Revealing the Development Patterns of the Mandibular Glands of Apis mellifera carnica Based on Transcriptomics and Morphology
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Microscopy
2.3. Library Construction and Transcriptomic Sequencing
2.4. DEG Identification and Analysis
2.5. RT-qPCR Validation of DEGs
3. Results
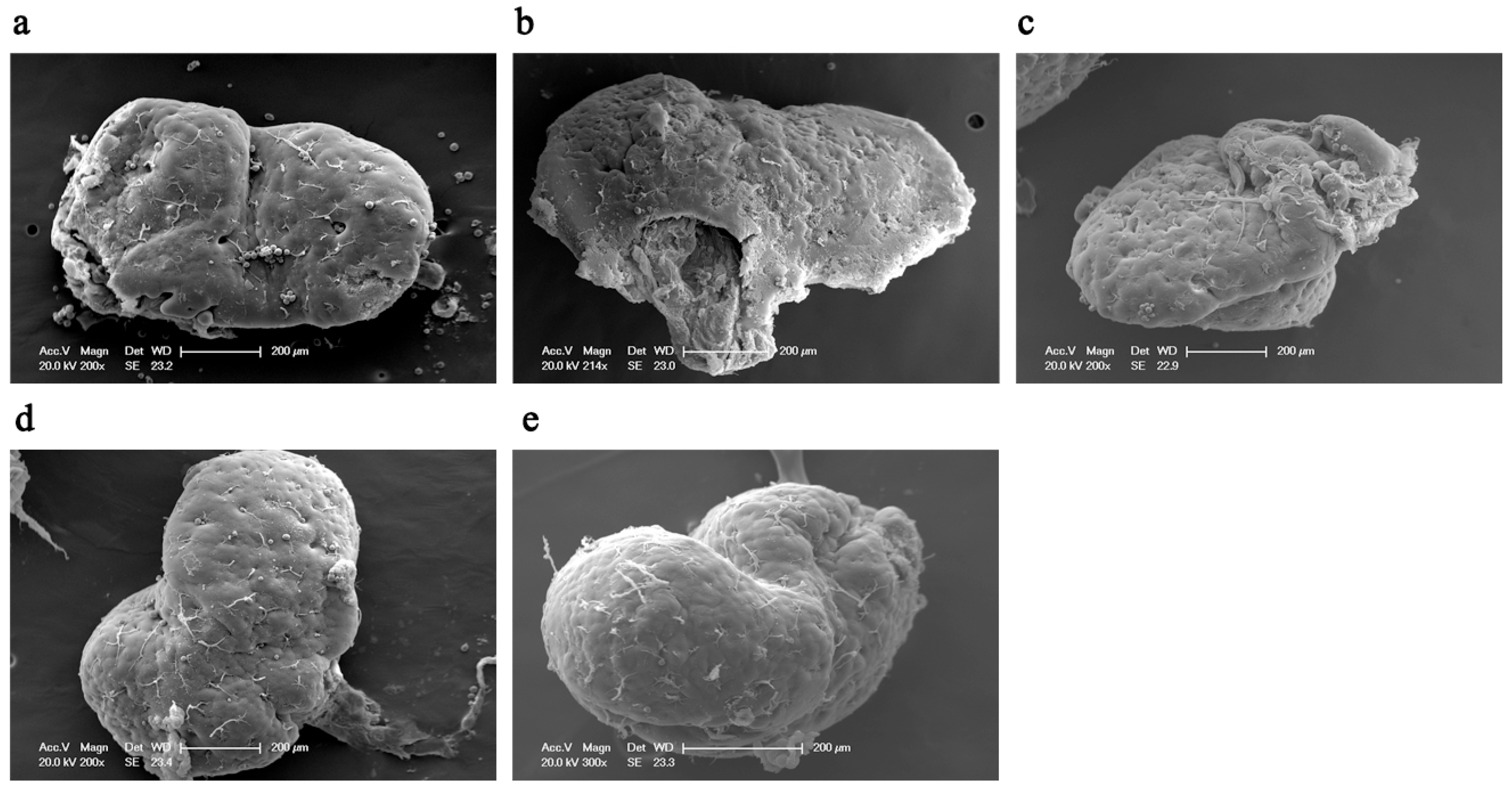
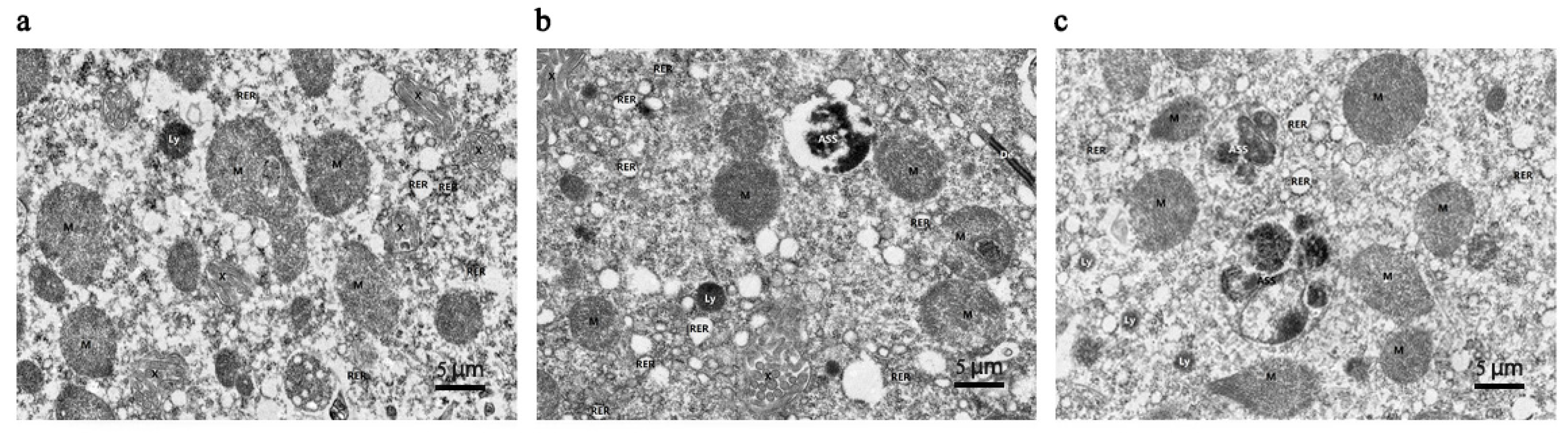
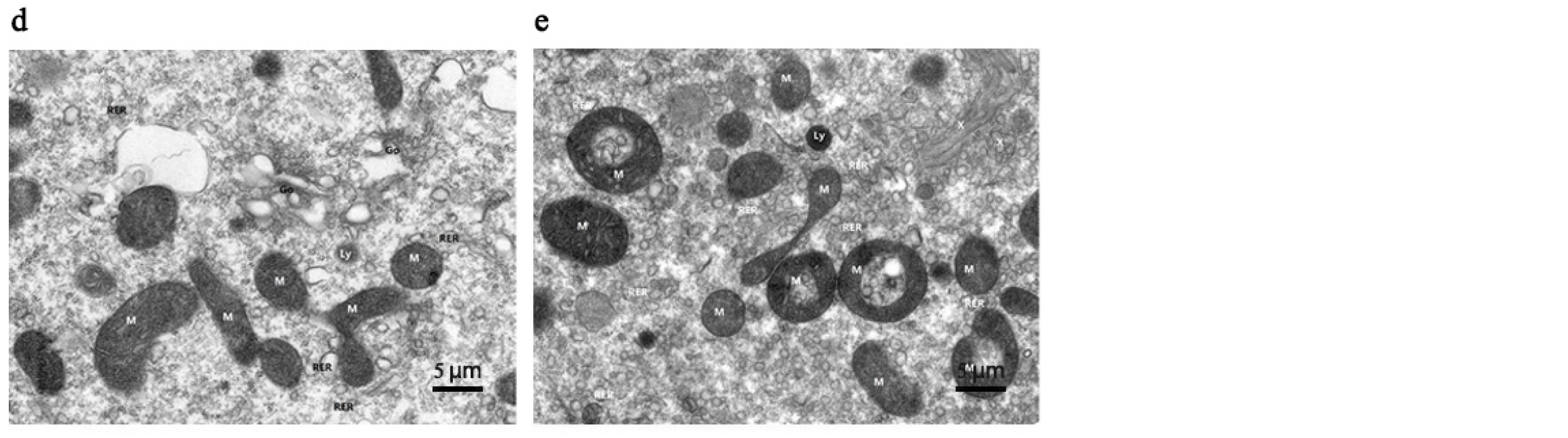
3.1. Mandibular Gland Morphology
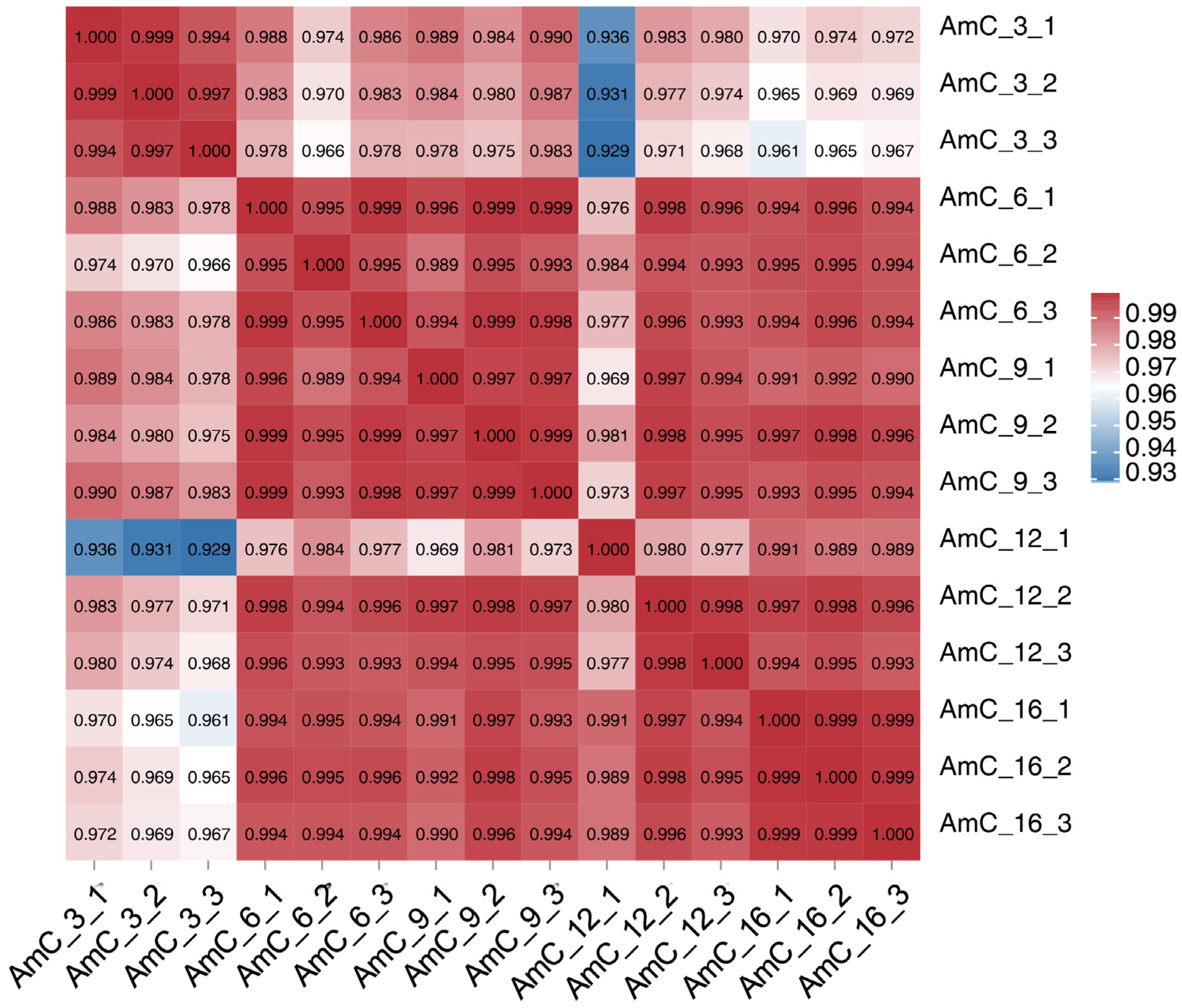
3.2. Quality Control and RNA Sequencing Data Alignment
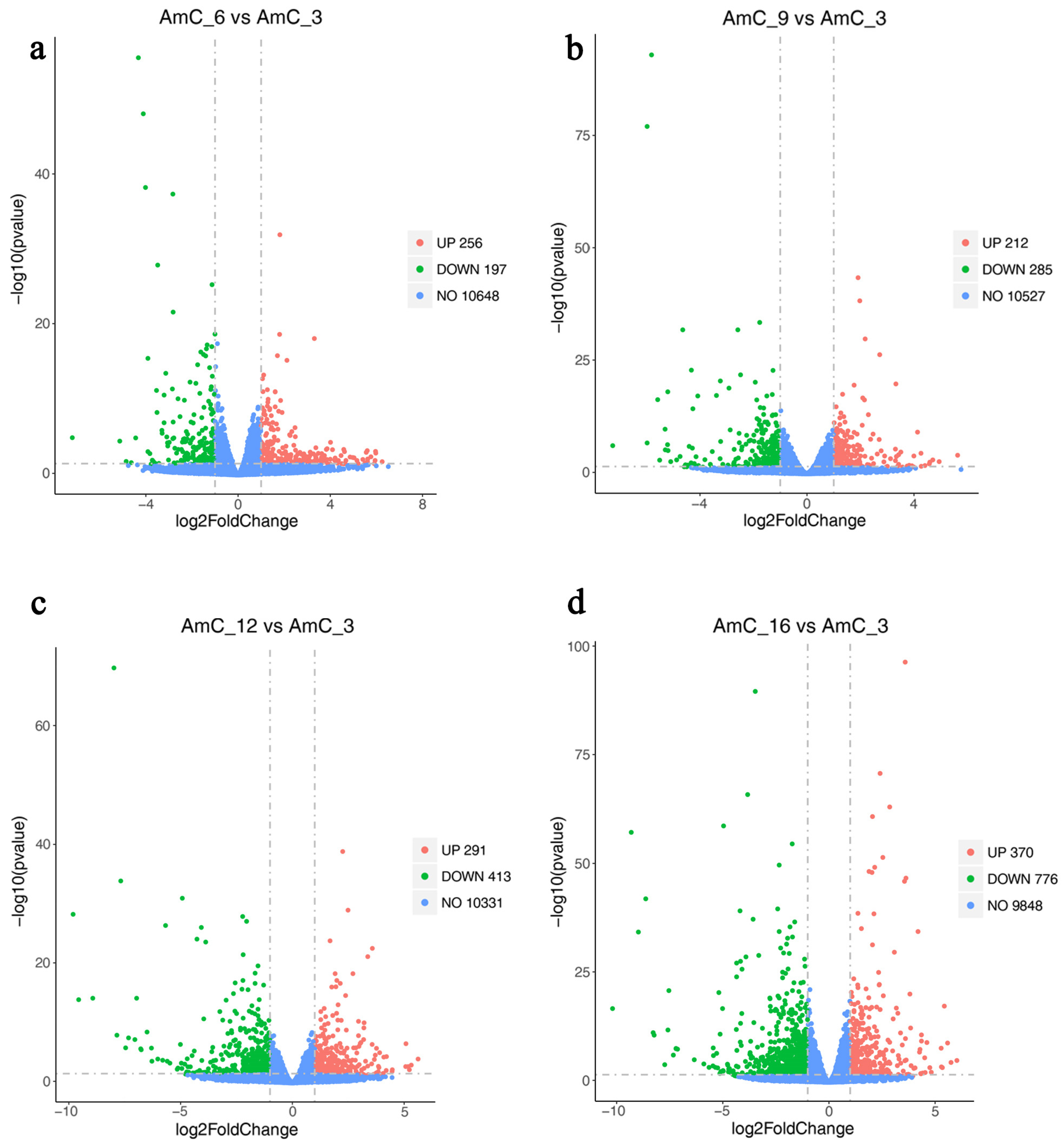
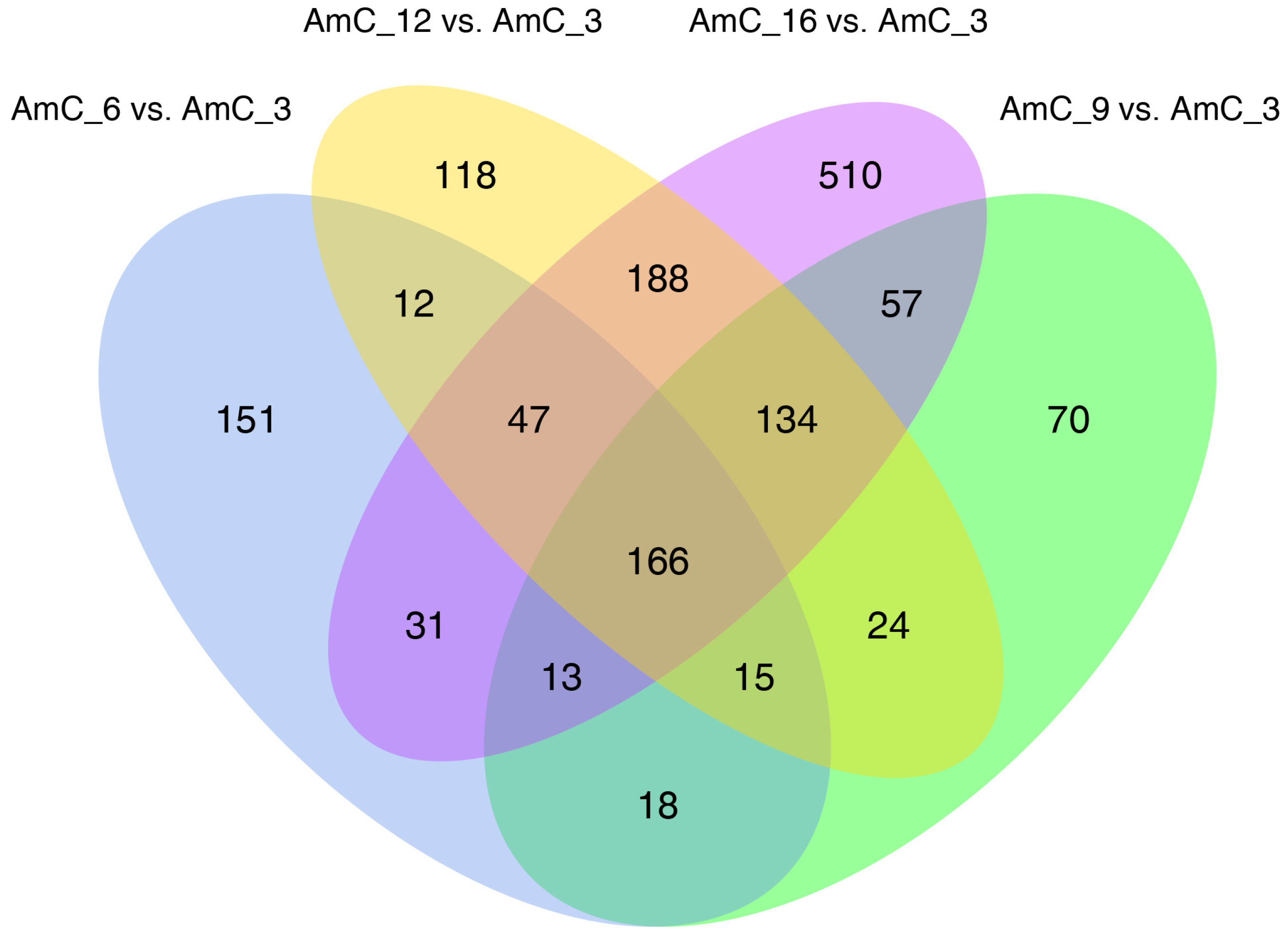
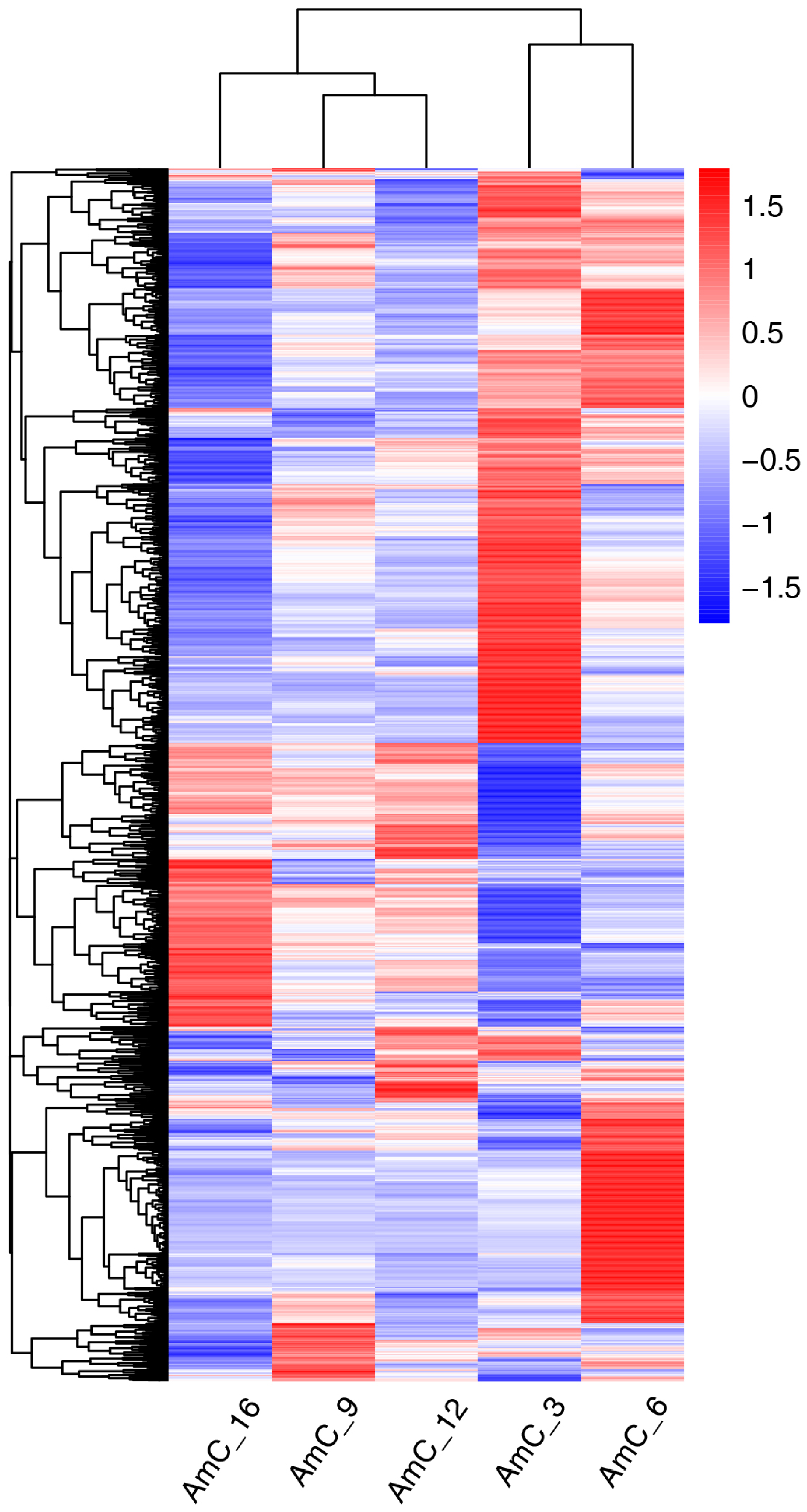
3.3. Differential Gene Expression
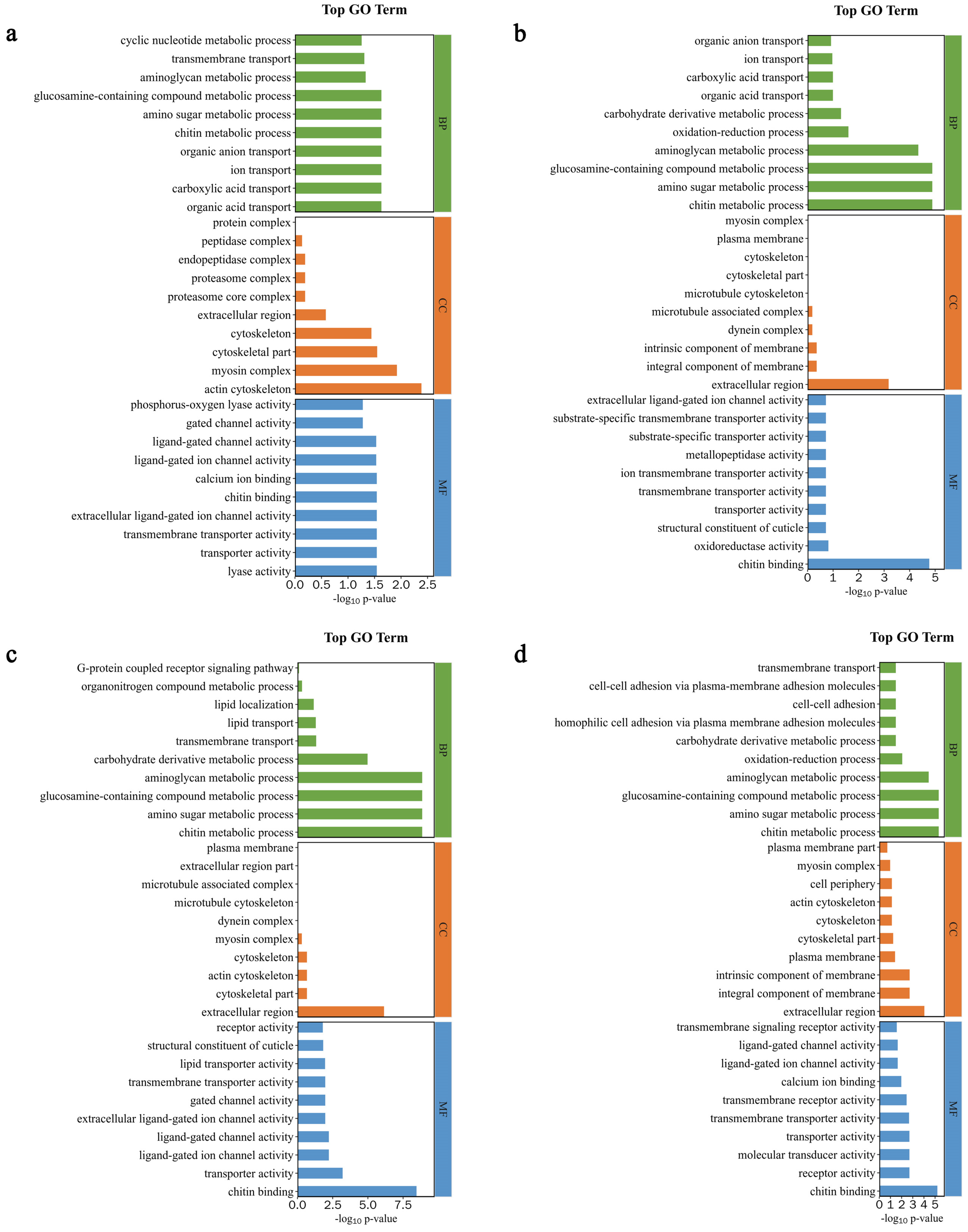
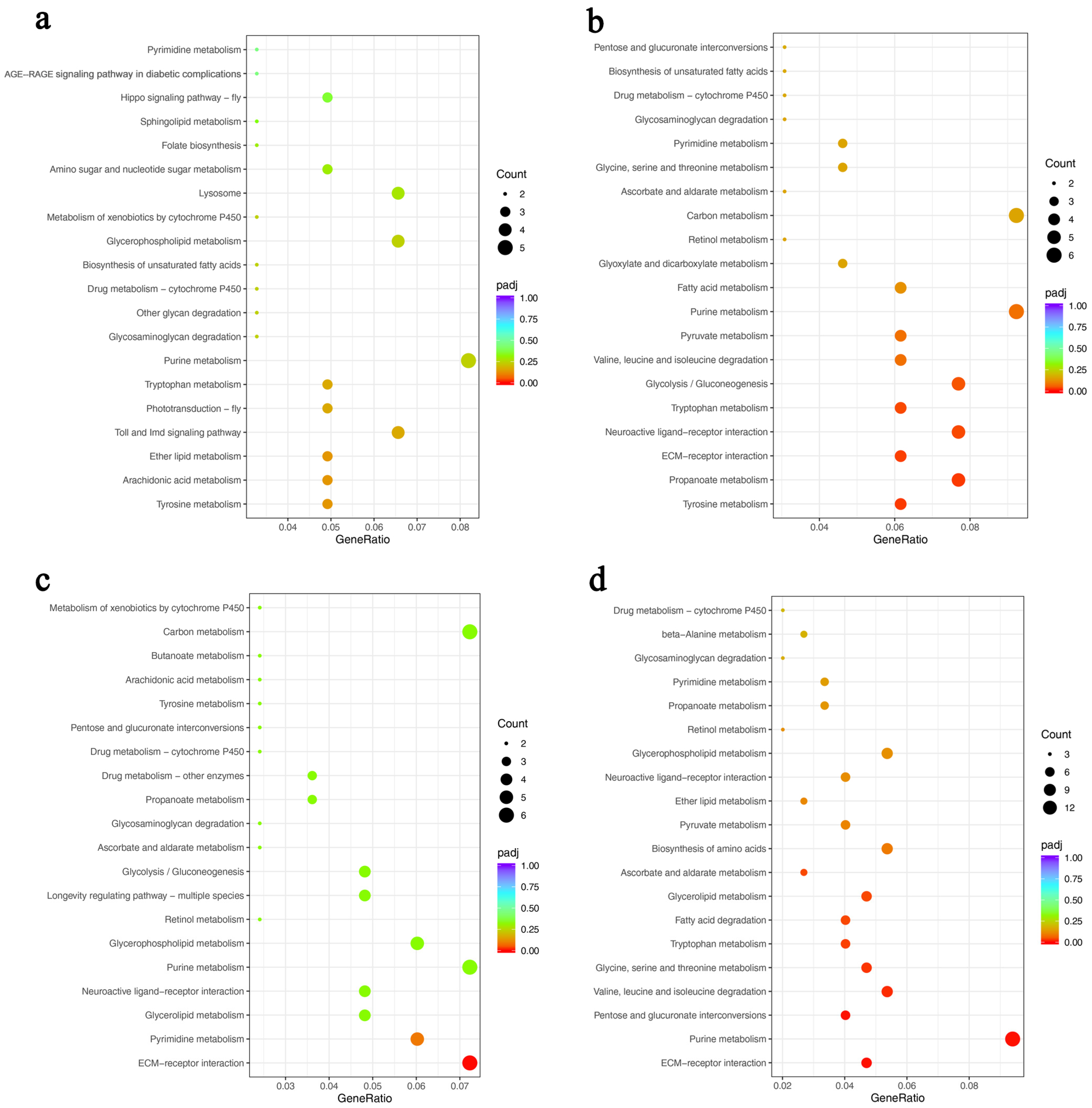
DEGs Function and Pathway Annotation
3.4. Verification of DEGS via RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fujita, T.; Kozuka-Hata, H.; Ao-Kondo, H.; Kunieda, T.; Oyama, M.; Kubo, T. Proteomic analysis of the royal jelly and characterization of the functions of its derivation glands in the honeybee. J. Proteome Res. 2013, 12, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, M. Royalactin induces queen differentiation in honeybees. Nature 2011, 473, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Takahashi, K.; Mori, H. Royal jelly acid, 10-hydroxy-trans-2-decenoic acid, as a modulator of the innate immune responses. Endocr. Metab. Immune