Northern Richness, Southern Dead End—Origin and Dispersal Events of Pseudolycoriella (Sciaridae, Diptera) between New Zealand’s Main Islands
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
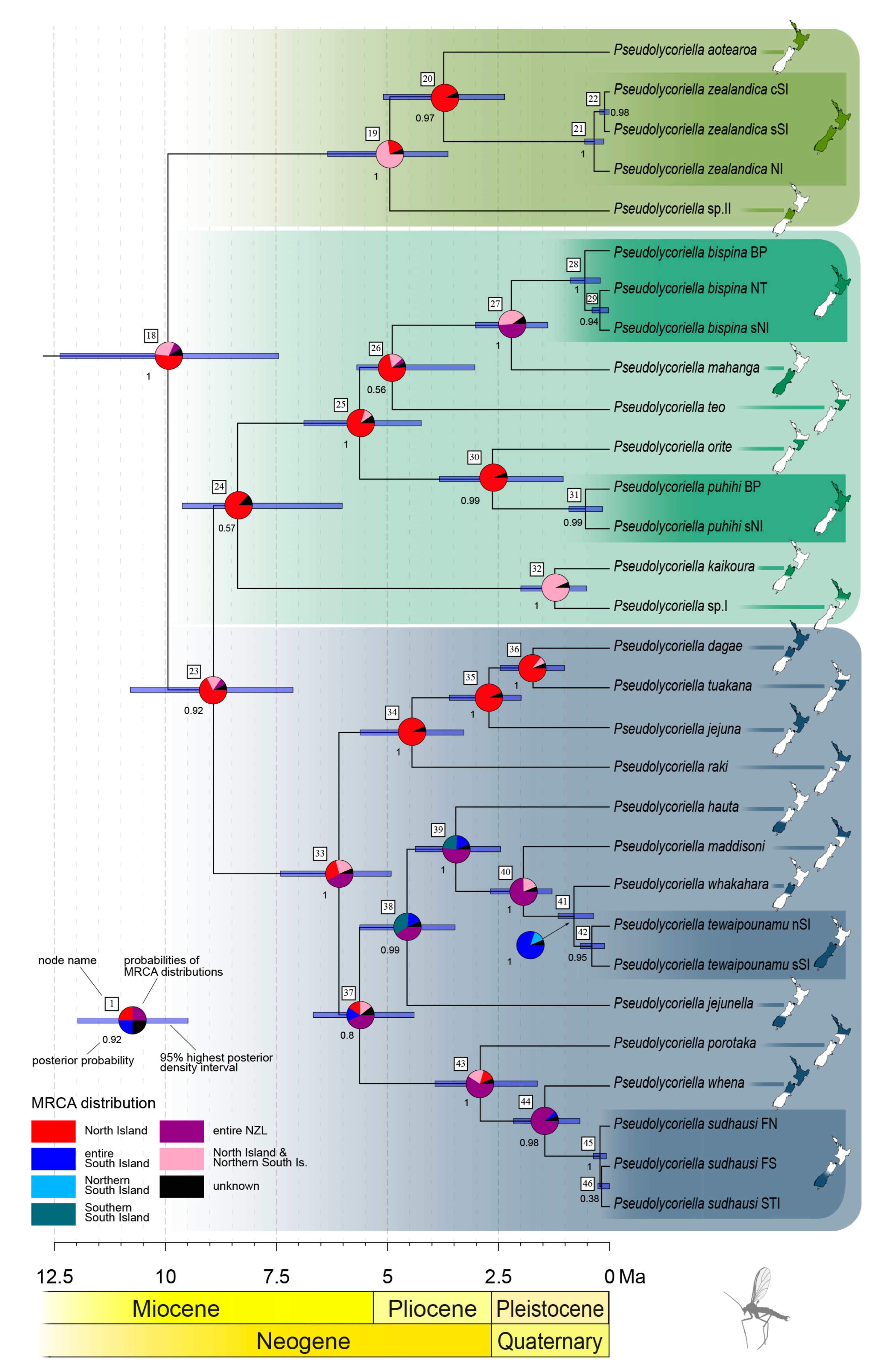
3. Results
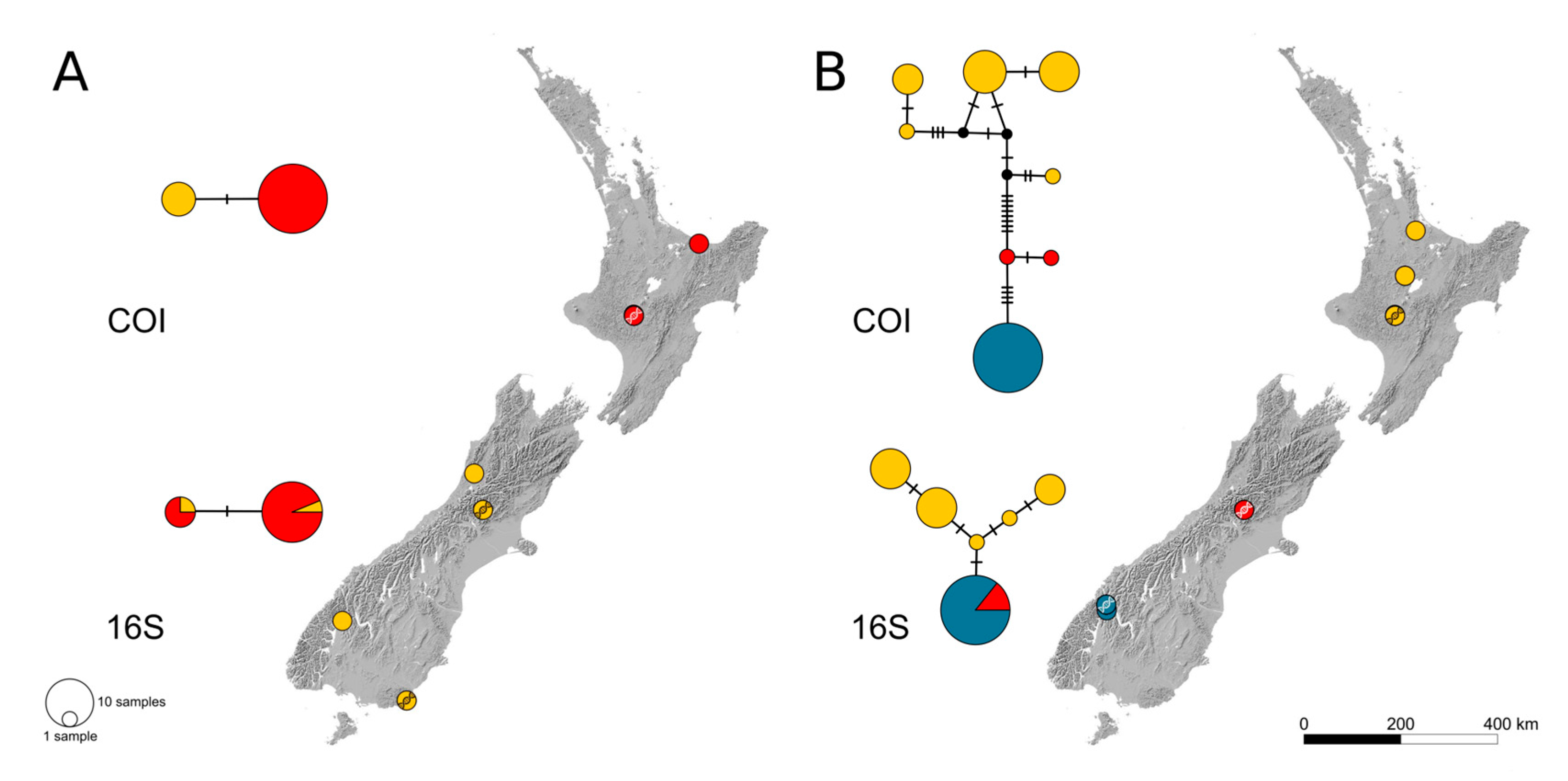
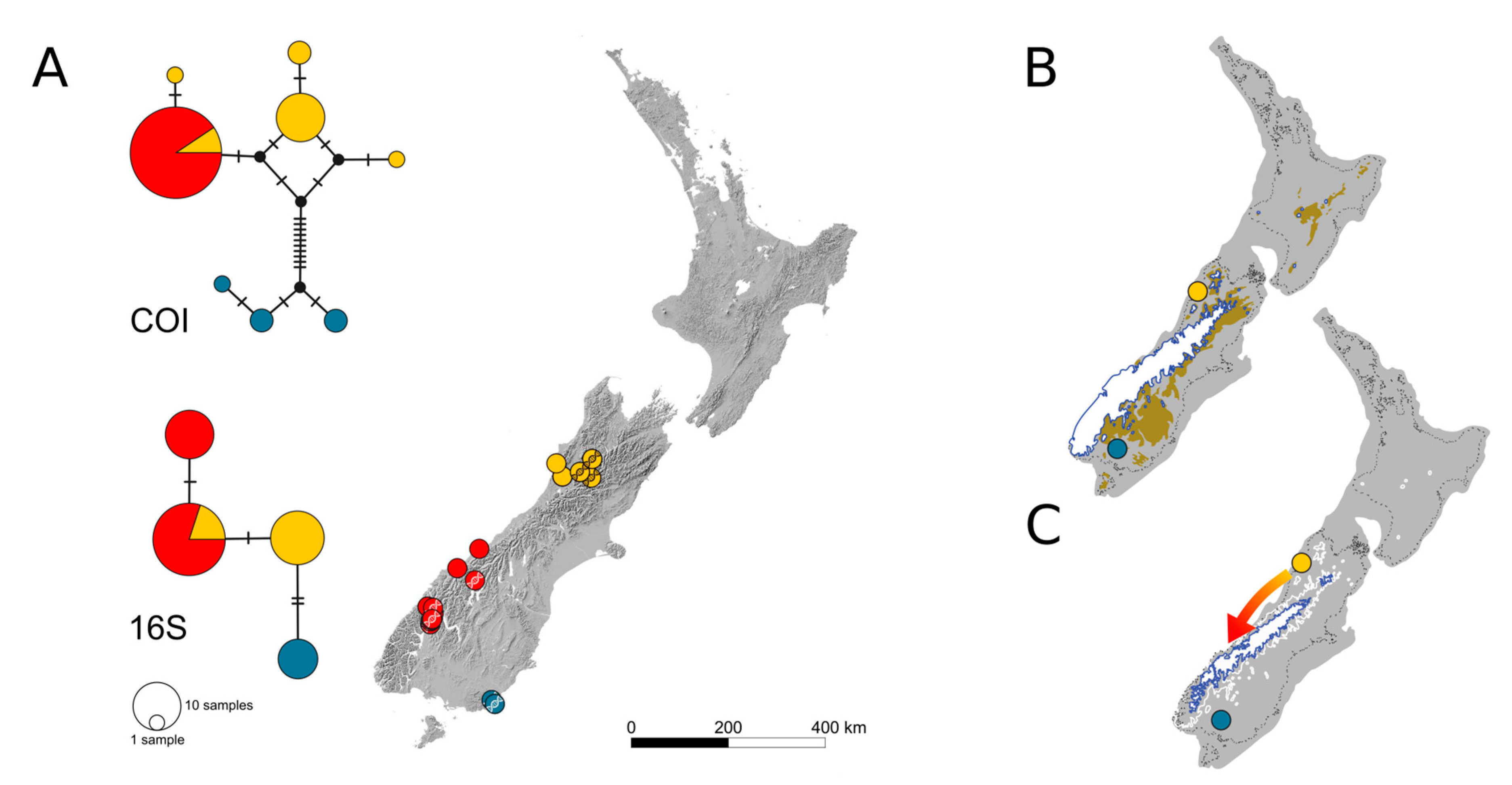
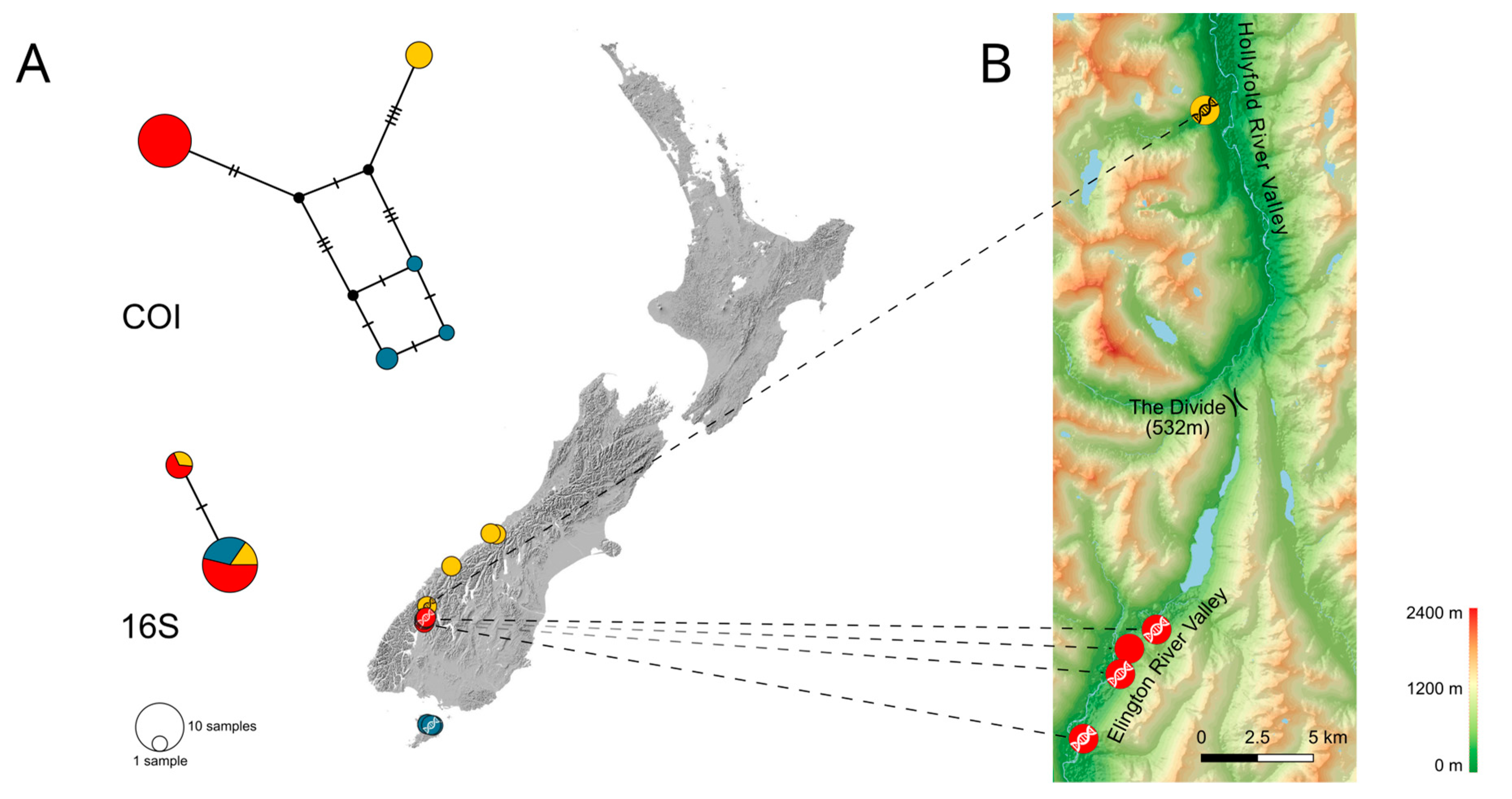
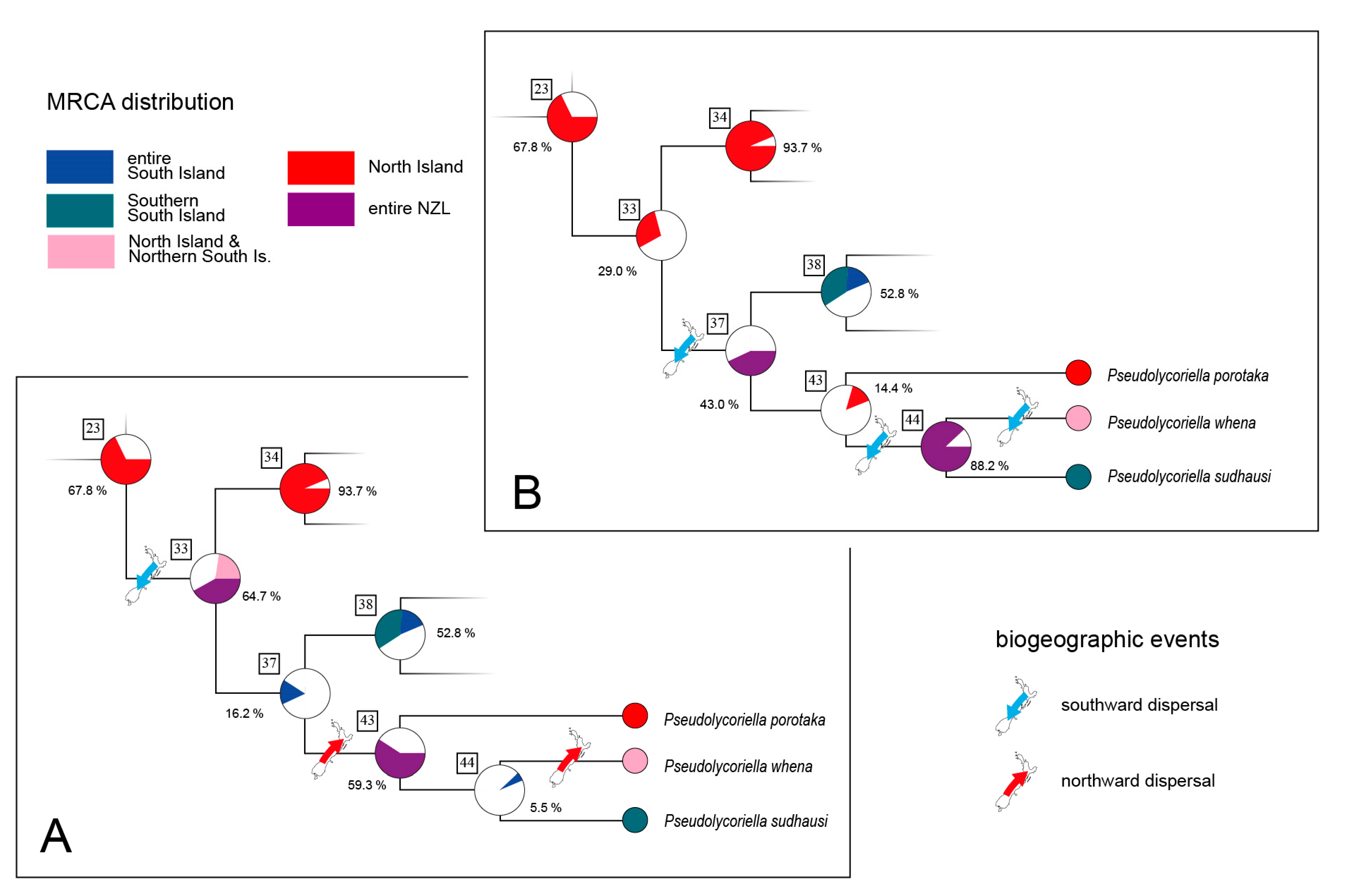
3.1. Intraspecific Biogeographic Patterns
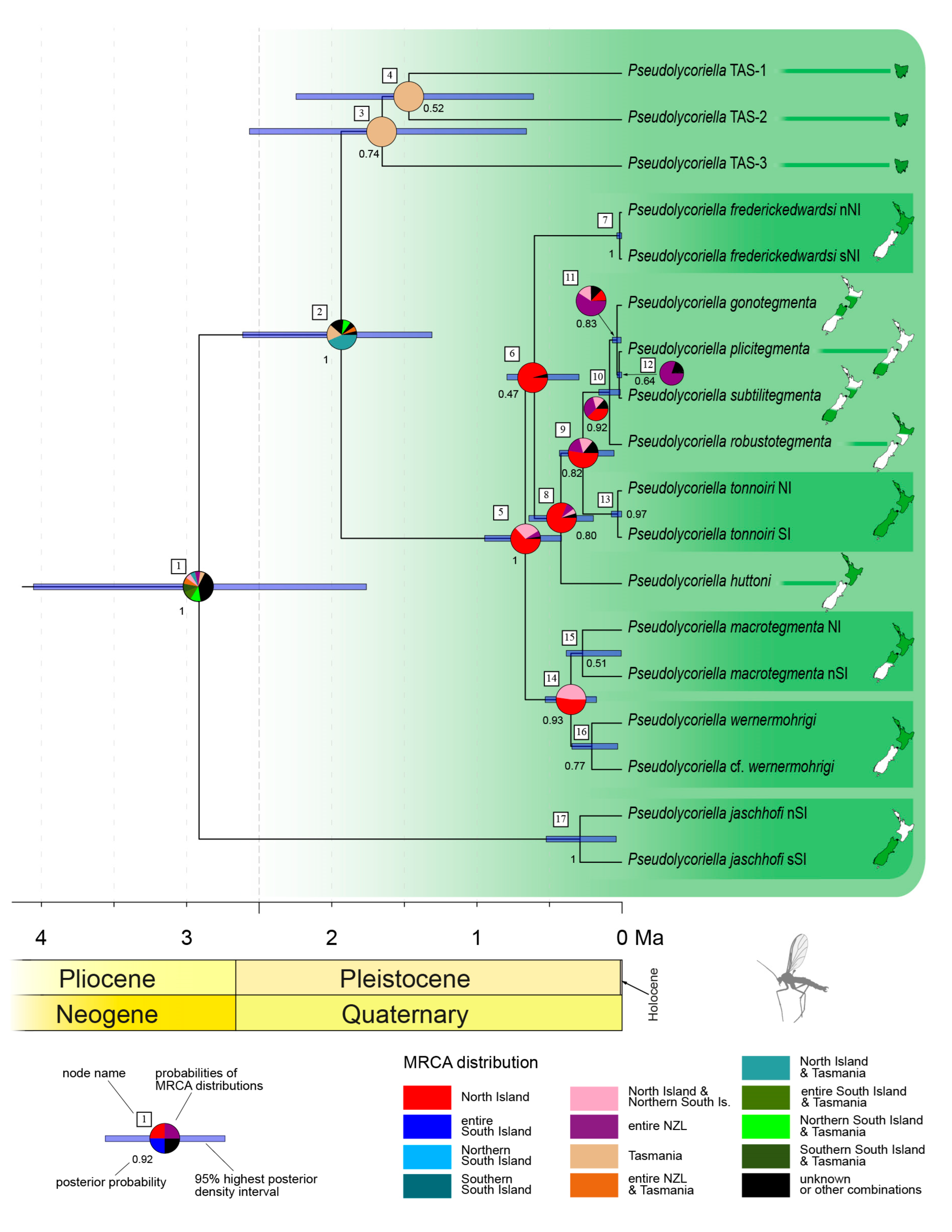
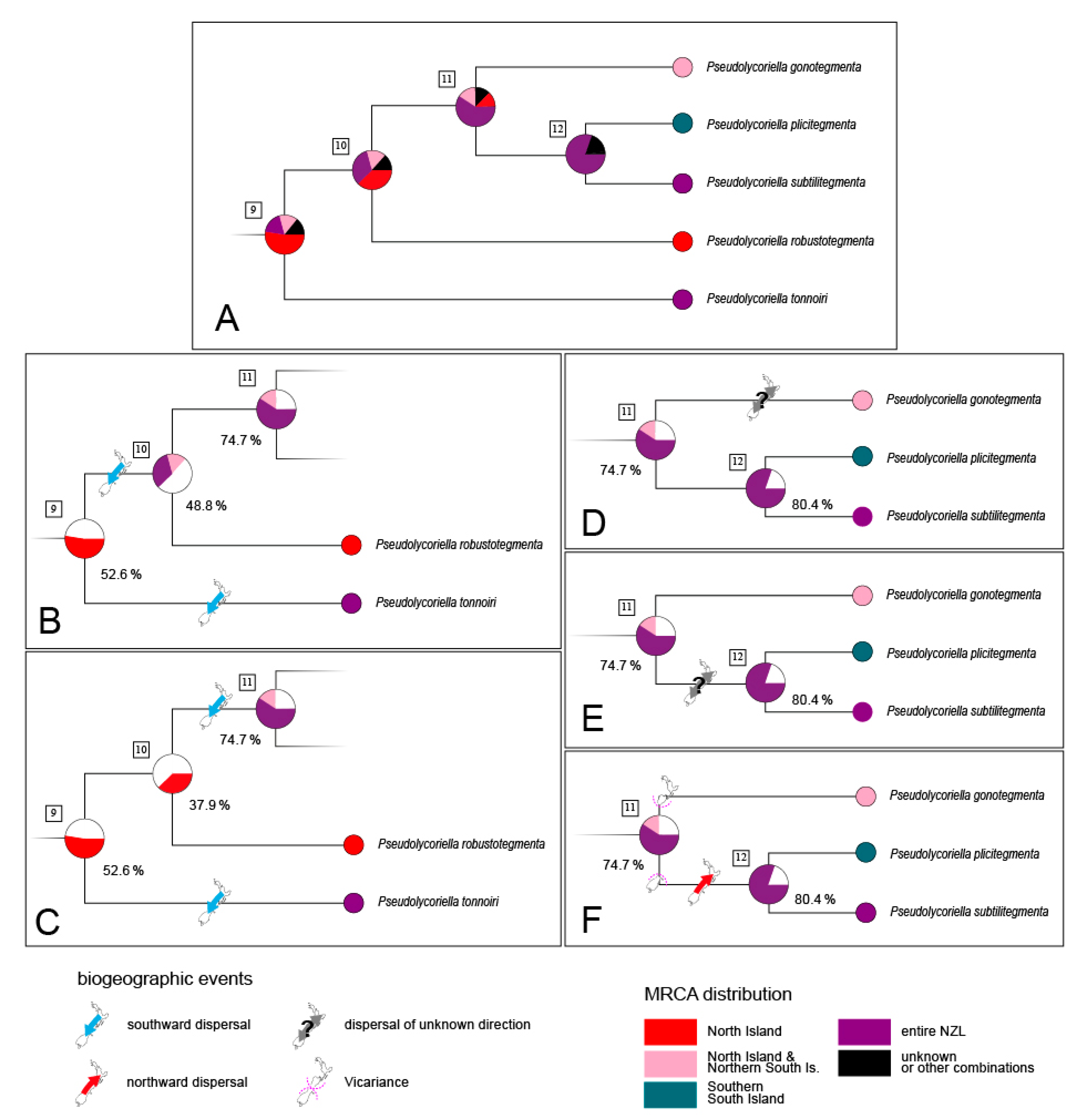
3.2. Interspecific Differentiation and Phylogeny
4. Discussion
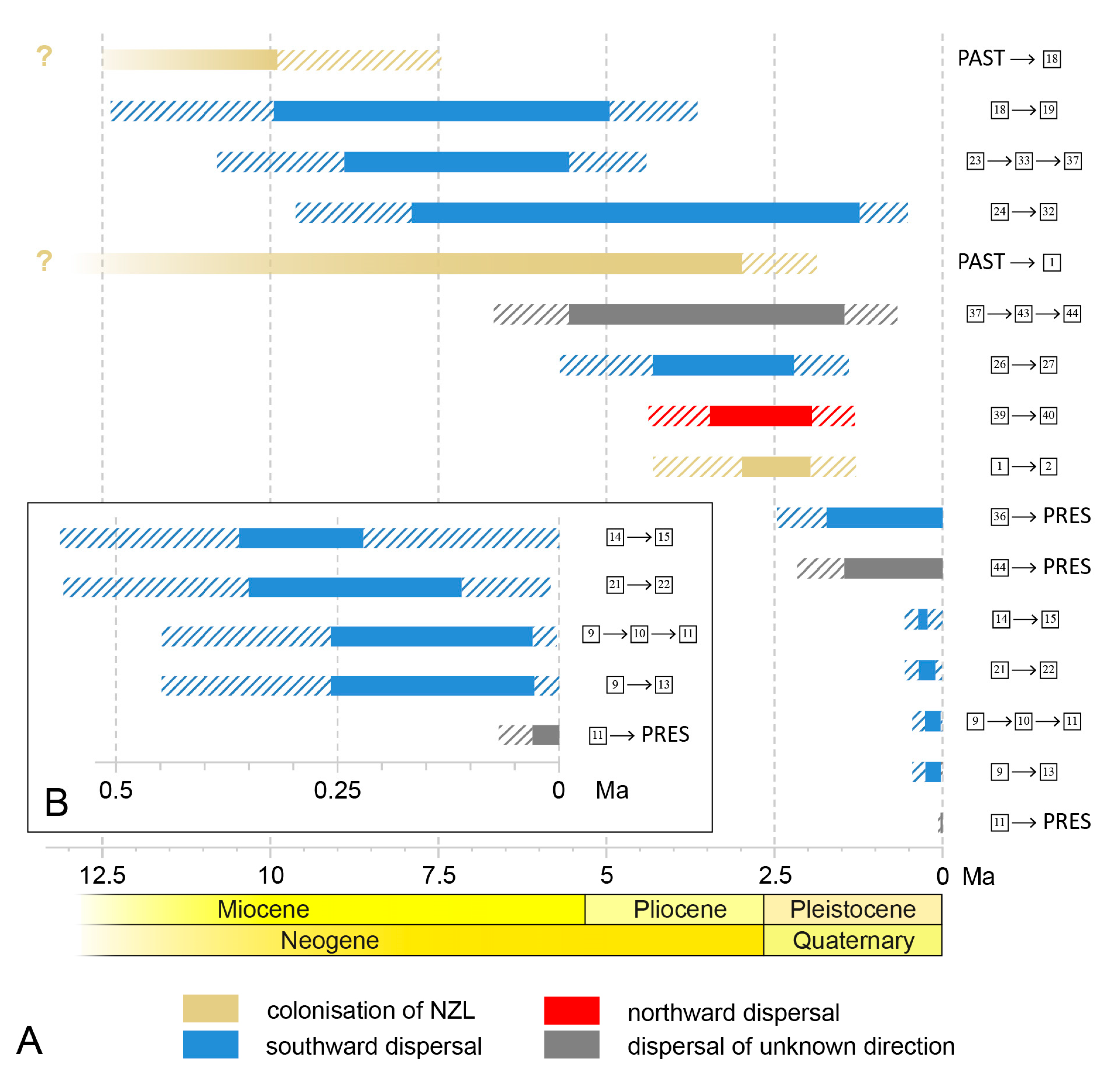
4.1. Ice Ages and Their Influence on New Zealand’s Scarids
4.2. Island Hopping and Speciation
4.3. Colonisation of New Zealand
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallis, G.P.; Trewick, S.A. New Zealand phylogeography: Evolution on a small continent. Mol. Ecol. 2009, 18, 3548–3580. [Google Scholar] [CrossRef] [PubMed]
- Buckley, T.R.; Krosch, M.; Leschen, R.A.B. Evolution of New Zealand insects: Summary and prospectus for future research. Austral Entomol. 2015, 54, 1–27. [Google Scholar] [CrossRef]
- Marske, K.A.; Boyer, S.L. Phylogeography reveals the complex impact of the Last Glacial Maximum on New Zealand’s terrestrial biota. J. R. Soc. N. Z. 2022, 1–22. [Google Scholar] [CrossRef]
- Wood, J.; Wilmshurst, J.; Newnham, R.; McGlone, M. Evolution and Ecological Change During the New Zealand Quaternary. In Landscape and Quaternary Environmental Change in New Zealand; Shulmeister, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 235–291. [Google Scholar] [CrossRef]
