Simple Summary
Increasing levels of insecticide resistance in disease-transmitting mosquitoes highlight the need for alternative strategies to control vector mosquito populations. RNA interference-based strategies have strong potential to become a major component of integrated vector management. To implement dsRNA-based insecticides, effective target genes need to be identified whose knockdown causes death or sterility in mosquitoes. Here, we show that the ribosomal protein P1 is a promising candidate as a target for mosquito birth control using RNA interference.
Abstract
After taking a blood meal, the fat body of the adult female yellow fever mosquito, Aedes aegypti, switches from a previtellogenic state of arrest to an active state of synthesizing large quantities of yolk protein precursors (YPPs) that are crucial for egg development. The synthesis of YPPs is regulated at both the transcriptional and translational levels. Previously, we identified the cytoplasmic protein general control nonderepressible 1 (GCN1) as a part of the translational regulatory pathway for YPP synthesis. In the current study, we used the C-terminal end of GCN1 to screen for protein–protein interactions and identified 60S acidic ribosomal protein P1 (P1). An expression analysis and RNAi-mediated knockdown of P1 was performed to further investigate the role of P1 in mosquito reproduction. We showed that in unfed (absence of a blood meal) adult A. aegypti mosquitoes, P1 was expressed ubiquitously in the mosquito organs and tissues tested. We also showed that the RNAi-mediated knockdown of P1 in unfed adult female mosquitoes resulted in a strong, transient knockdown with observable phenotypic changes in ovary length and egg deposition. Our results suggest that 60S acidic ribosomal protein P1 is necessary for mosquito reproduction and is a promising target for mosquito population control.
1. Introduction
In the last few decades, the yellow fever mosquito, Aedes aegypti, has become an important model organism for the study of nutrient-sensing and signaling pathways, disease transmission, and cellular metabolism and physiology [1,2,3,4,5]. This is due to A. aegypti’s capacity to vector diseases, specialized anatomy that is unique from other model organisms, and relatively quick life cycle. In addition, A. aegypti mosquitoes are anautogenous, meaning they require a blood meal to produce a clutch of eggs; as a result, their reproductive and nutrient processes are uniquely easy to study [6,7,8]. The uptake of a blood meal triggers several physiological processes including vitellogenesis. Vitellogenesis, the accumulation of yolk (vitellus) in the developing oocytes, starts with the synthesis of yolk protein precursors (YPPs) [9,10,11]. YPPs are synthesized exclusively by the mosquito’s fat body. After synthesis in the trophocytes of the fat body, YPPs are released into the hemolymph, taken up by the oocytes via receptor-mediated endocytosis, and then cleaved into yolk proteins [12,13,14].
Regulation of YPP gene expression. Several cellular signaling pathways have been linked to the regulation of vitellogenic gene expression (see Figure 1). Juvenile hormone (JH), ecdysone, target of rapamycin complex 1 (TORC1), insulin-like peptide, and general control nonderepressible (GCN) signaling pathways have all been shown to regulate YPP transcription and translation in the cells of the fat body [15,16,17,18]. TOR and GCN signaling both involve amino acid (AA) transporters at the top of their respective signaling cascades (see Figure 1c,e). The increase in AAs in a blood-fed mosquito triggers these pathways in the trophocytes of the fat body. This activation is due to the transport of AAs from the hemolymph into the cytoplasm of trophocytes.
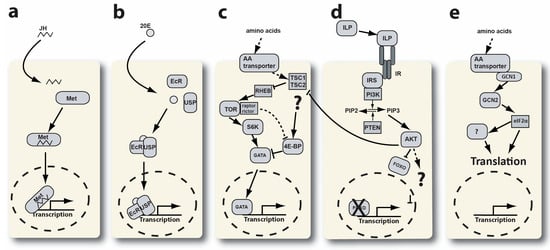
Figure 1.
The regulation of vitellogenic gene expression in mosquito fat body cells. This figure is modified from Hansen et al., 2014 [19]. (a) Juvenile hormone signaling. JH—juvenile hormone, Met—methoprene receptor. (b) Ecdysone signaling. 20E—20-hydroxy-ecdysone, EcR—ecdysone receptor, USP—ultraspiracle. (c) TOR signaling. AA—amino acid, TSC—tuberous sclerosis complex, RHEB—RAS homologue enriched in brain, S6K—S6 kinase, 4E-BP—4E—binding protein, GATA—GATA type transcription factor. (d) Insulin signaling. ILP—insulin-like peptide, IR—insulin receptor, IRS—insulin receptor substrate, PI3K—PI3 kinase, PTEN, AKT, FOXO—forkhead box transcription factor O. (e) GCN signaling. GCN—general control nonderepressible, eIF2α—eukaryotic initiation factor 2 alpha. The dotted circle represents the cell nucleus.
Cationic amino acid transporters. Membrane-bound AA transporter proteins of the SLC7 family transport AAs that are necessary for mosquito vitellogenesis [20,21,22,23]. The AAs histidine, lysine, and arginine are cationic AAs and require specialized transporters called cationic amino acid transporters (CATs) to cross cell membranes. CATs are membrane transporters that facilitate the massive flow of cationic AAs from one mosquito tissue to another [24,25]. The genome of A. aegypti encodes five CATs [1]. CAT1, CAT2, and CAT3 are expressed in the fat body of adult A. aegypti and the knockdown of these transporters significantly reduces YPP synthesis [20,26], demonstrating the importance of CATs for mosquito vitellogenesis.
Downstream of CATs, TOR, and GAAC signaling regulate mosquito egg development. The abovementioned studies showed that RNAi-mediated knockdown of specific SLC7 transporters reduced AA-induced TOR signaling and overall mosquito egg development [1,26]. Pinch and colleagues further investigated the mechanics and involvement of CATs in nutrient signaling pathways involved in the regulation of vitellogenesis. They performed a yeast two-hybrid (Y2H) screen using the C-terminal end of A. aegypti CAT1 and identified general control nonderepressible 1 (GCN1) as an interacting protein. GCN1 is a part of the general amino acid control (GAAC) signaling pathway that regulates the response to amino acid starvation in eukaryotic cells [27,28,29,30,31,32,33,34]. Next, they found that GCN1 was highly expressed in organs and structures associated with vitellogenesis, with the highest gene expression in the ovaries followed by the fat body. When they performed a knockdown of GCN1 in A. aegypti, they found no effect on YPP transcription but a significant reduction in fertility [35].
