The Genome of Arsenophonus sp. and Its Potential Contribution in the Corn Planthopper, Peregrinus maidis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Genome Sequencing, Assembly and Annotation
2.3. RNA Polymerase β Subunit Gene (rpoB) Identification and Phylogenetic Analysis
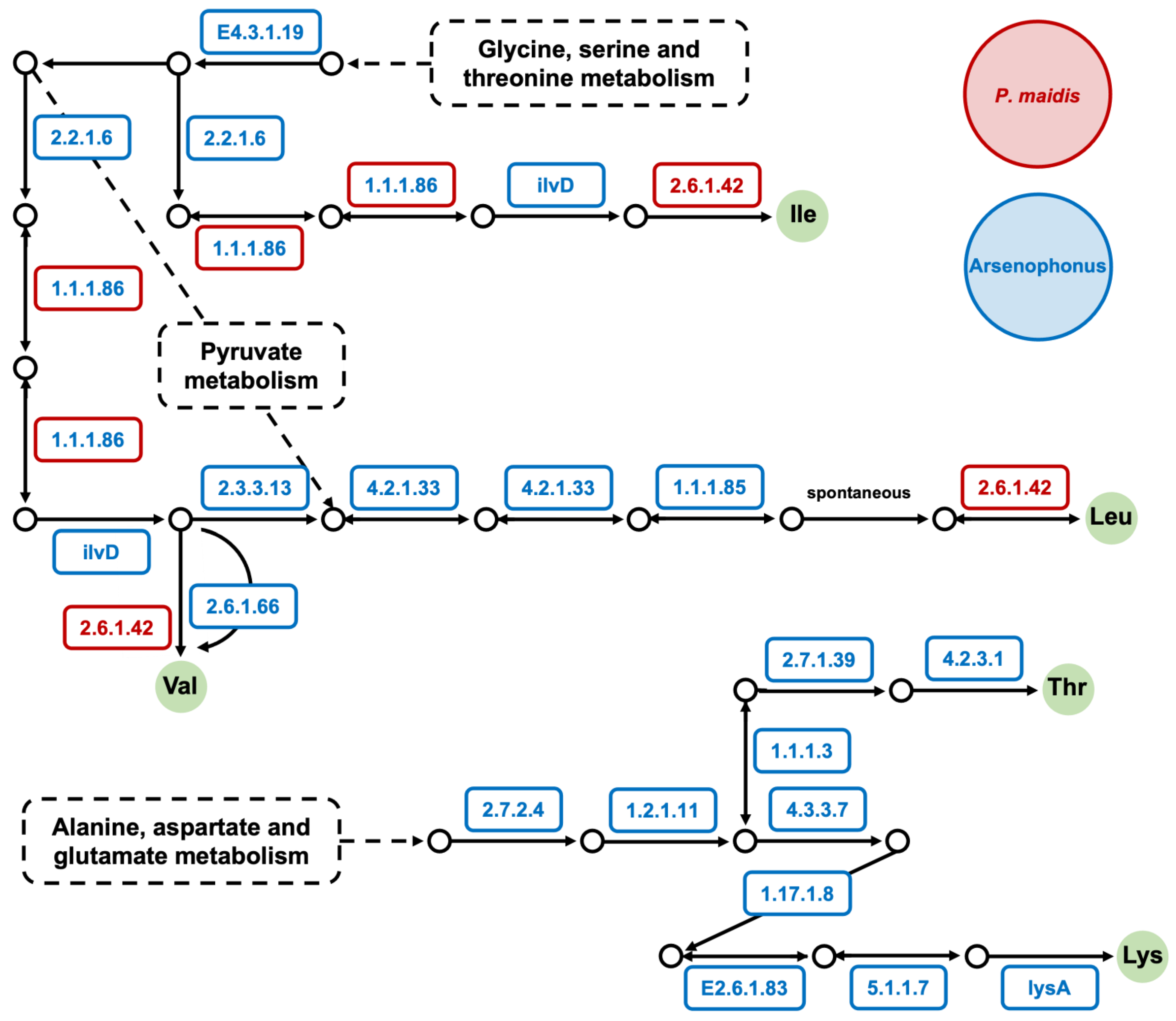
3. Results and Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, B.U.; Seetharama, N. Host plant interactions of the corn planthopper, Peregrinus maidis Ashm. (Homoptera: Delphacidae) in maize and sorghum agroecosystems. Arthropod Plant Interact. 2008, 2, 163–196. [Google Scholar] [CrossRef]
- Autrey, L. Maize mosaic virus and other maize virus diseases in the islands of the western Indian Ocean. In Proceedings of the International Maize Virus Diseases Colloquium and Workshop, Wooster, OH, USA, 2–6 August 1982. [Google Scholar]
- Gundersen, D.E.; Adrianos, S.L.; Allen, M.L.; Becnel, J.J.; Chen, Y.; Choi, M.Y.; Estep, A.; Evans, J.D.; Garczynski, S.F.; Geib, S.M.; et al. Arthropod genomics research in the United States Department of Agriculture-Agricultural Research Service: Applications of RNA interference and CRISPR gene editing technologies in pest control. Trends Entomol. 2017, 13, 109–137. [Google Scholar]
- Yao, J.; Rotenberg, D.; Afsharifar, A.; Barandoc-Alviar, K.; Whitfield, A.E. Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS ONE 2013, 8, e70243. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Klobasa, W.; Chu, F.-C.; Huot, O.; Whitfield, A.E.; Lorenzen, M. Structural and functional insights into the ATP-binding cassette transporter family in the corn planthopper, Peregrinus maidis. Insect Mol. Biol. 2023, 32, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.A.D.; Tyson, C.; Kerner, L.M.; Whitfield, A.E. Exportin 1 is required for the reproduction and maize mosaic virus accumulation in its insect vector Peregrinus maidis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Whitten, M.M.; Facey, P.D.; Del Sol, R.; Fernández-Martínez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-mediated RNA interference in insects. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160042. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef]
- Elston, K.M.; Maeda, G.P.; Perreau, J.; Barrick, J.E. Addressing the challenges of symbiont-mediated RNAi in aphids. PeerJ 2023, 11, e14961. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, X.; Zhang, C.X.; Yu, L.L.; Fan, H.W.; Wang, Z.; Xu, H.-J.; Xi, Y.; Zhu, Z.-R.; Zhou, W.-W.; et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014, 15, 521. [Google Scholar] [CrossRef]
- Dale, C.; Beeton, M.; Harbison, C.; Jones, T.; Pontes, M. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus”, an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 2006, 72, 2997–3004. [Google Scholar] [CrossRef]
