Effects of Gut Bacteria on the Fitness of Rice Leaf Folder Cnaphalocrocis medinalis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Gut Dissection and Bacteria Sampling
2.3. Bacteria Isolation and Identification
2.4. Effects of Each Bacterial Isolate on C. medinalis Fitness
2.5. Effects of Gut Bacterial Community on C. medinalis Fitness
2.6. Statistical Analyses
3. Results
3.1. Isolation and Identification of Cultured Bacteria from C. medinalis Gut
3.2. Effects of Single Bacterial Isolate on Development and Reproduction of C. medinalis
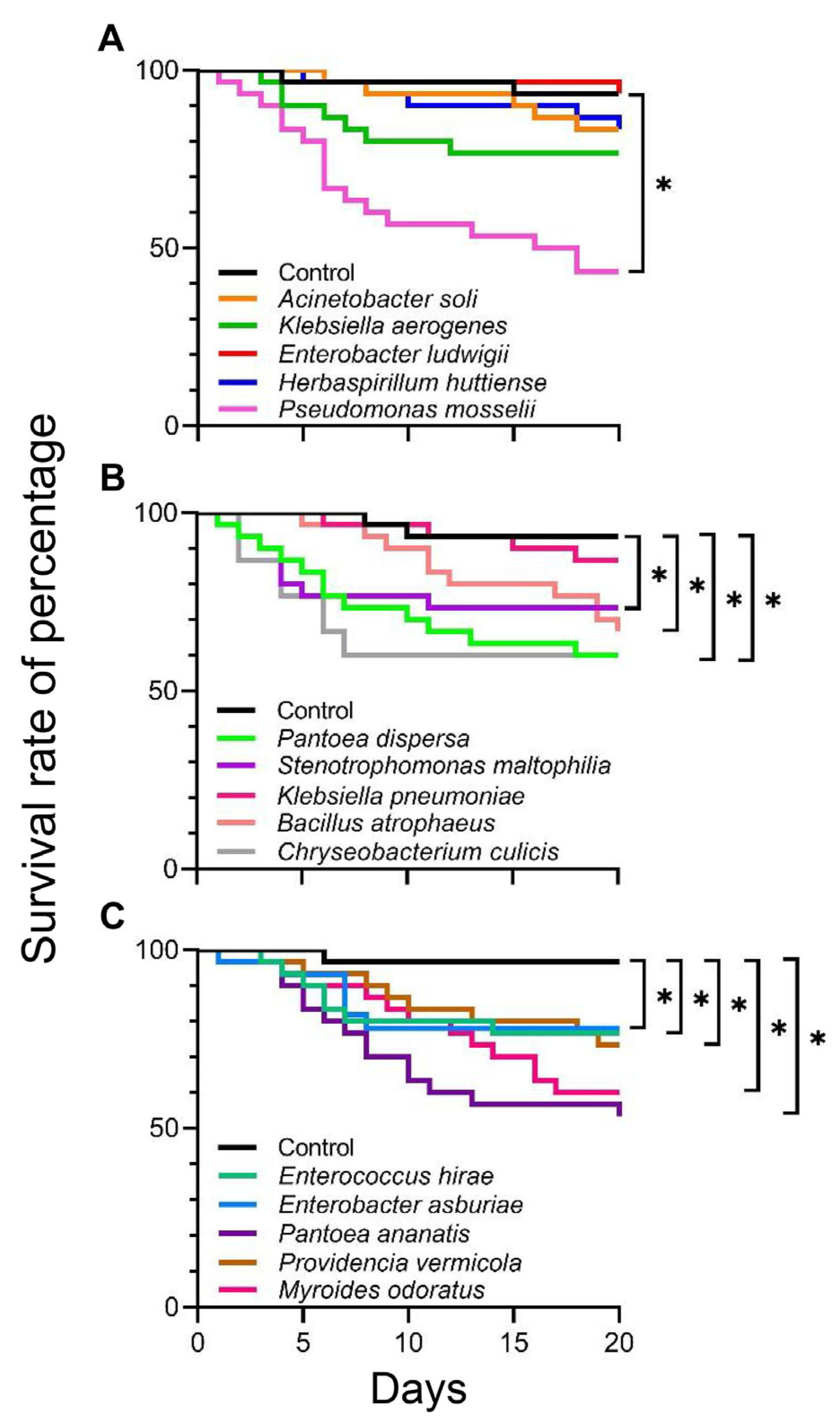
3.2.1. Survival of C. medinalis During Immature Stage
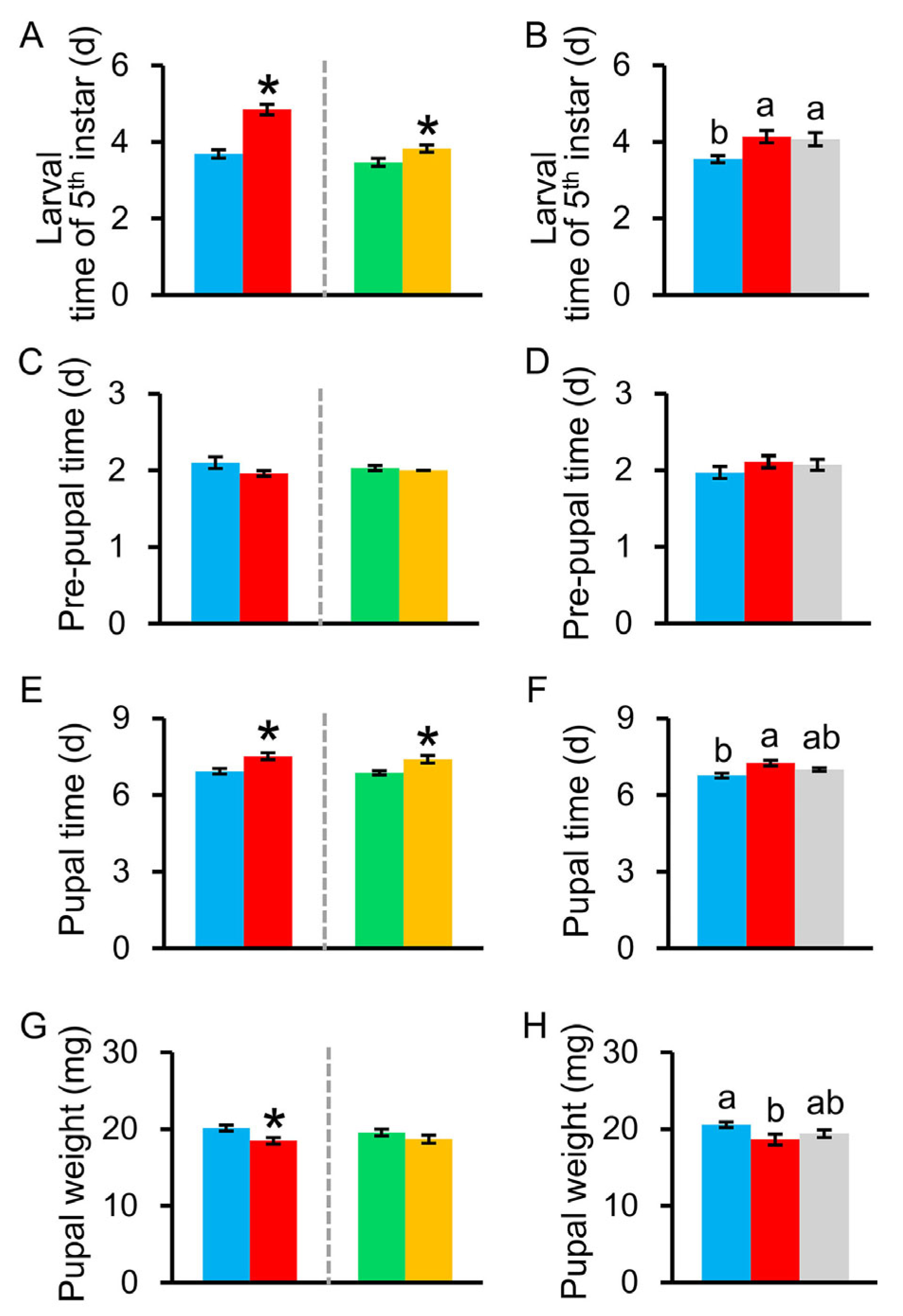
3.2.2. Development Time of C. medinalis Larvae
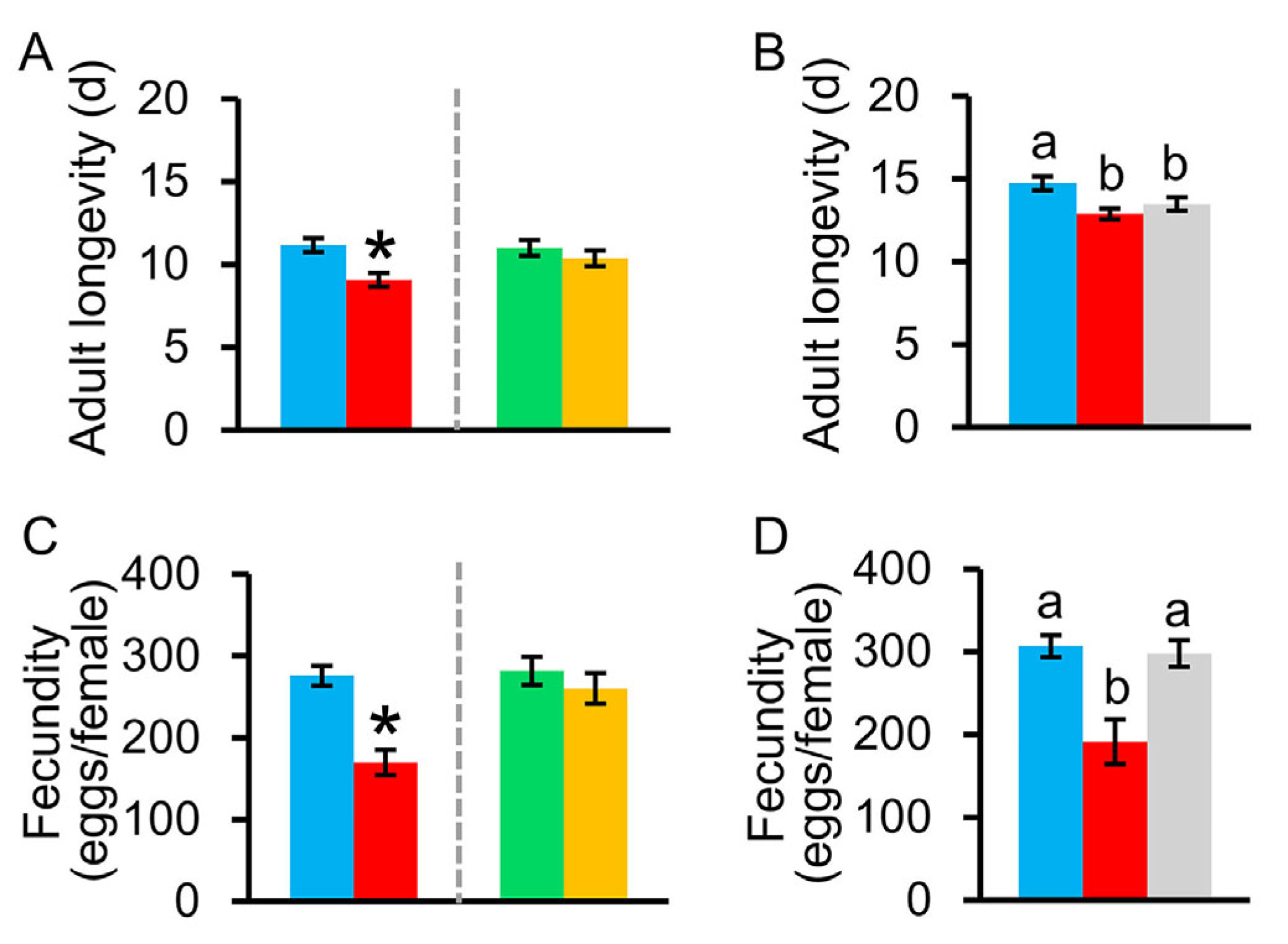
3.2.3. Pupal Time, Pupal Weight, Sex Ratio, Adult Longevity, and Fecundity of C. medinalis
3.3. Effects of Gut Bacterial Community on Development and Reproduction of C. medinalis
3.3.1. Immature Stage
3.3.2. Adult Stage
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engel, P.; Moran, N.A. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Mason, C.J.; Felton, G.W. Toward an integrated understanding of the lepidoptera microbiome. Annu. Rev. Phytopathol. 2024, 69, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Indiragandhi, P.; Anandham, R.; Madhaiyan, M.; Sa, T.M. Characterization of plant growth-promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr. Microbiol. 2008, 56, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Pilon, F.M.; da Rocha Silva, C.; Visôtto, L.E.; de Almeida Barros, R.; Rodrigues da Silva Júnior, N.; Campos, W.G.; de Almeida Oliveira, M.G. Purification and characterization of trypsin produced by gut bacteria from Anticarsia gemmatalis. Arch. Insect Biochem. Physiol. 2017, 96, e21407. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.F.; Gurr, G.M.; Vasseur, L.; Zheng, D.D.; Zhong, H.Z.; Qin, B.C.; Lin, J.H.; Wang, Y.; Song, F.Q.; Li, Y.; et al. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef]
- Bing, X.L.; Gerlach, J.; Loeb, G.; Buchon, N. Nutrient-dependent impact of microbes on Drosophila suzukii development. mBio 2018, 9, e02199-17. [Google Scholar] [CrossRef]
- Mason, C.J.; Lowe-Power, T.M.; Rubert-Nason, K.F.; Lindroth, R.L.; Raffa, K.F. Interactions between bacteria and aspen defense chemicals at the phyllosphere-herbivore interface. J. Chem. Ecol. 2016, 42, 193–201. [Google Scholar] [CrossRef]