Disord. Drug Targets 2012, 12, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Sha, F.; Ma, C.; Wei, Q.; Zhou, Y.; Shi, J.; Fu, J.; Zhang, L.; Han, B.; Li, J. 10-Hydroxydec-2-Enoic Acid Reduces Hydroxyl Free Radical-Induced Damage to Vascular Smooth Muscle Cells by Rescuing Protein and Energy Metabolism. Front. Nutr. 2022, 9, 873892. [Google Scholar] [CrossRef] [PubMed]
- Hamza, R.Z.; Al-Eisa, R.A.; El-Shenawy, N.S. Possible Ameliorative Effects of the Royal Jelly on Hepatotoxicity and Oxidative Stress Induced by Molybdenum Nanoparticles and/or Cadmium Chloride in Male Rats. Biology 2022, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhang, M.; Li, F.; Su, J.; Wang, R.; Li, G.; Yang, X. 10-hydroxy-2-decenoic acid alleviates lipopolysaccharide-induced intestinal mucosal injury through anti-inflammatory, antioxidant, and gut microbiota modulation activities in chickens. Front. Microbiol. 2023, 14, 1285299. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tao, R.; Zhou, J.; Qian, L.; Wu, J. Trans-10-Hydroxy-2-Decenoic Acid Alleviates Dextran Sulfate Sodium-Induced Colitis in Mice via Regulating the Inflammasome-Mediated Pyroptotic Pathway and Enhancing Colonic Barrier Function. Mol. Nutr. Food Res. 2022, 66, e2100821. [Google Scholar] [CrossRef]
- Gao, K.; Su, B.; Dai, J.; Li, P.; Wang, R.; Yang, X. Anti-Biofilm and Anti-Hemolysis Activities of 10-Hydroxy-2-decenoic Acid against Staphylococcus aureus. Molecules 2022, 27, 1485. [Google Scholar] [CrossRef]