- Wallis, G.P.; Jorge, F. Going under down under? Lineage ages argue for extensive survival of the Oligocene marine transgression on Zealandia. Mol. Ecol. 2018, 27, 4368–4396. [Google Scholar] [CrossRef]
- Buckley, T.R.; Lord, N.P.; Ramón-Laca, A.; Allwood, J.S.; Leschen, R.A.B. Multiple lineages of hyper-diverse Zopheridae beetles survived the New Zealand Oligocene Drowning. J. Biogeogr. 2020, 47, 927–940. [Google Scholar] [CrossRef]
- Bunce, M.; Worthy, T.H.; Phillips, M.J.; Holdaway, R.N.; Willerslev, E.; Haile, J.; Shapiro, B.; Scofield, R.P.; Drummond, A.; Kamp, P.J.J. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc. Natl. Acad. Sci. USA 2009, 106, 20646–20651. [Google Scholar] [CrossRef]
- Trewick, S.; Bland, K. Fire and slice: Palaeogeography for biogeography at New Zealand’s North Island/South Island juncture. J. R. Soc. N. Z. 2012, 42, 153–183. [Google Scholar] [CrossRef]
- Rawlence, N.J.; Potter, B.C.M.; Dussex, N.; Scarsbrook, L.; Orlovich, D.A.; Waters, J.M.; McGlone, M.; Knapp, M. Plio-Pleistocene environmental changes shape present day phylogeography of New Zealand’s southern beeches (Nothofagaceae). N. Z. J. Bot. 2021, 59, 55–71. [Google Scholar] [CrossRef]
- Menzel, F.; Mohrig, W. Revision der paläarktischen Trauermücken (Diptera: Sciaridae); Stark, A., Menzel, F., Eds.; Ampyx-Verlag: Halle (Saale), Germany, 2000; Volume 6, p. 761. [Google Scholar]
- Shin, S.; Jung, S.; Menzel, F.; Heller, K.; Lee, H.; Lee, S. Molecular phylogeny of black fungus gnats (Diptera: Sciaroidea: Sciaridae) and the evolution of larval habitats. Mol. Phylogenetics Evol. 2013, 66, 833–846. [Google Scholar] [CrossRef]