- Mogren, C.L.; Shikano, I. Microbiota, pathogens, and parasites as mediators of tritrophic interactions between insect herbivores, plants, and pollinators. J. Invertebr. Pathol. 2021, 186, 107589. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Sun, Z.; Xu, C.C.; Wang, J.; Mallik, A.U.; Gu, C.Z.; Chen, D.Q.; Lu, L.; Zeng, R.S.; Song, Y.Y. High nitrogen in maize enriches gut microbiota conferring insecticide tolerance in lepidopteran pest Spodoptera litura. iScience 2022, 25, 103726. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.L.; Maltz, M.; Aksoy, S. Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 2012, 188, 3395–3403. [Google Scholar] [CrossRef] [PubMed]
- Ridley, E.V.; Wong, A.C.; Westmiller, S.; Douglas, A.E. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS ONE 2012, 7, e36765. [Google Scholar] [CrossRef] [PubMed]
- Pitlik, S.D.; Koren, O. How holobionts get sick-toward a unifying scheme of disease. Microbiome 2017, 5, 64. [Google Scholar] [CrossRef]
- Mason, C.J.; Ray, S.; Shikano, I.; Peiffer, M.; Jones, A.G.; Luthe, D.S.; Hoover, K.; Felton, G.W. Plant defenses interact with insect enteric bacteria by initiating a leaky gut syndrome. Proc. Natl. Acad. Sci. USA 2019, 116, 15991–15996. [Google Scholar] [CrossRef]
- Mason, K.L.; Stepien, T.A.; Blum, J.E.; Holt, J.F.; Labbe, N.H.; Rush, J.S.; Raffa, K.F.; Handelsman, J. From commensal to pathogen: Translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. mBio 2011, 2, 1–7. [Google Scholar] [CrossRef]
- Mason, C.J.; Peiffer, M.; St Clair, A.; Hoover, K.; Felton, G.W. Concerted impacts of antiherbivore defenses and opportunistic Serratia pathogens on the fall armyworm (Spodoptera frugiperda). Oecologia 2022, 198, 167–178. [Google Scholar] [CrossRef]
- Deutscher, A.T.; Burke, C.M.; Darling, A.E.; Riegler, M.; Reynolds, O.L.; Chapman, T.A. Near full-length 16S rRNA gene next-generation sequencing revealed Asaia as a common midgut bacterium of wild and domesticated Queensland fruit fly larvae. Microbiome 2018, 6, 85. [Google Scholar] [CrossRef]
- Chintalapati, P.; Balakrishnan, D.; Nammi, T.V.V.G.; Javvaji, S.; Muthusamy, S.K.; Venkata, S.R.L.; Neelamraju, S.; Katti, G. Phenotyping and genotype × environment interaction of resistance to leaffolder, Cnaphalocrocis medinalis Guenee (Lepidoptera: Pyralidae) in Rice. Front. Plant Sci. 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, H.; Xue, L.; Wang, J. Assessment of yield loss caused by rice leaf folder (Cnaphalocrocis medinalis) in China. J. Integr. Agr. 2017, 16, 1745–1754. [Google Scholar]