- Nadal-Jimenez, P.; Griffin, J.S.; Davies, L.; Frost, C.L.; Marcello, M.; Hurst, G.D. Genetic manipulation allows in vivo tracking of the life cycle of the son-killer symbiont, Arsenophonus nasoniae, and reveals patterns of host invasion, tropism and pathology. Environ. Microbiol. 2019, 21, 3172–3182. [Google Scholar] [CrossRef]
- Gherna, R.L.; Werren, J.H.; Weisburg, W.; Côté, R.; Woese, C.R.; Mandelco, L.; Brenner, D.J. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int. J. Syst. Evol. Microbiol. 1991, 41, 563–565. [Google Scholar] [CrossRef]
- Nováková, E.; Hypša, V.; Moran, N.A. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Nováková, E.; Hypša, V.; Nguyen, P.; Husník, F.; Darby, A.C. Genome sequence of Candidatus Arsenophonus lipopteni, the exclusive symbiont of a blood sucking fly Lipoptena cervi (Diptera: Hippoboscidae). Stand. Genom. Sci. 2016, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Santos-Garcia, D.; Juravel, K.; Freilich, S.; Zchori-Fein, E.; Latorre, A.; Moya, A.; Morin, S.; Silva, F.J. To B or not to B: Comparative genomics suggests Arsenophonus as a source of B vitamins in whiteflies. Front. Microbiol. 2018, 9, 2254. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, T.E.; Duron, O.; Darby, A.C.; Hypša, V.; Nováková, E.; Hurst, G.D. The Genus Arsenophonus. In Manipulative Tenants: Bacteria Associated with Arthropods; Zchori-Fein, E., Bourtzis, K., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 225–244. [Google Scholar]
- Drew, G.C.; Budge, G.E.; Frost, C.L.; Neumann, P.; Siozios, S.; Yañez, O.; Hurst, G.D. Transitions in symbiosis: Evidence for environmental acquisition and social transmission within a clade of heritable symbionts. ISME J. 2021, 15, 2956–2968. [Google Scholar] [CrossRef] [PubMed]
- Bressan, A. Emergence and evolution of Arsenophonus bacteria as insect-vectored plant pathogens. Infect. Genet. Evol. 2014, 22, 81–90. [Google Scholar] [CrossRef]
- Barnett, D.W.; Garrison, E.K.; Quinlan, A.R.; Strömberg, M.P.; Marth, G.T. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 2011, 27, 1691–1692. [Google Scholar] [CrossRef]
- Miller, J.R.; Delcher, A.L.; Koren, S.; Venter, E.; Walenz, B.P.; Brownley, A.; Johnson, J.; Li, K.; Mobarry, C.; Sutton, G. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 2008, 24, 2818–2824. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Alonge, M.; Lebeigle, L.; Kirsche, M.; Jenike, K.; Ou, S.; Aganezov, S.; Wang, X.; Lippman, Z.B.; Schatz, M.C.; Soyk, S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022, 23, 258. [Google Scholar] [CrossRef] [PubMed]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. Available online: http://www.genome.org/cgi/doi/10.1101/gr.4086505 (accessed on 22 June 2022). [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. In Gene Prediction; Kollmar, M., Ed.; Humana: New York, NY, USA, 2019; pp. 227–245. Available online: https://link.springer.com/protocol/10.1007/978-1-4939-9173-0_14 (accessed on 22 June 2022).