- Asma, S.T.; Bobis, O.; Bonta, V.; Acaroz, U.; Shah, S.R.A.; Istanbullugil, F.R.; Arslan-Acaroz, D. General Nutritional Profile of Bee Products and Their Potential Antiviral Properties against Mammalian Viruses. Nutrients 2022, 14, 3579. [Google Scholar] [CrossRef]
- Jovanovic, M.M.; Seklic, D.S.; Rakobradovic, J.D.; Planojevic, N.S.; Vukovic, N.L.; Vukic, M.D.; Markovic, S.D. Royal Jelly and trans-10-Hydroxy-2-Decenoic Acid Inhibit Migration and Invasion of Colorectal Carcinoma Cells. Food Technol. Biotechnol. 2022, 60, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Salama, S.; Shou, Q.; Abd El-Wahed, A.A.; Elias, N.; Xiao, J.; Swillam, A.; Umair, M.; Guo, Z.; Daglia, M.; Wang, K.; et al. Royal Jelly: Beneficial Properties and Synergistic Effects with Chemotherapeutic Drugs with Particular Emphasis in Anticancer Strategies. Nutrients 2022, 14, 4166. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Wang, K.; Pan, Y.; Tao, L.; Ma, Q.; Zhang, G.; Hu, F. Combined royal jelly 10-hydroxydecanoic acid and aspirin has a synergistic effect against memory deficit and neuroinflammation. Food Funct. 2022, 13, 2336–2353. [Google Scholar] [CrossRef] [PubMed]
- Kunugi, H.; Mohammed Ali, A. Royal Jelly and Its Components Promote Healthy Aging and Longevity: From Animal Models to Humans. Int. J. Mol. Sci. 2019, 20, 4662. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, L.; Feng, M.; Fang, Y.; Hu, H.; Han, B.; Meng, L.; Li, J. Metabolic profiling unravels the effects of enhanced output and harvesting time on royal jelly quality. Food Res. Int. 2021, 139, 109974. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Tanga, B.M.; Bang, S.; Maigoro, A.Y.; Kang, H.; Cha, D.; Lee, S.; Lee, S.; Cho, J. MicroRNA profiling of royal jelly extracellular vesicles and their potential role in cell viability and reversing cell apoptosis. Funct. Integr. Genom. 2023, 23, 200. [Google Scholar] [CrossRef] [PubMed]
- Jianke, L.; Mao, F.; Begna, D.; Yu, F.; Aijuan, Z. Proteome comparison of hypopharyngeal gland development between Italian and royal jelly producing worker honeybees (Apis mellifera L.). J. Proteome Res. 2010, 9, 6578–6594. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bezabih, G.; Feng, M.; Wei, Q.H.; Zhang, X.F.; Hu, F.; Meng, L.F.; Fang, Y.; Han, B.; Ma, C.; et al. In-depth Proteome of the Hypopharyngeal Glands of Honeybee Workers Reveals Highly Activated Protein and Energy Metabolism in Priming the Secretion of Royal Jelly. Mol. Cell. Proteom. 2019, 18, 606–621. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhao, X.; Chen, Y.; Sun, C. Chromosome-scale genome assembly of the high royal jelly-producing honeybees. Sci. Data 2021, 8, 302. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Liu, X.; Pan, J.; Li, W.; Li, Z.; Zhang, S.; Chen, S.; Miao, X.; Zheng, N.; Su, S. Identification of genes related to high royal jelly production in the honey bee (Apis mellifera) using microarray analysis. Genet. Mol. Biol. 2017, 40, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Deseyn, J.; Billen, J. Age-dependent morphology and ultrastructure of the hypopharyngeal gland of Apis mellifera workers (Ymenoptera, Apidae). Apidologie 2005, 36, 49–57. [Google Scholar] [CrossRef]
- Hasegawa, M.; Asanuma, S.; Fujiyuki, T.; Kiya, T.; Sasaki, T.; Endo, D.; Morioka, M.; Kubo, T. Differential gene expression in the mandibular glands of queen and worker honeybees, Apis mellifera L.: Implications for caste-selective aldehyde and fatty acid metabolism. Insect Biochem. Mol. Biol. 2009, 39, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Holman, L.; Helantera, H.; Trontti, K.; Mikheyev, A.S. Comparative transcriptomics of social insect queen pheromones. Nat. Commun. 2019, 10, 1593. [Google Scholar] [CrossRef] [PubMed]
- Barker, S.A.; Foster, A.B.; Lamb, D.C. Biological origin and configuration of 10-hydroxy-2-decenoic acid. Nature 1959, 184 (Suppl. S9), 634. [Google Scholar] [CrossRef] [PubMed]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic Properties of Bioactive Compounds from Different Honeybee Products. Front. Pharmacol. 2017, 8, 412. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zheng, H.; Corona, M.; Pirk, C.; Meng, F.; Zheng, Y.; Hu, F. Comparative transcriptome analysis on the synthesis pathway of honey bee (Apis mellifera) mandibular gland secretions. Sci. Rep. 2017, 7, 4530. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Wu, B.; Feng, M.; Han, B.; Fang, Y.; Hao, Y.; Meng, L.; Wubie, A.J.; Fan, P.; Hu, H.; et al. Proteomic Analysis Reveals the Molecular Underpinnings of Mandibular Gland Development and Lipid Metabolism in Two Lines of Honeybees (Apis mellifera ligustica). J. Proteome Res. 2016, 15, 3342–3357. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhang, W.; Chi, X.; Wang, H.; Liu, Z.; Wang, Y.; Ma, L.; Xu, B. Non-targeted lipidomics and transcriptomics analysis reveal the molecular underpinnings of mandibular gland development in Apis mellifera ligustica. Dev. Biol. 2021, 479, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Fathian, M.; Amiri, B.; Maroosi, A. Application of honey-bee mating optimization algorithm on clustering. Appl. Math. Comput. 2007, 190, 1502–1513. [Google Scholar] [CrossRef]
- Kistler, T.; Basso, B.; Phocas, F. A simulation study of a honeybee breeding scheme accounting for polyandry, direct and maternal effects on colony performance. Genet. Sel. Evol. 2021, 53, 71. [Google Scholar] [CrossRef] [PubMed]
- Painter, T.S.; Biesele, J.J. The fine structure of the hypopharyngeal gland cell of the honey bee during development and secretion. Proc. Natl. Acad. Sci. USA 1966, 55, 1414–1419. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Xiang, Y.Q.; Wang, Y.J.; Baikati, K.; Cuervo, A.M.; Luu, Y.K.; Tang, Y.; Pessin, J.E.; Schwartz, G.J.; Czaja, M.J. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Investig. 2009, 119, 3329–3339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Goldman, S.; Baerga, R.; Zhao, Y.; Komatsu, M.; Jin, S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 19860–19865. [Google Scholar] [CrossRef] [PubMed]