- Padmavathi, C.; Katti, G.; Padmakumari, A.P.; Voleti, S.R.; Rao, L.V.S. The effect of leaffolder Cnaphalocrocis medinalis (Guenee) [Lepidoptera: Pyralidae] injury on the plant physiology and yield loss in rice. J. Appl. Entomol. 2012, 137, 249–256. [Google Scholar] [CrossRef]
- Wang, F.Y.; Yang, F.; Lu, M.H.; Luo, S.Y.; Zhai, B.P.; Lim, K.S.; McInerney, C.E.; Hu, G. Determining the migration duration of rice leaf folder (Cnaphalocrocis medinalis (Guenée)) moths using a trajectory analytical approach. Sci. Rep. 2017, 7, 39853. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Liu, X.G.; Xu, H.X.; Liu, Y.H.; Ali, P.; Bodlah, M.A.; Lu, Z.X. The abundance and diversity of gut bacteria of rice leaffolder Cnaphalocrocis medinalis (Guenée) across life stages. J. Asia-Pac. Entomol. 2020, 23, 430–438. [Google Scholar] [CrossRef]
- Li, C.M.; Han, G.J.; Sun, J.; Huang, L.X.; Lu, Y.R.; Xia, Y.; Liu, Q.; Xu, J. The gut microbiota composition of Cnaphalocrocis medinalis and their predicted contribution to larval nutrition. Front. Microbiol. 2022, 13, 909863. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, B.; Li, Q.; Li, X.W.; Guo, Q.T.; Chen, W.L.; Jiao, H.Y.; Akutse, K.S.; Zhu, X.Z. Gut bacterial community of Cnaphalocrocis medinalis (Guenée) (Lepidoptera: Crambidae) is driven by rice varieties. J. Appl. Entomol. 2024, 148, 434–446. [Google Scholar] [CrossRef]
- Pan, Q.J.; Shikano, I.; Liu, T.X.; Felton, G.W. Helicoverpa zea-associated gut bacteria as drivers in shaping plant anti-herbivore defense in tomato. Microb. Ecol. 2023, 86, 2173–2182. [Google Scholar] [CrossRef]
- Chen, L.; Pan, Q.J.; Waqas, M.S.; Liu, T.X. Morphological traits for sex identification of the oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae). J. Integr. Agr. 2020, 19, 1458–1463. [Google Scholar] [CrossRef]
- Bi, S.Y.; Wang, X.D.; Tang, Y.H.; Lei, K.X.; Guo, J.Y.; Yang, N.W.; Wan, F.H.; Lü, Z.C.; Liu, W.X. Bacterial communities of the internal reproductive and digestive tracts of virgin and mated Tuta absoluta. Insects 2023, 14, 779. [Google Scholar] [CrossRef]
- Wang, J.; Peiffer, M.; Hoover, K.; Rosa, C.; Zeng, R.S.; Felton, G.W. Helicoverpa zea gut-associated bacteria indirectly induce defenses in tomato by triggering a salivary elicitor(s). New Phytol. 2017, 214, 1294–1306. [Google Scholar] [CrossRef]