- Trinca, V.; Carli, S.; Uliana, J.V.C.; Garbelotti, C.V.; da Silva, M.M.; Kunes, V.; Meleiro, L.P.; Brancini, G.T.P.; Menzel, F.; Andrioli, L.P.M.; et al. Biocatalytic potential of Pseudolycoriella CAZymes (Sciaroidea, Diptera) in degrading plant and fungal cell wall polysaccharides. iScience 2023, 26, 106449. [Google Scholar] [CrossRef]
- Gressitt, J.L.; Sedlacek, J.; Wise, K.A.J.; Yoshimoto, C.M. A high speed airplane trap for air-borne organisms. Pac. Insects 1961, 3, 549–555. [Google Scholar]
- Ashmole, N.P.; Ashmole, M.J. Insect Dispersal on Tenerife, Canary Islands: High Altitude Fallout and Seaward Drift. Arct. Alp. Res. 1988, 20, 1–12. [Google Scholar] [CrossRef]
- Peck, S.B. Sea-Surface (Pleuston) Transport of Insects between Islands in the Galápagos Archipelago, Ecuador. Ann. Èntomol. Soc. Am. 1994, 87, 576–582. [Google Scholar] [CrossRef]
- Köhler, A. The genus Pseudolycoriella Menzel & Mohrig, 1998 (Diptera, Sciaridae) in New Zealand. Zootaxa 2019, 4707, 1–69. [Google Scholar] [CrossRef]
- Public Data Portal–BIN Page, BIN BOLD:ABW3602; BOLD, 2019. [CrossRef]
- Trewick, S.A.; Wallis, G.P.; Morgan-Richards, M. The Invertebrate Life of New Zealand: A Phylogeographic Approach. Insects 2011, 2, 297–325. [Google Scholar] [CrossRef] [PubMed]
- Bashford, R.; Taylor, R.; Driessen, M.; Doran, N.; Richardson, A. Research on invertebrate assemblages at the Warra LTER Site. Tasforests 2001, 13, 109–118. [Google Scholar]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Bouckaert, R.R. Bayesian Evolutionary Analysis with BEAST; Cambridge University Press: Cambridge, UK, 2015; p. 249. [Google Scholar]