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lerat, E.; Ochman, H. Ψ-Φ: Exploring the outer limits of bacterial pseudogenes. Genome Res. 2004, 14, 2273–2278. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef]
- Lo, W.S.; Huang, Y.Y.; Kuo, C.H. Winding paths to simplicity: Genome evolution in facultative insect symbionts. FEMS Microbiol. Rev. 2016, 40, 855–874. [Google Scholar] [CrossRef]
- Mira, A.; Ochman, H.; Moran, N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001, 17, 589–596. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, 633–642. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Case, R.J.; Boucher, Y.; Dahllöf, I.; Holmström, C.; Doolittle, W.F.; Kjelleberg, S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 2007, 73, 278–288. [Google Scholar] [CrossRef]
- Mollet, C.; Drancourt, M.; Raoult, D. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 1997, 26, 1005–1011. [Google Scholar] [CrossRef]
- Nadal-Jimenez, P.; Siozios, S.; Frost, C.L.; Court, R.; Chrostek, E.; Drew, G.C.; Evans, J.D.; Hawthorne, D.J.; Burritt, J.B.; Hurst, G.D. Arsenophonus apicola sp. nov. isolated from the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2022, 72, 005469. [Google Scholar] [CrossRef]
- Dittmer, J.; Van Opstal, E.J.; Shropshire, J.D.; Bordenstein, S.R.; Hurst, G.D.; Brucker, R.M. Disentangling a holobiont–recent advances and perspectives in Nasonia wasps. Front. Microbiol. 2016, 7, 1478. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.Y.; Lou, Y.H.; Fan, H.W.; Ye, Y.X.; Huang, H.J.; Hu, M.Q.; Zhu, Y.-N.; Zhang, C.X. Two endosymbiotic bacteria, Wolbachia and Arsenophonus, in the brown planthopper Nilaparvata lugens. Symbiosis 2013, 61, 47–53. [Google Scholar] [CrossRef]



| Name | Arsenophonus nasoniae | Arsenophonus sp. | Arsenophonus lipopteni | Arsenophonus triatominarum | |
|---|---|---|---|---|---|
| Accession No. | GCA_004768525.1 | GCA_020268605.1 | GCA_001534665.1 | GCA_001640365.1 | |
| Host | Nasonia vitripennis | Apis mellifera | Lipoptena fortisetosa | Triatoma infestans | |
| Sequencing | Oxford Nanopore MinION; PacBio RS II; Illumina MiSeq | Oxford Nanopore MinION; Illumina MiSeq | Illumina | PacBio | |
| Genome size (bp) | 4,987,107 | 3,639,254 | 836,724 | 3,858,720 | |
| GC content (%) | 38.1 1 | 37.7 1 | 24.9 1 | 38.3 | |
| Pseudogene 2 | 1086 | 720 | 104 | 523 | |
| Name | Arsenophonus sp. (ARAD) | Arsenophonus sp. (ARAF) | Arsenophonus sp. (ENCA) | Arsenophonus sp. | Arsenophonus sp. (this study) |
| Accession No. | GCA_900343015.1 | GCA_900343025.1 | GCA_002287155.1 | GCA_000757905.1 | |