- Pan, Q.J.; Shikano, I.; Hoover, K.; Liu, T.X.; Felton, G.W. Enterobacter ludwigii, isolated from the gut microbiota of Helicoverpa zea, promotes tomato plant growth and yield without compromising anti-herbivore defenses. Arthropod-Plant Interact. 2019, 13, 271–278. [Google Scholar] [CrossRef]
- Acevedo, F.E.; Peiffer, M.; Tan, C.W.; Stanley, B.A.; Stanley, A.; Wang, J.; Jones, A.G.; Hoover, K.; Rosa, C.; Luthe, D.; et al. Fall armyworm-associated gut bacteria modulate plant defense responses. Mol. Plant-Microbe Interact. 2017, 30, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, F.; Hussain, A.; Manzoor, M.; Younis, T.; Rasul, A.; Qazi, J.I. Potential of bacterial chitinolytic, Stenotrophomonas maltophilia, in biological control of termites. Egypt. J. Biol. Pest Control. 2018, 28, 86. [Google Scholar] [CrossRef]
- Soleimani, H.; Jamshidi, M.; Khakvar, R.; Hasheminia, S.M. Stenotrophomonas maltophilia a new biocontrol agent against the cowpea weevil Callosobruchus maculatus F. (Coleoptera: Bruchidae). Idesia 2022, 40, 17–23. [Google Scholar] [CrossRef]
- Al-qwabah, A.A.; Al-limoun, M.O.; Al-Mustafa, A.H.; Al-Zereini, W.A. Bacillus atrophaeus A7 crude chitinase: Characterization and potential role against Drosophila melanogaster larvae. Jordan J. Biol. Sci. 2018, 11, 451–459. [Google Scholar]
- Yi, Y.K.; Park, H.W.; Shrestha, S.; Seo, J.; Kim, Y.O.; Shin, C.S.; Kim, Y.G. Identification of two entomopathogenic bacteria from a nematode pathogenic to the Oriental beetle, Blitopertha orientalis. J. Microbiol. Biotechnol. 2007, 17, 968–978. [Google Scholar]
- Salas, B.; Conway, H.E.; Vacek, D.C.; Vitek, C.; Schuenzel, E.L. Pathogenicity of multiple Providencia species (Enterobacteriales: Morganellaceae) to the mass-reared Mexican fruit fly (Diptera: Tephritidae). J. Insect Sci. 2023, 23, 4. [Google Scholar] [CrossRef]
- Voirol, L.R.P.; Frago, E.; Kaltenpoth, M.; Hilker, M.; Fatouros, N.E. Bacterial symbionts in lepidoptera: Their diversity, transmission, and impact on the host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, L.Y.; Zhao, Q.Y.; Fu, D.Y.; Yu, H.; Xu, J.; Yang, S. Antibiotics ingestion altered the composition of gut microbes and affected the development and reproduction of the fall armyworm. J. Pest Sci. 2024, 97, 2187–2201. [Google Scholar] [CrossRef]
- Hammer, T.J.; Janzen, D.H.; Hallwachs, W.; Fierer, N. Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA 2017, 114, 9641–9646. [Google Scholar] [CrossRef]