- Schmidt, A.R.; Kaulfuss, U.; Bannister, J.M.; Baranov, V.; Beimforde, C.; Bleile, N.; Borkent, A.; Busch, A.; Conran, J.G.; Engel, M.S.; et al. Amber inclusions from New Zealand. Gondwana Res. 2018, 56, 135–146. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Anastasiou, I.; Vogler, A.P. Revisiting the Insect Mitochondrial Molecular Clock: The Mid-Aegean Trench Calibration. Mol. Biol. Evol. 2010, 27, 1659–1672. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Blair, C.; Harris, A.J.; He, X. A Rough Guide to RASP 4.2; 2019. Available online: http://www.lmse.org/assets/workshop/2017/YGX/A-Rough-Guide-to-RASP.pdf (accessed on 26 April 2022).
- Yu, Y.; Blair, C.; He, X. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Mol. Biol. Evol. 2019, 37, 604–606. [Google Scholar] [CrossRef] [PubMed]
- McGlone, M.S.; Newnham, R.M.; Moar, N.T. The vegetation cover of New Zealand during the Last Glacial Maximum: Do pollen records under-represent woody vegetation. Terra Aust. 2010, 32, 49–68. [Google Scholar]
- Golledge, N.R.; Mackintosh, A.N.; Anderson, B.M.; Buckley, K.M.; Doughty, A.M.; Barrell, D.J.; Denton, G.H.; Vandergoes, M.J.; Andersen, B.G.; Schaefer, J.M. Last Glacial Maximum climate in New Zealand inferred from a modelled Southern Alps icefield. Quat. Sci. Rev. 2012, 46, 30–45. [Google Scholar] [CrossRef]
- Broadley, A.; Mohrig, W.; Kauschke, E.; Menzel, F. Revision of Black fungus gnats (Diptera: Sciaridae) of the Antarctic region. Zootaxa. in preparation.