General control nonderepressible 1. GCN1 is a conserved protein in eukaryotic organisms [31,36]. It was first described in yeast as an effector protein of GCN2 in the GAAC pathway. In AA-starved cells, ribosomes collide during translation, creating a disome. GCN1 binds to both ribosomes in this disome and subsequently activates GCN2, starting the GCN signaling pathway [31,37,38]. The N-terminal domain of GCN1 is necessary for robust ribosome binding [39,40,41]. Downstream of this N-terminal domain is a region containing several HEAT repeats, repetitive arrays of short amphiphilic α-helices, that are involved in the formation of protein–protein interactions (see Figure 2A) [42,43]. The C-terminal domain of GCN1 also interacts with colliding ribosomes and mediates the interaction between GCN1 and GCN2 [41,44].
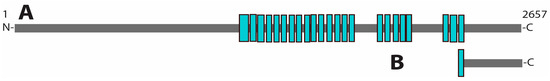
Figure 2.
Predicted domain structure of A. aegypti GCN1. (A) Structure of GCN1 complete protein. (B) C-terminal domain of GCN1 that was used for Y2H screening. HEAT domains are shown in blue.
The role of GCN1 in mosquito vitellogenesis has only been recently discovered and there is a significant gap in our knowledge of GCN1’s functions within the mosquito fat body. To fill this gap and uncover novel components in the GCN pathway, we performed a yeast two-hybrid (Y2H) screen using a C-terminal fragment of A. aegypti GCN1 (see Figure 2B). In this study, we identified 60S acidic ribosomal protein P1 (P1) as a protein interactor of GCN1 and demonstrated that the knockdown of P1 results in female A. aegypti sterility.
2. Materials and Methods
Yeast two-hybrid assay: A yeast two-hybrid (Y2H) screen was performed using the Matchmaker® Gold Yeast Two-Hybrid System (Takara Bio, San Jose, CA, USA Cat. Nos. 630466, 630489, 630498, 630,499 PT4084-1 (071519)), following the manufacturer’s protocol. The GCN1 protein sequence (2656 amino acids) was retrieved from GenBank (accession number: XP_001651826) and a 906-nucleotide C-terminal sequence was amplified using PCR and cloned into a pGBKT7 plasmid.
This sequence includes a HEAT domain at the C-terminal end:
AELIVIHTRPDPLFVEMQNGIKNTDDSTIRETMLQALRGILTPAGDKMTEPLKKQ IYSMLSGMLGHPEDVTRAAAAGCLGALCRWLNPDQLDDALNSHMLNEDYGDDAA LRHGRTAALFVALKEYPSAIFTDKYEAKICKTIASSLISDKIPVALNGVRAAGYLLQYG MCSDDVKLPQQIIGPFVKSMNHTSNEVKQLLAKTCLYLAKTVPAEKTAPEYLRLVIPM LVNGTKEKNGYVKSNSEIALVYVLRLREGDEVHQKCIALLEPGARDSLSEVVSKVLRK VAMQPVGKEEELDDTILT
The following primer set was used for PCR amplification of the bait cDNA:
GCN1 bait forward:
5′CATGGAGGCCGAATTCGCAGAACTGATCGTAATCCACACC3′
GCN1 bait reverse:
5′GCAGGTCGACGGATCCCGTAAGAATCGTATCGTCTAGTTC3′
To clone the GCN1 bait PCR fragments into the pGBKT7 plasmid, we used the In-Fusion® HD Cloning Kit (Takara Bio USA Inc., Orchard Pkwy, San Jose, CA , USA). To transform the bait pGBKT7 plasmid into competent yeast cells (Y2HGold strain), the Yeastmaker™ Yeast Transformation System 2 Kit (Takara Bio USA Inc.) was used. For prey library construction, the yeast transformation protocol from Yeastmaker™ Yeast Transformation System 2 (Takara Bio USA Inc.) was used to prepare competent yeast cells of the prey strain Y187. The Make Your Own “Mate & Plate™” Library System (Takara Bio USA Inc.) was used to construct the two-hybrid library strain, following the manufacturer’s instructions. Library strain construction is described in detail elsewhere [35].
Briefly, whole female mosquitoes, 24 h after a blood meal, were decapitated and their legs and wings were removed. Total RNA was isolated and purified, and mRNA was subsequently enriched using a commercially available purification kit (Takara Bio USA Inc.). cDNA was synthesized using SMART MMLV reverse transcriptase, followed by In-Fusion® cloning into the prey vector pGADT7 AD. Library plasmids were transformed into the yeast strain Y187.
The bait and prey strains were mated, and zygote formation was confirmed using a phase contrast microscope. After screening for yeast colonies that displayed reporter phenotypes, indicating that they contained interacting proteins, individual library plasmids were recovered from these colonies and sent for Sanger sequencing (MCLAB, South San Francisco, CA). DNA sequences were analyzed using NCBI BLASTX.
Mosquito rearing: Five- to seven-day-old female A. aegypti mosquitoes (Black Eye Liverpool Strain, BEI resources, Catalog No. NR-48921) were used for all experiments in this study and reared under standard conditions described before [43]. Larvae were reared in pans and fed ad libitum with cat food (Special Kitty, Walmart Stores Inc., Bentonville, AR, USA). Pupae were transferred to Bug Dorm-1 insect rearing cages (30 × 30 × 30 cm; BugDorm, Taichung, Taiwan) and stored in a room with 80% humidity and 28 °C temperature with a 12:12 light:dark cycle. Adult mosquitoes were fed ad libitum on a 20% sucrose solution.
Protein isolation: Mosquitoes were ice-anesthetized, and their wings and legs were removed. Organs and tissues (head, thorax, midgut, fat bodies, Malpighian tubules, and ovaries) were dissected in Aedes physiological saline (APS) [45]. The dissected organs and tissues were transferred into separate Eppendorf tubes with lysis buffer (100 µL IGEPAL, 1 µL protease inhibitor, Thermo Fisher Scientific, Waltham, MA, USA Lot # VA293119) and 1 µL phosphatase inhibitor (Thermo Fisher Scientific Lot # UH288468) and homogenized using a handheld homogenizer (VWR, Avantor; Radnor, PA, USA; Cat. No. 4774-370) with disposable polybutylene terephthalate pestles (VWR, Avantor; Cat. No. 4774-358). The samples were subsequently centrifuged at 12,000 RPM for 10 min at 4 °C. The supernatant was collected in 1.5 mL Eppendorf tubes and protein concentrations were measured using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific Inc.). Protein concentration measurements were performed with a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific Inc.) using the protein BCA program.