- Hammer, T.J.; Sanders, J.G.; Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019, 369, fnz117. [Google Scholar] [CrossRef]
- Mason, C.J.; Jones, A.G.; Felton, G.W. Co-option of microbial associates by insects and their impact on plant-folivore interactions. Plant Cell Environ. 2018, 42, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.J. Complex relationships at the intersection of insect gut microbiomes and plant defenses. J. Chem. Ecol. 2020, 46, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.J.; Bowers, M.D. Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia 2015, 179, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.J.; Couture, J.J.; Raffa, K.F. Plant-associated bacteria degrade defense chemicals and reduce their adverse effects on an insect defoliator. Oecologia 2014, 175, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.J.; Clair, A.S.; Peiffer, M.; Gomez, E.; Jones, A.G.; Felton, G.W.; Hoover, K. Diet influences proliferation and stability of gut bacterial populations in herbivorous lepidopteran larvae. PLoS ONE 2020, 15, e0229848. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.G.; Mason, C.J.; Felton, G.W.; Hoover, K. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 2019, 9, 2792. [Google Scholar] [CrossRef]
- Priya, N.G.; Ojha, A.; Kajla, M.K.; Raj, A.; Rajagopal, R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS ONE 2012, 7, e30768. [Google Scholar] [CrossRef]
- Staudacher, H.; Kaltenpoth, M.; Breeuwer, J.A.J.; Menken, S.B.J.; Heckel, D.G.; Groot, A.T. Variability of bacterial communities in the moth Heliothis virescens indicates transient association with the host. PLoS ONE 2016, 11, e0154514. [Google Scholar] [CrossRef]
| Sample | NCBI Closest Match | Identity | Identification | GenBank No. |
|---|---|---|---|---|
| 1 | MT081629.1 | 99.88% | Acinetobacter soli | PP955231 |
| 2 | MN173944.1 | 99.52% | Klebsiella aerogenes | PP955232 |
| 3 | KF938661.1 | 99.76% | Enterobacter ludwigii | PP955233 |
| 4 | MK064226.1 | 99.64% | Herbaspirillum huttiense | PP955234 |
| 5 | MT598025.1 | 99.88% | Pseudomonas mosselii | PP955235 |
| 6 | KF542916.1 | 99.79% | Pantoea dispersa | PP955236 |
| 7 | MG905289.1 | 99.93% | Stenotrophomonas maltophilia | PP955237 |
| 8 | MT081681.1 | 99.76% | Klebsiella pneumoniae | PP955238 |
| 9 | KX225450.1 | 99.76% | Bacillus atrophaeus | PP955239 |