- Shulmeister, J.; Fink, D.; Winkler, S.; Thackray, G.; Borsellino, R.; Hemmingsen, M.; Rittenour, T. Evidence for slow late-glacial ice retreat in the upper Rangitata Valley, South Island, New Zealand. Quat. Sci. Rev. 2018, 185, 102–112. [Google Scholar] [CrossRef]
- Waters, J.M.; Fraser, C.I.; Hewitt, G.M. Founder takes all: Density-dependent processes structure biodiversity. Trends Ecol. Evol. 2013, 28, 78–85. [Google Scholar] [CrossRef]
- Marshall, D.C.; Hill, K.B.R.; Fontaine, K.M.; Buckley, T.R.; Simon, C. Glacial refugia in a maritime temperate climate: Cicada (Kikihia subalpina) mtDNA phylogeography in New Zealand. Mol. Ecol. 2009, 18, 1995–2009. [Google Scholar] [CrossRef]
- Goldberg, J.; Trewick, S.A. Exploring Phylogeographic Congruence in a Continental Island System. Insects 2011, 2, 369–399. [Google Scholar] [CrossRef]
- Marske, K.A.; Leschen, R.A.; Buckley, T.R. Reconciling phylogeography and ecological niche models for New Zealand beetles: Looking beyond glacial refugia. Mol. Phylogenetics Evol. 2011, 59, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, T. Molekulare Biogeographie—Gene in Raum und Zeit; Uni-Taschenbücher GmbH; Haupt Verlag: Bern, Switzerland, 2020. [Google Scholar]
- Williams, P.W.; McGlone, M.; Neil, H.; Zhao, J.-X. A review of New Zealand palaeoclimate from the Last Interglacial to the global Last Glacial Maximum. Quat. Sci. Rev. 2015, 110, 92–106. [Google Scholar] [CrossRef]
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Baker, C.H.; Graham, G.C.; Scott, K.D.; Cameron, S.; Yeates, D.K.; Merritt, D.J. Distribution and phylogenetic relationships of Australian glow-worms Arachnocampa (Diptera, Keroplatidae). Mol. Phylogenetics Evol. 2008, 48, 506–514. [Google Scholar] [CrossRef]