Western Blot: Twenty micrograms of protein from each sample was diluted in a 1:1 ratio of Laemmli buffer and 1% beta-mercaptoethanol. The mixture was incubated for 5 min at 95 °C to denature the proteins. Samples were size-separated on a 7.5% acrylamide gel at 100V (Bio-Rad Laboratories, Irvine, CA, USA) for 1 h. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane using a wet-blotting apparatus and the protocol described in the manufacturer’s manual (Bio-Rad Laboratories). The membrane was then washed four times in five-minute intervals on a rocking table using a wash buffer (TBST + 0.05% Tween-20). The membrane was then blocked with a blocking buffer (3% BSA) for an hour at room temperature with a gentle rocking motion. A diluted GCN1 primary antibody (polyclonal RPLP1 antibody, Catalog # PA5-77002, Thermo Fisher Scientific Inc., 1:500 in blocking buffer) was added to the membrane and allowed to incubate at 4 °C overnight with gentle shaking. The membrane was then washed four times, and a secondary antibody (anti-rabbit secondary Ab, Millipore Sigma, Burlington, MA, USA; 1:1000 dilution) was added and incubated at room temperature for one hour. The membrane was washed again four times. A chemiluminescence assay was performed (WesternSure PREMIUM Chemiluminescent Substrate, Li-Cor, Lincoln, NE, USA). The membrane was analyzed with a C-Digit imager using the Image Studio software v5.5.4. (LI-COR, Lincoln, NB, USA).
RNA isolation: RNA was isolated using the Qiagen RNeasy Kit (Qiagen, Germantown, MD, USA), following the manufacturer’s protocol. For qPCR expression analyses, various numbers of organs/tissues were dissected and combined for each biological replicate in order to isolate at least 1 µg of RNA for each tissue replicate. For the knockdown qPCR experiments, three whole mosquito females were pooled for each of four biological replicates. Wings, legs, and heads were removed. The pools were homogenized in a 1.5 mL Eppendorf tube in 350 µL of lysis buffer (Buffer RLT, Qiagen, Germantown, MD, USA) using a VWR cordless motor (VWR, Avantor; Radnor, PA, USA; Cat. No. 4774-370) with disposable polybutylene terephthalate pestles (VWR, Avantor; Cat. No. 4774-358). RNA concentration was determined using a Nanodrop™ 1000 spectrophotometer (Thermo Fisher Scientific Inc.).
RNAi-mediated gene knockdown: For RNAi-mediated knockdown, double-stranded RNA (dsRNA) targeting P1 and green fluorescent protein (GFP) as a control was synthesized. Primers were designed using PrimerBlast (see Table 1) [46]. To facilitate in vitro dsRNA synthesis, T7 promoter sequences were added to the 5′ ends of both forward and reverse primers. dsRNA was synthesized using the Megascript™ RNAi Kit (Thermo Fisher Scientific Inc., Lot No. 01130674), following the manufacturer’s instructions. Synthesized dsRNA was diluted in APS to a concentration of 1 µg/µL. Sets of A. aegypti females (5–7 days post-eclosion) were injected intrathoracically with 1 µL of either the GFP dsRNA (control) or P1 dsRNA (knockdown) [47]. Injected mosquitoes were transferred into Bug Dorm-1 insect rearing cages to recover. Mosquitoes were fed ad libitum on a 20% sucrose solution and stored in the rearing environment described above. Mosquitoes that did not recover from the injections were discarded.

Table 1.
dsRNA synthesis primers. T7 promoter regions are in bold.
Quantitative reverse transcription PCR (qRT-PCR) analysis: For the qRT-PCR analysis, qRT-PCR primers were designed for P1 and β-actin, a “housekeeping” gene used as a reference. β-actin has been validated as a suitable reference gene for qPCR during different life stages and in different tissues of A. aegypti [48]. qRT-PCR primers were designed using PrimerBlast (see Table 2). The primer sequences for β-actin qRT-PCR primers were previously described. cDNA synthesis and qRT-PCR reactions were performed as described previously [35]. Three repeats were performed, and relative expression levels were either calculated in comparison to whole mosquitoes (for tissue/organ expression analyses) or GFP-injected control mosquitoes (for knockdown experiments).

Table 2.
qRT-PCR primers.
Ovary morphology assay: GFP-dsRNA- and P1-dsRNA-injected mosquitoes, 24 h post-injection (PI), were fed defibrinated bovine blood (HemoStat Laboratories, Dixon, CA, USA) for one hour. Mosquitoes that did not take a blood meal during this time were subsequently removed from the cages. Prior to the blood meal, 10 mosquitoes were collected from each treatment cage to study the morphology of unfed mosquito ovaries. In total, ten ovaries from each of the four time points (0, 24, 48, and 72 h post-blood meal (PBM)) were collected. All ovaries were imaged using an Olympus CX41 stereo microscope (Olympus Life Science, Waltham, MA, USA). Ovary lengths were determined using ImageJ software Version 1.54d [49].
Egg number assay: For this assay, four groups of twenty GFP- and four groups of twenty P1-dsRNA-injected mosquitoes were blood fed, as described above. A wet filter paper was placed in the treatment and control cages 72 h PBM and collected 96 h later. Egg numbers were counted using a stereomicroscope.
Statistical analysis: All statistical analyses were performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA, Version 9.5.1). A Shapiro–Wilk test was performed to test if the data were normally distributed. An unpaired t-test was performed to determine differences in ovary length between the P1-dsRNA-injected and GFP-dsRNA-injected mosquitoes. An unpaired t-test with Welch’s correction was used for each time point to analyze the qRT-PCR data. A simple survival analysis (Kaplan–Meier) was used to make the survival curves. A log-rank (Mantel–Cox) test was used to determine differences between the survival of GFP- and P1-dsRNA-injected mosquitoes. p-values < 0.05 were considered statistically significant for all tests.
3. Results
3.1. Protein–Protein Interactions of GCN1
The Y2H screen we performed using the C-terminal end of A. aegypti GCN1 resulted in the isolation of six separate library plasmids (see Table S1). All six plasmids encoded the C-terminus of the same protein: 60S acidic ribosomal protein P1 (GenBank accession number: XP_001663779).