| 10 | MN889357.1 | 99.76% | Chryseobacterium culicis | PP955240 |
| 11 | KF040095.1 | 99.52% | Enterococcus hirae | PP955241 |
| 12 | MK641844.1 | 99.88% | Enterobacter asburiae | PP955242 |
| 13 | KX161909.1 | 99.76% | Pantoea ananatis | PP955243 |
| 14 | KU870748.1 | 99.58% | Providencia vermicola | PP955244 |
| 15 | MN833504.1 | 99.57% | Myroides odoratus | PP955245 |
| Isolates | Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|---|
| Acinetobacter soli | Proteobacteria | Gammproteobacteria | Pseudomonadales | Moraxellacea | Acinetobacter |
| Klebsiella aerogenes | Proteobacteria | Gammproteobacteria | Enterobacteriales | Enterobacteriaceae | Unclassified |
| Enterobacter ludwigii | Proteobacteria | Gammproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter |
| Herbaspirillum huttiense | Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Herbaspirillum |
| Pseudomonas mosselii | Proteobacteria | Gammproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| Pantoea dispersa | Proteobacteria | Gammproteobacteria | Enterobacteriales | Erwiniaceae | Pantoea |
| Stenotrophomonas maltophilia | Proteobacteria | Gammproteobacteria | Enterobacteriales | Erwiniaceae | Pantoea |
| Klebsiella pneumoniae | Proteobacteria | Gammproteobacteria | Enterobacteriales | Enterobacteriaceae | Klebsiella |
| Bacillus atrophaeus | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| Chryseobacterium culicis | Bacteroidetes | Flavobacteriaceae | Flavobacteriales | Flavobacteriaceae | Chryseobacterium |
| Enterococcus hirae | Firmicutes | Bacilli | Lactobacillale | Enterococcaceae | Enterococcus |
| Enterobacter asburiae | Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus |
| Pantoea ananatis | Proteobacteria | Gammproteobacteria | Enterobacteriales | Enterobacteriaceae | Pantoea |
| Providencia vermicola | Proteobacteria | Gammproteobacteria | Enterobacteriales | Morganellaceae | Providencia |
| Myroides odoratus | Bacteroidetes | Flavobacteriaceae | Flavobacteriales | Flavobacteriaceae | Myroides |
| Groups | Treatments | 1st Instar Days ± SE | 2nd Instar Days ± SE | 3rd Instar Days ± SE | 4th Instar Days ± SE | 5th Instar Days ± SE | Total Larva Days ± SE |
|---|---|---|---|---|---|---|---|
| 1 | Control | 3.68 ± 0.10 | 2.14 ± 0.10 | 2.07 ± 0.07 | 2.21 ± 0.12 | 3.86 ± 0.15 | 14.00 ± 0.19 |
| Acinetobacter soli | 3.56 ± 0.12 | 2.16 ± 0.08 | 2.12 ± 0.07 | 2.24 ± 0.12 | 3.64 ± 0.14 | 13.72 ± 0.20 | |