- Arensburger, P.; Buckley, T.R.; Simon, C.; Moulds, M.; Holsinger, K.E. Biogeography and phylogeny of the New Zealand cicada genera (Hemiptera: Cicadidae) based on nuclear and mitochondrial DNA data. J. Biogeogr. 2004, 31, 557–569. [Google Scholar] [CrossRef]
- Mohrig, W.; Kauschke, E.; Broadley, A. Black fungus gnats (Diptera: Sciaridae) of Queensland, Australia. Part II. Genus Pseudolycoriella Menzel & Mohrig, 1998. Zootaxa 2020, 4751, 487–506. [Google Scholar] [CrossRef]
- Mohrig, W.; Kauschke, E.; Broadley, A. New black fungus gnats (Diptera: Sciaridae) from Eastern Australia. Zootaxa 2018, 4450, 203–241. [Google Scholar] [CrossRef]
- Mohrig, W.; Kauschke, E.; Broadley, A. Pseudolycoriella skusei sp. nov. (Diptera: Sciaridae), a new dark-winged fungus gnat from Norfolk Island and Australia. Zootaxa 2016, 4097, 139–142. [Google Scholar] [CrossRef]
- Broadley, A.; Kauschke, E.; Mohrig, W. Revision of the types of male Sciaridae (Diptera) described from Australia by F.A.A. Skuse. Zootaxa 2016, 4193, 401–450. [Google Scholar] [CrossRef]
- Köhler, A.; Menzel, F. New records of Black Fungus Gnats (Diptera: Sciaridae) from New Caledonia, with the description of two new Bradysia species and an updated checklist. Zootaxa 2013, 3718, 63–72. [Google Scholar] [CrossRef]
- Vilkamaa, P.; Hippa, H.; Mohrig, W. The genus Pseudolycoriella Menzel & Mohrig (Diptera, Sciaridae) in New Caledonia, with the description of thirteen new species. Zootaxa 2012, 3207, 1–21. [Google Scholar] [CrossRef]
- Sturman, A.P.; Tapper, N.J. The Weather and Climate of Australia and New Zealand; Oxford University Press: Oxford, UK, 2006; p. 541. [Google Scholar]