3.2. Gene Expression of P1 in Different Mosquito Organs and Structures
In unfed, 5–7-day-old adult female A. aegypti mosquitoes, we profiled the expression of P1 within six different mosquito organs and tissues: thorax (TH), fat body (FB), midgut (MG), Malpighian tubules (MT), ovary (OV), and head (H). The qRT-PCR analysis revealed a ubiquitous and uniform expression of P1 across the different organs and tissues tested (see Figure 3a). To investigate the protein expression patterns of P1, we performed a Western blot using an antibody against human P1 (see Figure 3b). The predicted molecular weight of A. aegypti P1 protein is 15 kDa. A band at this size was observed in fat body, midgut, and ovary samples. Various other nonspecific bands of higher molecular weight were observed in different patterns in different organs/structures. This included a band at 38 kDa, the predicted size of the P1/P2 dimer of A. aegypti.
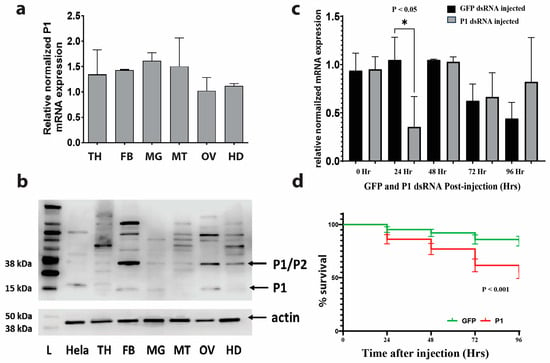
Figure 3.
P1 gene expression and mosquito survival after knockdown. Variation between replicates is displayed as the standard error of the mean (SEM). (a) P1 is expressed at uniform levels in various organs/structures of unfed female mosquitoes. Relative expression levels were determined via qRT-PCR (thorax, TH; fat body, FB; midgut, MG; Malpighian tubule, MT; ovary, OV; head, HD). (b) Antibody against human P1 detects bands of various sizes in different mosquito tissues. The upper panel shows the results of a Western blot using P1 antibody. The lower panel shows the results of a Western blot using an antibody against actin as a loading control (L, protein molecular weight marker; Hela—HeLa cell protein extract; see Figures S1–S3). (c) Transient knockdown of P1. Shown are qRT-PCR results of P1 expression at different time points after injection of dsRNA. The “0 Hr” time point represents uninjected control mosquitoes. β-actin gene expression was used to normalize the data. Three pools of three mosquitoes were used for each time point. An unpaired t-test with Welch’s correction was used for each time point to determine significant differences in P1 expression between GFP- and P1-dsRNA-injected mosquitoes. (d) Significant increase in mosquito mortality after P1 dsRNA injection. Shown is the average proportion of survival for mosquitoes injected with GFP (n = 192) or P1 dsRNA (n = 196) measured at different time points after injection (see Table S2 and Figure S4 for individual replicate survival curves). A log-rank (Mantel–Cox) test was used to determine differences between the survival of GFP- and P1-dsRNA-injected mosquitoes.
3.3. RNAi-Mediated Knockdown of P1 Is Transient
Knockdown efficacy was measured at five different time points post-injection using qRT-PCR. We found a strong knockdown effect in P1-injected mosquitoes 24 h after injection (see Figure 3c).
3.4. Mosquito Mortality after RNAi-Mediated P1 Knockdown
Survival was measured over four days after dsRNA injection. P1-dsRNA-injected mosquitoes had significantly (p < 0.001) lower survival rates after injection compared to the control (see Figure 3d). Twenty-four hours post-injection, the percentage of survival for GFP-injected mosquitoes and P1-injected mosquitoes was 94.8% and 86.2%, respectively. Forty-eight hours post-injection, there was a 91.3% and 77.6% survival, respectively. At 72 h post-injection, it was 85.2% and 61.8%, respectively, and at 96 h post-injection, it was 83.7% and 56.3%, respectively.
3.5. RNAi-Mediated P1 Knockdown Results in Ovary Phenotype in Mosquitoes
The ovary length of GFP- and P1-dsRNA-injected mosquitoes was measured at four different time points after a blood meal (see Figure 4). The ovaries of GFP-dsRNA-injected mosquitoes had an average length of 0.78 mm before a blood meal. Seventy-two hours after a blood meal, the ovaries had lengthened to an average of 2.86 mm. The ovaries of P1-dsRNA-injected mosquitoes had an average length of 0.85 mm before a blood meal. Seventy-two hours after a blood meal, the ovaries had lengthened to an average of 1.47 mm (see Figure 4a,b). The ovary lengths of P1-dsRNA-injected mosquitoes were significantly shorter compared to the ovary lengths from GFP-injected control mosquitoes at all time points PBM (p < 0.001) (see Figure 4b). P1-knockdown mosquitoes did not lay any eggs after a blood meal (see Figure 4c). Egg trays were left in cages for up to nine days (216 h PBM).
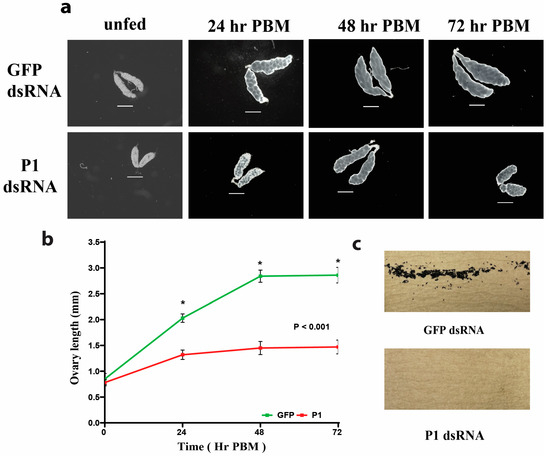
Figure 4.
RNAi-mediated P1 knockdown affects ovary length and egg numbers. (a) P1-knockdown mosquitoes show a pronounced ovary phenotype. Shown are ovaries from GFP- and P1-dsRNA-injected mosquitoes at different time points PBM. The top row shows ovaries from GFP-dsRNA-injected mosquitoes. The bottom row shows ovaries from P1-dsRNA-injected mosquitoes. The scale bar represents 500 µm. (b) Ovary lengths at different times PBM are significantly shorter in P1-knockdown mosquitoes. Ovary lengths of GFP- and P1-dsRNA-injected mosquitoes sampled at 0 h PBM (unfed), 24 h PBM, 48 h PBM, and 72 h PBM. The plotted data represent the average length of ovaries (n = 10) at each time point. Error bars represent the standard error of the mean (SEM), and an unpaired t-test was performed to determine the statistical significance of the difference between groups. (see Table S3) (c) RNAi-mediated P1-knockdown mosquitoes do not lay eggs after a blood meal. Shown are images of egg deposition papers from GFP- (top image) or P1- (bottom image) dsRNA-injected mosquitoes. Note the complete absence of eggs produced by P1-knockdown mosquitoes. Two replicate trials with 20 mosquitoes for both control and knockdown groups were performed, with consistent results (see Figure S5).