| Host | Aleurodicus disperses | Aleurodicus floccissimus | Entylia carinata | Nilaparvata lugens | Peregrinus maidis |
| Sequencing | Illumina HiSeq 2000 | Illumina HiSeq 2000 | Illumina MiSeq | Illumina HiSeq 2000 | PacBio |
| Genome size (bp) | 663,125 | 3,001,875 | 3,228,533 | 2,953,863 | 4,888,380 |
| GC content (%) | 32.2 1 | 37 | 39.5 | 37.6 | 41.3 |
| Pseudogene 2 | 29 | 382 | 430 | 469 | 1123 |
| Name | Host | Role and Function 1 | Transmission | Reference |
|---|---|---|---|---|
| Arsenophonus phytopathogenicus | Pentastiridius leporinus | Plant pathogen of sugar beets vectored by P. leporinus | Maternal and horizontal (major) transmission by infecting the same sugar beet | [19] |
| Arsenophonus nasoniae | Nasonia vitripennis Nasonia longicornis | Reproductive parasite Killing sons, 80% of sons die | Maternal and horizontal transmission by infecting the same pupa host | [46] |
| Arsenophonus sp. | Apis mellifera | Providing B vitamins (B2, B6, B7, B9) | Horizontal transmission by social interactions (trophallaxis and/or general contact) and environmental acquisition | [18,45] |
| Arsenophonus lipopteni | Lipoptena cervi | Primary endosymbiont Providing B vitamins (B2, B6, B7) | Unclear | [15] |
| Arsenophonus triatominarum | triatomine bugs | Secondary endosymbiont No apparent effect on host fitness or reproduction | Vertical transmission (transovarially) | [17] |
| Arsenophonus sp. (ARAD) | Aleurodicus disperses | ARAD as primary endosymbiont ARAF and ARTV as secondary endosymbiont ARAD providing cofactors and B vitamins (B1, B2, B6, B7, B9) | Unclear | [16] |
| Arsenophonus sp. (ARAF) | Aleurodicus floccissimus | |||
| Arsenophonus sp. (ARTV) | Trialeurodes vaporariorum | |||
| Arsenophonus sp. | Nilaparvata lugens | Secondary endosymbiont Providing B vitamins | Maternal transmission | [10,47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-H.; Mikaelyan, A.; Coates, B.S.; Lorenzen, M. The Genome of Arsenophonus sp. and Its Potential Contribution in the Corn Planthopper, Peregrinus maidis. Insects 2024, 15, 113. https://doi.org/10.3390/insects15020113
Wang Y-H, Mikaelyan A, Coates BS, Lorenzen M. The Genome of Arsenophonus sp. and Its Potential Contribution in the Corn Planthopper, Peregrinus maidis. Insects. 2024; 15(2):113. https://doi.org/10.3390/insects15020113
Chicago/Turabian StyleWang, Yu-Hui, Aram Mikaelyan, Brad S. Coates, and Marcé Lorenzen. 2024. "The Genome of Arsenophonus sp. and Its Potential Contribution in the Corn Planthopper, Peregrinus maidis" Insects 15, no. 2: 113. https://doi.org/10.3390/insects15020113
APA StyleWang, Y.-H., Mikaelyan, A., Coates, B. S., & Lorenzen, M. (2024). The Genome of Arsenophonus sp. and Its Potential Contribution in the Corn Planthopper, Peregrinus maidis. Insects, 15(2), 113. https://doi.org/10.3390/insects15020113