- Oke, P.R.; Pilo, G.S.; Ridgway, K.; Kiss, A.; Rykova, T. A search for the Tasman Front. J. Mar. Syst. 2019, 199, 103217. [Google Scholar] [CrossRef]
- Bostock, H.C.; Hayward, B.W.; Neil, H.L.; Sabaa, A.T.; Scott, G.H. Changes in the position of the Subtropical Front south of New Zealand since the last glacial period. Paleoceanography 2015, 30, 824–844. [Google Scholar] [CrossRef]
- Li, X.; Jiang, D.; Zhang, Z.; Zhang, R.; Tian, Z.; Yan, Q. Mid-Pliocene westerlies from PlioMIP simulations. Adv. Atmos. Sci. 2015, 32, 909–923. [Google Scholar] [CrossRef]
- Scher, H.D.; Martin, E.E. Timing and Climatic Consequences of the Opening of Drake Passage. Science 2006, 312, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Steinthorsdottir, M.; Jardine, P.E.; Rember, W.C. Near-Future pCO2 During the Hot Miocene Climatic Optimum. Paleoceanogr. Paleoclimatology 2020, 36, e2020PA003900. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Pole, M. The Miocene climate in New Zealand: Estimates from paleobotanical data. Palaeontol. Electron. 2014, 17, 1–79. [Google Scholar] [CrossRef]
- Prebble, J.G.; Reichgelt, T.; Mildenhall, D.C.; Greenwood, D.R.; Raine, J.I.; Kennedy, E.M.; Seebeck, H.C. Terrestrial climate evolution in the Southwest Pacific over the past 30 million years. Earth Planet. Sci. Lett. 2017, 459, 136–144. [Google Scholar] [CrossRef]
- Martin, H. Cenozoic climatic change and the development of the arid vegetation in Australia. J. Arid. Environ. 2006, 66, 533–563. [Google Scholar] [CrossRef]
- Public Data Portal–BIN Page, BIN BOLD:ACP1302. [CrossRef]