4. Discussion
Mosquitoes are medically important vectors of diseases like dengue, chikungunya, yellow fever, and malaria [50,51,52,53,54]. Mosquito population control is an effective strategy used to mitigate the transmission of these diseases [55]. Disease transmission is strongly correlated with the reproductive life cycle of anautogenous mosquitoes because they require a blood meal for the development and maturation of eggs [56,57]. In the last few decades, there has been emphasis on the study of regulatory pathways that are involved in vitellogenesis, the synthesis of YPPs, because of the prospect of identifying novel high-value targets for mosquito population control. There has been significant pursuit and progress in our understanding of the mechanisms that regulate vitellogenesis in mosquitoes, particularly in the yellow fever mosquito A. aegypti [58,59].
Earlier studies from our laboratory and others have investigated the mTOR and GAAC signaling pathways and their role in the regulation of mosquito vitellogenesis (see Figure 1c,e) [58,59,60]. A crucial upstream component of these pathways is cationic amino acid transporters (CATs). These transporters facilitate the movement of essential amino acids between midgut and fat body tissues during vitellogenesis and are hypothesized to be transceptors—functioning as both an amino acid transporter and receptor [57]. A recent Y2H screen using the C-terminus of CAT1 identified a protein–protein interaction with GCN1 [35]. In the present study, we performed a follow-up Y2H screen for potential protein–protein interactors of the C-terminus of GCN1 and identified an interaction with the 60S acidic ribosomal protein P1 (P1) (see Figure 5a). P1 is a component of the P-stalk region on the large subunit of eukaryotic ribosomes and has been shown to play an important role during the elongation stage of protein synthesis [61,62,63].
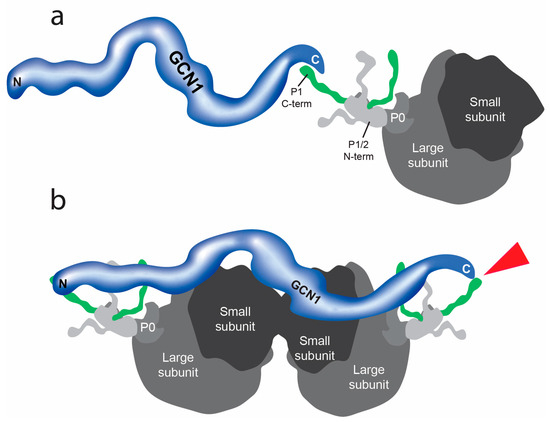
Figure 5.
Schematic of the interaction of GCN1 with ribosomal protein P1. (a) The C-terminal of GCN1 interacts with ribosomal protein P1. Proteins P2 and P0 are part of the ‘stalk’ region of the larger subunit of eukaryotic ribosomes. (b) Hypothetical model of GCN1/P1 interactions with a colliding ribosome (left) and leading ribosome (right). The red arrowhead points to the protein/protein interaction between GCN1 and P1.
In eukaryotes, the P-stalk is made up of three acidic ribosomal proteins, P0, P1, and P2, which together form a pentameric P-complex with a ratio of 1:2:2 [64,65]. During translation, P1 and P2 are responsible for binding translation factors [66,67,68]. P1 and P2 proteins are present in the cytoplasm in both ribosome-bound and free forms. The exchange between these two pools regulates the activity of ribosomes [69].
Interestingly, it has been shown that in vitro, the presence of isolated P-stalks is sufficient to trigger GCN2-mediated phosphorylation of the eukaryotic initiation factor—2α (eIF2α) even in the absence of the rest of the ribosome [39]. We have shown in an earlier study that eIF2α is phosphorylated after a blood meal in the mosquito fat body [35].
Under amino acid starvation conditions or in the presence of other cellular stressors, ribosomes collide during mRNA translation [70,71,72]. A recently published cryo-electron microscopy study revealed the structure of GCN1 bound to two colliding ribosomes [73]. In this model, the N-terminus of GCN1 interacts with the P-stalk of the colliding ribosome while the C-terminus is closely located near the P-stalk of the leading ribosome. Considering the strong interaction between the C-terminus of GCN1 and P1 we found in the Y2H screen, we hypothesize that the N-terminal and C-terminal regions of GCN1 bind to the P-stalks of both the leading and the colliding ribosomes during amino acid starvation (see Figure 5b).
In an interesting finding, it has been shown that P1/P2 coupled to ribosomes is necessary for flaviviruses like dengue viruses (DENV) and yellow fever virus (YFV) to replicate [74]. The substantial reduction in early DENV protein accumulation caused by a P1/P2 knockdown suggests that these P-stalk proteins could be necessary for viral protein translation.
Our study showed that P1 was expressed in all mosquito organs tested (see Figure 3a). This suggests that P1 is a ‘housekeeping gene’ with a constant level of expression [75]. Therefore, P1 has the potential to be used as a control gene in quantitative PCR for unfed adult A. aegypti.
Our Western blot results concerning the protein expression of P1 in different mosquito tissues showed bands at the predicted sizes of mosquito P1 (15 kDa) and the mosquito P1/P2 complex (38 kDa). The Western blot shown in Figure 3b also includes several bands at higher molecular weights in unique patterns in different tissues. We speculate that these bands represent either larger protein complexes that involve P1 or cross-reaction of the antibody with other mosquito proteins. To further investigate our findings, we plan to raise alternative antibodies against specific mosquito epitopes of P1.
The phenotypes we observed after an RNAi-mediated knockdown of P1 were severe. Knockdown females laid no eggs after ingesting a blood meal, and they had significantly lower survival rates. This decrease in survival time in P1-knockdown mosquitoes may be due to the disruption in protein synthesis after injection. This likely temporarily impacts wound healing from the injection and diminishes their immunological defenses against infection. Development of ovaries and eggs of P1-dsRNA-injected mosquitoes was arrested, and those eggs did not mature (see Figure 4a,c). Our previous study on the CAT1 and GCN1 interaction showed that GCN1 knockdown affected mosquito fecundity but did not affect vitellogenin expression [35]. Earlier knockdown studies of proteins associated with the mTOR signaling pathway and A. aegypti CATs often resulted in a reduction in egg numbers but never total sterility [57,59,60,76]. P1 knockdown also caused a significant reduction in the survival of female mosquitoes; in contrast, knockdown of GCN1 did not cause any mortality.