- Free, J.B. A social insect: The biology of the honey bee. Science 1987, 238, 1591–1592. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Fang, Y.; Li, J. Proteomic analysis of honeybee worker (Apis mellifera) hypopharyngeal gland development. BMC Genom. 2009, 10, 645. [Google Scholar] [CrossRef]
- Mariman, E.C.; Wang, P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010, 67, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Romao, J.M. Adipogenic transcriptome profiling using high throughput technologies. J. Genom. 2013, 1, 22–28. [Google Scholar] [CrossRef] [PubMed]

| Sample | Raw Reads | Clean Reads | Clean Bases | Q20 | Q30 |
|---|---|---|---|---|---|
| AmC_3_1 | 43,925,926 | 43,006,096 | 6.45 G | 97.73 | 93.48 |
| AmC_3_2 | 57,984,684 | 57,254,280 | 8.59 G | 97.80 | 93.54 |
| AmC_3_3 | 60,166,614 | 58,746,146 | 8.81 G | 97.73 | 93.43 |
| AmC_6_1 | 59,208,756 | 57,797,970 | 8.67 G | 97.80 | 93.69 |
| AmC_6_2 | 57,649,114 | 56,128,514 | 8.42 G | 97.82 | 93.71 |
| AmC_6_3 | 63,687,014 | 61,954,796 | 9.29 G | 97.80 | 93.67 |
| AmC_9_1 | 58,043,056 | 56,465,200 | 8.47 G | 97.76 | 93.54 |
| AmC_9_2 | 52,090,762 | 50,420,902 | 7.56 G | 97.69 | 93.46 |
| AmC_9_3 | 54,290,204 | 49,824,086 | 7.47 G | 97.73 | 93.59 |
| AmC_12_1 | 57,338,498 | 56,223,708 | 8.43 G | 97.77 | 93.30 |
| AmC_12_2 | 50,767,738 | 49,313,272 | 7.40G | 97.54 | 93.12 |