| Widely Distributed on Both Islands | Distributed on North Island and Northern South Island | Endemic to North Island | Endemic to South Island |
|---|---|---|---|
| P. cavatica * | P. dagae | P. aoteraoa | P. fiordlandia |
| P. subtilitegmenta | P. gonotegmenta | P. bispina | P. jaschhofi |
| P. tonnoiri | P. macrotegmenta | P. breviseta | P. jejunella |
| P. zealandica | P. whena | P. frederickedwardsi | P. mahanga |
| P. jejuna | P. sudhausi | ||
| P. maddisoni | P. tewaipounamu | ||
| P. orite | |||
| P. porotaka | |||
| P. puhihi | |||
| P. raki | |||
| P. robustotegmenta | |||
| P. wernermohrigi |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Köhler, A.; Schmitt, T. Northern Richness, Southern Dead End—Origin and Dispersal Events of Pseudolycoriella (Sciaridae, Diptera) between New Zealand’s Main Islands. Insects 2023, 14, 548. https://doi.org/10.3390/insects14060548
Köhler A, Schmitt T. Northern Richness, Southern Dead End—Origin and Dispersal Events of Pseudolycoriella (Sciaridae, Diptera) between New Zealand’s Main Islands. Insects. 2023; 14(6):548. https://doi.org/10.3390/insects14060548
Chicago/Turabian StyleKöhler, Arne, and Thomas Schmitt. 2023. "Northern Richness, Southern Dead End—Origin and Dispersal Events of Pseudolycoriella (Sciaridae, Diptera) between New Zealand’s Main Islands" Insects 14, no. 6: 548. https://doi.org/10.3390/insects14060548
APA StyleKöhler, A., & Schmitt, T. (2023). Northern Richness, Southern Dead End—Origin and Dispersal Events of Pseudolycoriella (Sciaridae, Diptera) between New Zealand’s Main Islands. Insects, 14(6), 548. https://doi.org/10.3390/insects14060548