5. Conclusions
We identified P1 as an interactor of GCN1 in the mosquito fat body and found that it was necessary for mosquito egg development. A. aegypti females, when injected with P1 dsRNA, lost their capacity to produce eggs after a blood meal. Our results show that P1 is a housekeeping gene that is necessary for mosquito reproduction.
6. Future Directions
Our results point towards P1 and other ribosomal stalk proteins as potential targets for the development of dsRNA-based mosquito birth control. The next steps for this novel research include investigating the duration of the ovary phenotype after P1 knockdown, studying the effects of knockdown of the other P-stalk proteins, testing for larvicidal activity of dsRNA against P1, and expanding our studies to other pest arthropods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects15020084/s1, Figure S1: P1 expression in different mosquito organs and tissues tested (Figure 3b); Figure S2: Image for Actin control (Figure 3b); Figure S3: P1 expression western blot for different mosquito organs tested (Thorax, Fat Body, Mid Gut, Malpighian Tubules, Ovary, Head); Figure S4: Survival Curve Replicates (R1–R3), (Supplemental figure for Figure 3); Figure S5. Egg-laying paper with eggs and without eggs; Table S1. List of proteins identified after Yeast-Two Hybrid screen; Table S2: Mosquito survival data; Table S3. Measurements from mosquito ovaries post-injection and PBM.
Author Contributions
Conceptualization, M.P. and I.A.H.; methodology, M.L.; formal analysis, M.L. and H.A.L.; writing—original draft preparation, M.L. and H.A.L.; writing—review and editing, M.L., H.A.L. and I.A.H.; project administration, M.L. and M.P.; funding acquisition, I.A.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of General Medical Sciences, grant numbers R35GM144049 and T32GM148394.
Data Availability Statement
The data presented in this study are available in the main text of the article.
Acknowledgments
The authors would like to acknowledge Yashoda Kandel for training authors on mosquito dissections and injections; the NIH/NIAID Filariasis Research Reagent Resource Center for distribution by BEI Resources, NIAID, NIH: Aedes aegypti, Strain Black Eye Liverpool, Eggs NR-48921; and all colleagues from the Molecular Vector Physiology Laboratory at New Mexico State University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Carpenter, V.K.; Drake, L.L.; Aguirre, S.E.; Price, D.P.; Rodriguez, S.D.; Hansen, I.A. SLC7 amino acid transporters of the yellow fever mosquito Aedes aegypti and their role in fat body TOR signaling and reproduction. J. Insect Physiol. 2012, 58, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.J.; Vosshall, L.B. How to turn an organism into a model organism in 10 ‘easy’ steps. J. Exp. Biol. 2020, 223, jeb218198. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Chen, K.; Brown, M.R.; Strand, M.R. Multiple endocrine factors regulate nutrient mobilization and storage in Aedes aegypti during a gonadotrophic cycle. Insect Sci. 2023, 30, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Pinch, M.; Mitra, S.; Rodriguez, S.D.; Li, Y.; Kandel, Y.; Dungan, B.; Holguin, F.O.; Attardo, G.M.; Hansen, I.A. Fat and happy: Profiling mosquito fat body lipid storage and composition post-blood meal. Front. Insect Sci. 2021, 1, 693168. [Google Scholar] [CrossRef]
- Clemons, A.; Haugen, M.; Flannery, E.; Tomchaney, M.; Kast, K.; Jacowski, C.; Le, C.; Mori, A.; Holland, W.S.; Sarro, J. Aedes aegypti: An emerging model for vector mosquito development. Cold Spring Harb. Protoc. 2010, 2010, pdb.emo141. [Google Scholar] [CrossRef] [PubMed]
- Gulia-Nuss, M.; Elliot, A.; Brown, M.R.; Strand, M.R. Multiple factors contribute to anautogenous reproduction by the mosquito Aedes aegypti. J. Insect Physiol. 2015, 82, 8–16. [Google Scholar] [CrossRef]
- Attardo, G.M.; Higgs, S.; Klingler, K.A.; Vanlandingham, D.L.; Raikhel, A.S. RNA interference-mediated knockdown of a GATA factor reveals a link to anautogeny in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2003, 100, 13374–13379. [Google Scholar] [CrossRef]
- Ariani, C.V.; Smith, S.C.; Osei-Poku, J.; Short, K.; Juneja, P.; Jiggins, F.M. Environmental and genetic factors determine whether the mosquito Aedes aegypti lays eggs without a blood meal. Am. J. Trop. Med. Hyg. 2015, 92, 715. [Google Scholar] [CrossRef]
- Valle, D. Vitellogenesis in insects and other groups: A review. Mem. Inst. Oswaldo Cruz 1993, 88, 1–26. [Google Scholar] [CrossRef]
- Roth, T.F.; Porter, K.R. Yolk protein uptake in the oocyte of the mosquito Aedes aegypti. L. J. Cell Biol. 1964, 20, 313–332. [Google Scholar] [CrossRef]
- Snigirevskaya, E.S.; Hays, A.R.; Raikhel, A.S. Secretory and internalization pathways of mosquito yolk protein precursors. Cell Tissue Res. 1997, 290, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Sappington, T.W.; Kokoza, V.A.; Cho, W.-L.; Raikhel, A.S. Molecular characterization of the mosquito vitellogenin receptor reveals unexpected high homology to the Drosophila yolk protein receptor. Proc. Natl. Acad. Sci. USA 1996, 93, 8934–8939. [Google Scholar] [CrossRef] [PubMed]
- Bownes, M.; Shirras, A.; Blair, M.; Collins, J.; Coulson, A. Evidence that insect embryogenesis is regulated by ecdysteroids released from yolk proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 1554–1557. [Google Scholar] [CrossRef] [PubMed]
- Townson, H. The biology of mosquitoes. Volume 1. Development, nutrition and reproduction. By A.N. Clements. (London: Chapman & Hall, 1992). viii + 509 pp. Hard cover £50. ISBN 0-412-40180-0. Bull. Entomol. Res. 1993, 83, 307–308. [Google Scholar] [CrossRef]
- Roy, S.G.; Raikhel, A.S. The small GTPase Rheb is a key component linking amino acid signaling and TOR in the nutritional pathway that controls mosquito egg development. Insect Biochem. Mol. Biol. 2011, 41, 62–69. [Google Scholar] [CrossRef]