| Klebsiella aerogenes | 3.48 ± 0.15 | 2.35 ± 0.10 | 2.22 ± 0.14 | 2.70 ± 0.12 * | 3.70 ± 0.15 | 14.39 ± 0.29 | |
| Enterobacter ludwigii | 3.82 ± 0.09 | 2.25 ± 0.08 | 2.32 ± 0.09 | 2.36 ± 0.12 | 3.82 ± 0.13 | 14.57 ± 0.17 * | |
| Herbaspirillum huttiense | 3.96 ± 0.11 | 2.20 ± 0.08 | 2.16 ± 0.11 | 2.44 ± 0.13 | 3.96 ± 0.17 | 14.72 ± 0.21 * | |
| Pseudomonas mosselii | 4.00 ± 0.16 | 2.46 ± 0.18 | 2.92 ± 0.14 * | 2.23 ± 0.20 | 4.38 ± 0.14 * | 15.77 ± 0.46 * | |
| 2 | Control | 3.82 ± 0.15 | 2.50 ± 0.13 | 2.29 ± 0.10 | 2.39 ± 0.14 | 3.46 ± 0.16 | 14.46 ± 0.23 |
| Pantoea dispersa | 3.50 ± 0.20 | 2.28 ± 0.11 | 2.56 ± 0.17 | 2.72 ± 0.18 | 4.56 ± 0.19 * | 15.61 ± 0.38 | |
| Stenotrophomonas maltophilia | 3.95 ± 0.14 | 2.64 ± 0.16 | 2.36 ± 0.14 | 3.05 ± 0.14 * | 4.05 ± 0.14 * | 16.10 ± 0.27 * | |
| Klebsiella pneumoniae | 3.77 ± 0.14 | 2.42 ± 0.10 | 2.38 ± 0.10 | 2.65 ± 0.16 | 3.73 ± 0.14 | 15.00 ± 0.30 | |
| Bacillus atrophaeus | 4.14 ± 0.22 | 2.52 ± 0.11 | 2.48 ± 0.15 | 2.62 ± 0.13 | 3.86 ± 0.14 | 15.62 ± 0.38 * | |
| Chryseobacterium culicis | 3.76 ± 0.16 | 2.71 ± 0.17 | 2.18 ± 0.10 | 2.65 ± 0.17 | 3.71 ± 0.14 | 15.00 ± 0.35 | |
| 3 | Control | 3.34 ± 0.11 | 2.24 ± 0.10 | 2.17 ± 0.09 | 2.48 ± 0.15 | 3.90 ± 0.10 | 14.14 ± 0.17 |
| Enterococcus hirae | 4.35 ± 0.12 * | 2.57 ± 0.11 * | 2.61 ± 0.18 | 2.74 ± 0.17 | 4.26 ± 0.13 * | 16.52 ± 0.36 * | |
| Enterobacter asburiae | 3.79 ± 0.18 * | 2.47 ± 0.12 | 2.26 ± 0.17 | 2.58 ± 0.16 | 3.89 ± 0.15 | 15.00 ± 0.43 | |
| Pantoea ananatis | 3.94 ± 0.25 * | 2.63 ± 0.18 | 2.44 ± 0.16 | 2.44 ± 0.13 * | 4.44 ± 0.18 | 15.88 ± 0.41 * | |
| Providencia vermicola | 3.27 ± 0.12 | 2.59 ± 0.18 | 2.91 ± 0.19 * | 3.18 ± 0.17 * | 4.09 ± 0.16 | 16.05 ± 0.37 * | |
| Myroides odoratus | 3.72 ± 0.16 | 2.28 ± 0.14 | 2.44 ± 0.12 | 2.44 ± 0.15 | 4.28 ± 0.14 * | 15.17 ± 0.29 * |
| Groups | Treatments | Pre–Pupa Days ± SE | Pupa Days ± SE | Pupal Weight mg ± SE | Female Ratio | Adult Longevity Days ± SE | Fecundity Eggs/Female |
|---|---|---|---|---|---|---|---|
| 1 | Control | 2.11 ± 0.08 | 7.07 ± 0.13 | 21.21 ± 0.46 | 0.43 | 13.18 ± 0.51 | 305.33 ± 22.90 |
| Acinetobacter soli | 2.08 ± 0.06 | 6.88 ± 0.12 | 20.84 ± 0.47 | 0.48 | 10.76 ± 0.50 * | 194.57 ± 23.58 * | |
| Klebsiella aerogenes | 2.04 ± 0.04 | 7.04 ± 0.12 | 22.27 ± 0.45 | 0.48 | 11.39 ± 0.66 * | 354.25 ± 32.42 | |
| Enterobacter ludwigii | 2.21 ± 0.15 | 7.07 ± 0.17 | 21.65 ± 0.48 | 0.46 | 12.21 ± 0.35 | 348.18 ± 16.80 | |