| AmC_12_3 | 66,511,356 | 62,514,040 | 9.38 G | 97.59 | 93.27 |
| AmC_16_1 | 56,240,840 | 54,560,816 | 8.18 G | 97.66 | 93.36 |
| AmC_16_2 | 60,768,102 | 58,984,280 | 8.85 G | 97.59 | 93.19 |
| AmC_16_3 | 51,563,118 | 50,354,212 | 7.55 G | 96.89 | 91.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, C.; Zhang, Y.; Liu, C.; Zhang, Z.; Tao, L.; Wang, K.; Lin, Z.; Ji, T.; Gao, F. Revealing the Development Patterns of the Mandibular Glands of Apis mellifera carnica Based on Transcriptomics and Morphology. Insects 2024, 15, 176. https://doi.org/10.3390/insects15030176
Pan C, Zhang Y, Liu C, Zhang Z, Tao L, Wang K, Lin Z, Ji T, Gao F. Revealing the Development Patterns of the Mandibular Glands of Apis mellifera carnica Based on Transcriptomics and Morphology. Insects. 2024; 15(3):176. https://doi.org/10.3390/insects15030176
Chicago/Turabian StylePan, Chunlei, Yi Zhang, Chunguang Liu, Zhihao Zhang, Liang Tao, Kang Wang, Zheguang Lin, Ting Ji, and Fuchao Gao. 2024. "Revealing the Development Patterns of the Mandibular Glands of Apis mellifera carnica Based on Transcriptomics and Morphology" Insects 15, no. 3: 176. https://doi.org/10.3390/insects15030176
APA StylePan, C., Zhang, Y., Liu, C., Zhang, Z., Tao, L., Wang, K., Lin, Z., Ji, T., & Gao, F. (2024). Revealing the Development Patterns of the Mandibular Glands of Apis mellifera carnica Based on Transcriptomics and Morphology. Insects, 15(3), 176. https://doi.org/10.3390/insects15030176