- Avruch, J.; Long, X.; Ortiz-Vega, S.; Rapley, J.; Papageorgiou, A.; Dai, N. Amino acid regulation of TOR complex 1. Am. J. Physiol.-Endocrinol. Metab. 2009, 296, E592–E602. [Google Scholar] [CrossRef]
- Smykal, V.; Raikhel, A.S. Nutritional control of insect reproduction. Curr. Opin. Insect Sci. 2015, 11, 31–38. [Google Scholar] [CrossRef]
- Ling, L.; Raikhel, A.S. Cross-talk of insulin-like peptides, juvenile hormone, and 20-hydroxyecdysone in regulation of metabolism in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2021, 118, e2023470118. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Rodriguez, S.D.; Drake, L.L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 2014, 5, 103. [Google Scholar] [CrossRef]
- Hundal, H.S.; Taylor, P.M. Amino acid transceptors: Gate keepers of nutrient exchange and regulators of nutrient signaling. Am. J. Physiol.-Endocrinol. Metab. 2009, 296, E603–E613. [Google Scholar] [CrossRef]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D.; Kanai, Y.; Palacin, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. CATs and HATs: The SLC7 family of amino acid transporters. Pflug. Arch. 2004, 447, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Attardo, G.M.; Hansen, I.A.; Shiao, S.-H.; Raikhel, A.S. Identification of two cationic amino acid transporters required for nutritional signaling during mosquito reproduction. J. Exp. Biol. 2006, 209, 3071–3078. [Google Scholar] [CrossRef]
- Boudko, D.Y.; Tsujimoto, H.; Rodriguez, S.D.; Meleshkevitch, E.A.; Price, D.P.; Drake, L.L.; Hansen, I.A. Substrate specificity and transport mechanism of amino-acid transceptor Slimfast from Aedes aegypti. Nat. Commun. 2015, 6, 8546. [Google Scholar] [CrossRef] [PubMed]
- Hansen, I.A.; Boudko, D.Y.; Shiao, S.-H.; Voronov, D.A.; Meleshkevitch, E.A.; Drake, L.L.; Aguirre, S.E.; Fox, J.M.; Attardo, G.M.; Raikhel, A.S. AaCAT1 of the yellow fever mosquito, Aedes aegypti: A novel histidine-specific amino acid transporter from the SLC7 family. J. Biol. Chem. 2011, 286, 10803–10813. [Google Scholar] [CrossRef] [PubMed]
- Staschke, K.A.; Dey, S.; Zaborske, J.M.; Palam, L.R.; McClintick, J.N.; Pan, T.; Edenberg, H.J.; Wek, R.C. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 2010, 285, 16893–16911. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, P.T.; Hay, N. The two TORCs and AKT. Dev. Cell 2007, 12, 487–502. [Google Scholar] [CrossRef]
- Battu, S.; Minhas, G.; Mishra, A.; Khan, N. Amino acid sensing via general control nonderepressible-2 kinase and immunological programming. Front. Immunol. 2017, 8, 1719. [Google Scholar] [CrossRef]
- Kubota, H.; Ota, K.; Sakaki, Y.; Ito, T. Budding yeast GCN1 binds the GI domain to activate the eIF2α kinase GCN2. J. Biol. Chem. 2001, 276, 17591–17596. [Google Scholar] [CrossRef]
- Gottfried, S.; Koloamatangi, S.M.; Daube, C.; Schiemann, A.H.; Sattlegger, E. A genetic approach to identify amino acids in Gcn1 required for Gcn2 activation. PLoS ONE 2022, 17, e0277648. [Google Scholar] [CrossRef] [PubMed]
- Sattlegger, E.; Hinnebusch, A.G. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2α kinase GCN2 during amino acid starvation. J. Biol. Chem. 2005, 280, 16514–16521. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zou, Y.; Mao, D.; Sun, D.; Gao, G.; Shi, J.; Liu, X.; Zhu, C.; Yang, M.; Ye, W. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 2014, 206, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Raina, M.; Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 2014, 5, 171. [Google Scholar] [CrossRef] [PubMed]
- Pinch, M.; Muka, T.; Kandel, Y.; Lamsal, M.; Martinez, N.; Teixeira, M.; Boudko, D.Y.; Hansen, I.A. General control nonderepressible 1 interacts with cationic amino acid transporter 1 and affects Aedes aegypti fecundity. Parasites Vectors 2022, 15, 383. [Google Scholar] [CrossRef] [PubMed]
- Castilho, B.A.; Shanmugam, R.; Silva, R.C.; Ramesh, R.; Himme, B.M.; Sattlegger, E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 1948–1968. [Google Scholar] [CrossRef]
- Marton, M.; Crouch, D.; Hinnebusch, A. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 1993, 13, 3541–3556. [Google Scholar]
- Marton, M.J.; Vazquez de Aldana, C.R.; Qiu, H.; Chakraburtty, K.; Hinnebusch, A.G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol. Cell. Biol. 1997, 17, 4474–4489. [Google Scholar] [CrossRef]
- Inglis, A.J.; Masson, G.R.; Shao, S.; Perisic, O.; McLaughlin, S.H.; Hegde, R.S.; Williams, R.L. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. USA 2019, 116, 4946–4954. [Google Scholar] [CrossRef]
- Lee, S.J.; Swanson, M.J.; Sattlegger, E. Gcn1 contacts the small ribosomal protein Rps10, which is required for full activation of the protein kinase Gcn2. Biochem. J. 2015, 466, 547–559. [Google Scholar] [CrossRef]
- Sattlegger, E.; Hinnebusch, A.G. Separate domains in GCN1 for binding protein kinase GCN2 and ribosomes are required for GCN2 activation in amino acid-starved cells. EMBO J. 2000, 19, 6622–6633. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.H.; Hirano, T. HEAT repeats–versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 2016, 129, 3963–3970. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, D.; Marintchev, A.; Arthanari, H. The metaphorical swiss army knife: The multitude and diverse roles of HEAT domains in eukaryotic translation initiation. Nucleic Acids Res. 2022, 50, 5424–5442. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, R.; Krishnan, R.; Sattlegger, E.; Srinivasan, N. Recognition of a structural domain (RWDBD) in Gcn1 proteins that interacts with the RWD domain containing proteins. Biol. Direct 2017, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Hayes, E. Determination of a physiological saline solution for Aedes aegypti (L.). J. Econ. Entomol. 1953, 46, 624–627. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Drake, L.L.; Price, D.P.; Aguirre, S.E.; Hansen, I.A. RNAi-mediated gene knockdown and in vivo diuresis assay in adult female Aedes aegypti mosquitoes. J. Vis Exp. 2012, e3479. [Google Scholar] [CrossRef]