| Herbaspirillum huttiense | 2.12 ± 0.07 | 6.96 ± 0.11 | 21.91 ± 0.42 | 0.40 | 10.84 ± 0.57 * | 300.67 ± 12.48 | |
| Pseudomonas mosselii | 2.00 ± 0.16 | 7.15 ± 0.27 | 21.88 ± 0.88 | 0.54 | 12.31 ± 0.38 | 318.50 ± 45.21 | |
| 2 | Control | 2.00 ± 0.05 | 7.14 ± 0.15 | 21.85 ± 0.69 | 0.50 | 12.64 ± 0.46 | 316.79 ± 21.00 |
| Pantoea dispersa | 2.11 ± 0.08 | 7.11 ± 0.08 | 17.42 ± 0.60 * | 0.50 | 10.39 ± 0.63 * | 281.50 ± 16.43 | |
| Stenotrophomonas maltophilia | 2.05 ± 0.08 | 7.41 ± 0.13 | 19.79 ± 0.44 * | 0.50 | 10.64 ± 0.47 * | 208.38 ± 38.54 * | |
| Klebsiella pneumoniae | 2.04 ± 0.04 | 7.50 ± 0.28 | 18.71 ± 0.63 * | 0.50 | 11.50 ± 0.63 | 321.73 ± 17.34 | |
| Bacillus atrophaeus | 2.00 ± 0.00 | 7.33 ± 0.13 | 18.73 ± 0.49 * | 0.62 | 11.76 ± 0.64 | 203.91 ± 30.87 * | |
| Chryseobacterium culicis | 2.06 ± 0.10 | 7.29 ± 0.17 | 19.05 ± 0.73 * | 0.47 | 12.76 ± 0.60 | 277.43 ± 35.85 | |
| 3 | Control | 2.03 ± 0.06 | 6.86 ± 0.08 | 19.74 ± 0.42 | 0.48 | 11.97 ± 0.46 | 281.00 ± 19.49 |
| Enterococcus hirae | 2.09 ± 0.11 | 7.22 ± 0.11 * | 21.48 ± 0.60 * | 0.48 | 10.61 ± 0.63 | 279.64 ± 8.49 | |
| Enterobacter asburiae | 1.89 ± 0.07 | 7.21 ± 0.15 * | 19.62 ± 0.49 | 0.47 | 10.68 ± 0.57 | 286.67 ± 24.00 | |
| Pantoea ananatis | 2.06 ± 0.06 | 7.44 ± 0.16 * | 19.14 ± 0.37 | 0.44 | 10.81 ± 0.48 | 292.00 ± 18.23 | |
| Providencia vermicola | 2.14 ± 0.08 | 7.95 ± 0.31 * | 19.05 ± 0.43 | 0.45 | 11.36 ± 0.54 | 210.89 ± 26.47 * | |
| Myroides odoratus | 2.11 ± 0.08 | 6.89 ± 0.18 | 18.84 ± 0.520 | 0.50 | 12.17 ± 0.40 | 294.50 ± 19.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Q.; Wang, Q.; Shikano, I.; Liu, F.; Yao, Z. Effects of Gut Bacteria on the Fitness of Rice Leaf Folder Cnaphalocrocis medinalis. Insects 2024, 15, 947. https://doi.org/10.3390/insects15120947
Pan Q, Wang Q, Shikano I, Liu F, Yao Z. Effects of Gut Bacteria on the Fitness of Rice Leaf Folder Cnaphalocrocis medinalis. Insects. 2024; 15(12):947. https://doi.org/10.3390/insects15120947
Chicago/Turabian StylePan, Qinjian, Qingpeng Wang, Ikkei Shikano, Fang Liu, and Zhichao Yao. 2024. "Effects of Gut Bacteria on the Fitness of Rice Leaf Folder Cnaphalocrocis medinalis" Insects 15, no. 12: 947. https://doi.org/10.3390/insects15120947
APA StylePan, Q., Wang, Q., Shikano, I., Liu, F., & Yao, Z. (2024). Effects of Gut Bacteria on the Fitness of Rice Leaf Folder Cnaphalocrocis medinalis. Insects, 15(12), 947. https://doi.org/10.3390/insects15120947







