| α-Terpineol | 41.46 | >36 | – | ||||||
| Aldehydes | |||||||||
| Hexanal | 21.08 | >20 | – | ||||||
| (E)-2-Hexenal | 33.76 | >20 | – | ||||||
| Octanal | 22.75 | >20 | – | ||||||
| Nonanal | 29.30 | >20 | – | ||||||
| Decanal | 28.74 | >20 | – | ||||||
| Undecanal | 27.70 | >20 | – | ||||||
| Benzaldehyde | 36.91 | >20 | – | ||||||
| Phenylacetaldehyde | 27.32 | >20 | – | ||||||
| Syringaldehyde | 43.39 | 31.02 ± 0.97 | 20.44 ± 0.64 | 42.68 | 28.09 ± 1.33 | 17.66 ± 0.84 | 42.18 | 28.58 ± 1.12 | 18.82 ± 0.74 |
| Citronellal | 34.20 | >20 | – | ||||||
| Citral | 39.68 | >20 | – | ||||||
| Esters | |||||||||
| Ethyl butyrate | 28.67 | >20 | – | ||||||
| Ethyl acetate | 37.65 | >20 | – | ||||||
| Geranyl acetate | 33.33 | >20 | – | ||||||
| Others | |||||||||
| Methyl o-toluate | 37.28 | >20 | – | ||||||
| Indole | 39.05 | >20 | – | ||||||
| Carvacrol | 37.40 | >20 | – | ||||||
| Synthetic insecticides | |||||||||
| Benzoylureas | |||||||||
| Chlorbenzuron | 42.75 | >20 | – | ||||||
| Chlorfluazuron | 60.51 | 11.88 ± 1.04 | 7.83 ± 0.68 | 54.70 | 12.72 ± 1.49 | 7.99 ± 0.94 | 57.01 | 13.49 ± 0.60 | 8.88 ± 0.40 |
| Diflubenzuron | 26.96 | >20 | – | ||||||
| Hexaflumuron | 59.55 | 12.15 ± 2.36 | 8.01 ± 1.56 | 56.46 | 12.37 ± 0.73 | 7.78 ± 0.46 | 59.61 | 12.18 ± 1.56 | 8.02 ± 1.03 |
| Triflumuron | 27.99 | >20 | – | ||||||
| Pyrethroids | |||||||||
| α-Cypermethrin | 5.65 | >20 | – | ||||||
| Deltamethrin | 39.31 | >20 | – | ||||||
| Diamide | |||||||||
| Chlorantraniliprole | 43.56 | >20 | – | ||||||
| Organophosphates | |||||||||
| Acephate | 37.07 | >20 | – | ||||||
| Chlorpyrifos | 75.06 | 5.52 ± 0.16 | 3.64 ± 0.11 | 70.28 | 5.87 ± 0.47 | 3.69 ± 0.29 | 73.23 | 5.32 ± 0.40 | 3.51 ± 0.27 |
| Phoxim | 87.27 | 2.63 ± 0.12 | 1.73 ± 0.08 | 83.13 | 2.66 ± 0.20 | 1.67 ± 0.13 | 84.75 | 3.16 ± 0.35 | 2.08 ± 0.23 |
| Profenofos | 41.04 | >20 | – | ||||||
| Trichlorphon | 33.61 | >20 | – | ||||||
| Pyrazole | |||||||||
| Fipronil | 37.47 | >20 | – | ||||||
| Chloronicotinyls | |||||||||
| Acetamiprid | 42.89 | >20 | – | ||||||
| Imidacloprid | 41.72 | >20 | – | ||||||
| Thiamethoxam | 44.56 | >20 | – | ||||||
| Carbamates | |||||||||
| Thiodicarb | 37.39 | >20 | – | ||||||
| Methomyl | 45.40 | >20 | – | ||||||
| Indoxacarb | 61.54 | 10.83 ± 0.65 | 7.14 ± 0.43 | 59.12 | 11.27 ± 0.41 | 7.08 ± 0.26 | 62.53 | 12.11 ± 0.56 | 7.98 ± 0.37 |
| Nereistoxin | |||||||||
| Monosultap | 53.60 | 16.80 ± 1.01 | 11.07 ± 0.66 | 54.78 | 17.58 ± 0.73 | 11.05 ± 0.46 | 61.01 | 17.88 ± 1.02 | 11.77 ± 0.67 |
| Ecdysome agonist | |||||||||
| Tebufenozide | 48.54 | >20 | – | ||||||
| Pyrrole | |||||||||
| Chlorfenapyr | 58.72 | 10.71 ± 0.09 | 7.06 ± 0.06 | 63.82 | 10.29 ± 0.43 | 6.47 ± 0.27 | 64.33 | 10.14 ± 0.68 | 6.68 ± 0.45 |
| Antibiotic | |||||||||
| Emamectin benzoate | 46.98 | >20 | – | ||||||
| Biopesticides | |||||||||
| Rotenone | 67.63 | 9.15 ± 0.76 | 6.03 ± 0.50 | 61.84 | 11.12 ± 0.04 | 6.99 ± 0.03 | 64.39 | 11.35 ± 0.58 | 7.43 ± 0.39 |
| Matrine | 39.95 | >20 | – | ||||||
| Azadirachtin | 34.50 | >20 | – | ||||||
| Rhodojaponin III | 38.52 | >20 | – | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, N.; Shen, D.; Liang, Y.; Wang, P.; Li, Y.; Liu, N. A Female-Biased Chemosensory Protein PxutCSP19 in the Antennae of Papilio xuthus Tuned to Host Volatiles and Insecticides. Insects 2024, 15, 501. https://doi.org/10.3390/insects15070501
Yin N, Shen D, Liang Y, Wang P, Li Y, Liu N. A Female-Biased Chemosensory Protein PxutCSP19 in the Antennae of Papilio xuthus Tuned to Host Volatiles and Insecticides. Insects. 2024; 15(7):501. https://doi.org/10.3390/insects15070501
Chicago/Turabian StyleYin, Ningna, Dan Shen, Yinlan Liang, Pengfei Wang, Yonghe Li, and Naiyong Liu. 2024. "A Female-Biased Chemosensory Protein PxutCSP19 in the Antennae of Papilio xuthus Tuned to Host Volatiles and Insecticides" Insects 15, no. 7: 501. https://doi.org/10.3390/insects15070501
APA StyleYin, N., Shen, D., Liang, Y., Wang, P., Li, Y., & Liu, N. (2024). A Female-Biased Chemosensory Protein PxutCSP19 in the Antennae of Papilio xuthus Tuned to Host Volatiles and Insecticides. Insects, 15(7), 501. https://doi.org/10.3390/insects15070501