- Dzaki, N.; Ramli, K.N.; Azlan, A.; Ishak, I.H.; Azzam, G. Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci. Rep. 2017, 7, 43618. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Calzolari, M. Mosquito-borne diseases in Europe: An emerging public health threat. Rep. Parasitol. 2016, 5, 1–12. [Google Scholar] [CrossRef]
- Patterson, J.; Sammon, M.; Garg, M. Dengue, Zika and chikungunya: Emerging arboviruses in the New World. West. J. Emerg. Med. 2016, 17, 671. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, D. Outbreak of locally acquired mosquito-transmitted (autochthonous) malaria—Florida and Texas, May–July 2023. MMWR. Morb. Mortal. Wkly. Rep. 2023, 72, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Anoopkumar, A.; Aneesh, E.M. A critical assessment of mosquito control and the influence of climate change on mosquito-borne disease epidemics. Environ. Dev. Sustain. 2022, 24, 8900–8929. [Google Scholar] [CrossRef]
- Jones, R.T.; Ant, T.H.; Cameron, M.M.; Logan, J.G. Novel control strategies for mosquito-borne diseases. Philos. Trans. R. Soc. B 2021, 376, 20190802. [Google Scholar] [CrossRef] [PubMed]
- Benelli, G.; Jeffries, C.L.; Walker, T. Biological control of mosquito vectors: Past, present, and future. Insects 2016, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.; Cardoso, J.C.; Felix, R.C.; Santana, R.A.; Guerra, M.d.G.B.; Power, D.; Silveira, H. Fresh-blood-free diet for rearing malaria mosquito vectors. Sci. Rep. 2018, 8, 17807. [Google Scholar] [CrossRef]
- Clements, A.N. The Biology of Mosquitoes; Chapman & Hall: London, UK, 1992; Volume 1. [Google Scholar]
- Perdomo, H.D.; Hussain, M.; Parry, R.; Etebari, K.; Hedges, L.M.; Zhang, G.; Schulz, B.L.; Asgari, S. Human blood microRNA hsa-miR-21-5p induces vitellogenin in the mosquito Aedes aegypti. Commun. Biol. 2021, 4, 856. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Park, J.H.; Peng, Q.; Raikhel, A.S. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc. Natl. Acad. Sci. USA 2004, 101, 10626–10631. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Roy, S.G.; Raikhel, A.S. Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. J. Biol. Chem. 2005, 280, 20565–20572. [Google Scholar] [CrossRef]
- Remacha, M.; Jimenez-Diaz, A.; Santos, C.; Briones, E.; Zambrano, R.; Gabriel, M.R.; Guarinos, E.; Ballesta, J.P. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 1995, 73, 959–968. [Google Scholar] [CrossRef]
- Tchórzewski, M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002, 34, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Hinnebusch, A.G. Differential requirements for P stalk components in activating yeast protein kinase Gcn2 by stalled ribosomes during stress. Proc. Natl. Acad. Sci. USA 2023, 120, e2300521120. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-M.; Yu, C.W.-H.; Chan, D.S.-B.; Chiu, T.Y.-H.; Zhu, G.; Sze, K.-H.; Shaw, P.-C.; Wong, K.-B. Solution structure of the dimerization domain of ribosomal protein P2 provides insights for the structural organization of eukaryotic stalk. Nucleic Acids Res. 2010, 38, 5206–5216. [Google Scholar] [CrossRef]
- Kulczyk, A.W.; Sorzano, C.O.S.; Grela, P.; Tchórzewski, M.; Tumer, N.E.; Li, X.-P. Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction. J. Biol. Chem. 2023, 299, 102795. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Wong, B.; Xie, X.; Lu, Y.-F.; Shi, P.-Y.; Pompon, J.; Garcia-Blanco, M.A.; Bradrick, S.S. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J. Virol. 2017, 91, e01706-16. [Google Scholar] [CrossRef]
- Tchórzewski, M.; Boldyreff, B.; Issinger, O.-G.; Grankowski, N. Analysis of the protein–protein interactions between the human acidic ribosomal P-proteins: Evaluation by the two hybrid system. Int. J. Biochem. Cell Biol. 2000, 32, 737–746. [Google Scholar] [CrossRef]
- Wahl, M.C.; Moller, W. Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Pept. Sci. 2002, 3, 93–106. [Google Scholar] [CrossRef]
- Ballesta, J.P.; Remacha, M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acid Res. Mol. Biol. 1996, 55, 157–193. [Google Scholar]
- Wu, C.C.-C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 2020, 182, 404–416.e414. [Google Scholar] [CrossRef]
- Ryder, L.; Arendrup, F.S.; Martínez, J.F.; Snieckute, G.; Pecorari, C.; Shah, R.A.; Lund, A.H.; Blasius, M.; Bekker-Jensen, S. Nitric oxide-induced ribosome collision activates ribosomal surveillance mechanisms. Cell Death Dis. 2023, 14, 467. [Google Scholar] [CrossRef]
- Vind, A.C.; Snieckute, G.; Bekker-Jensen, S.; Blasius, M. Run, Ribosome, Run: From compromised translation to human health. Antioxidants Redox Signal. 2023, 39, 336–350. [Google Scholar] [CrossRef] [PubMed]
- Pochopien, A.A.; Beckert, B.; Kasvandik, S.; Berninghausen, O.; Beckmann, R.; Tenson, T.; Wilson, D.N. Structure of Gcn1 bound to stalled and colliding 80S ribosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2022756118. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Wijeratne, H.S.; Shah, P.; Garcia-Blanco, M.A.; Bradrick, S.S. Ribosomal stalk proteins RPLP1 and RPLP2 promote biogenesis of flaviviral and cellular multi-pass transmembrane proteins. Nucleic Acids Res. 2020, 48, 9872–9885. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef]
- Martinez, N.P.; Pinch, M.; Kandel, Y.; Hansen, I.A. Knockdown of the Sodium/Potassium ATPase Subunit Beta 2 Reduces Egg Production in the Dengue Vector. Aedes aegypti. Insects 2023, 14, 50. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).