Serpentine and Vermiform Are Produced Autonomously to Fulfill Their Function in Drosophila Wings
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks
2.2. RNAi of Serp and Verm
2.3. Adult Wing Observation
2.4. Reverse Transcription Quantitative PCR (RT-PCR)
2.5. Cell Apoptosis Detection in Wing Discs
2.6. Eosin Y Penetration Assay
2.7. FB28 Staining
3. Results
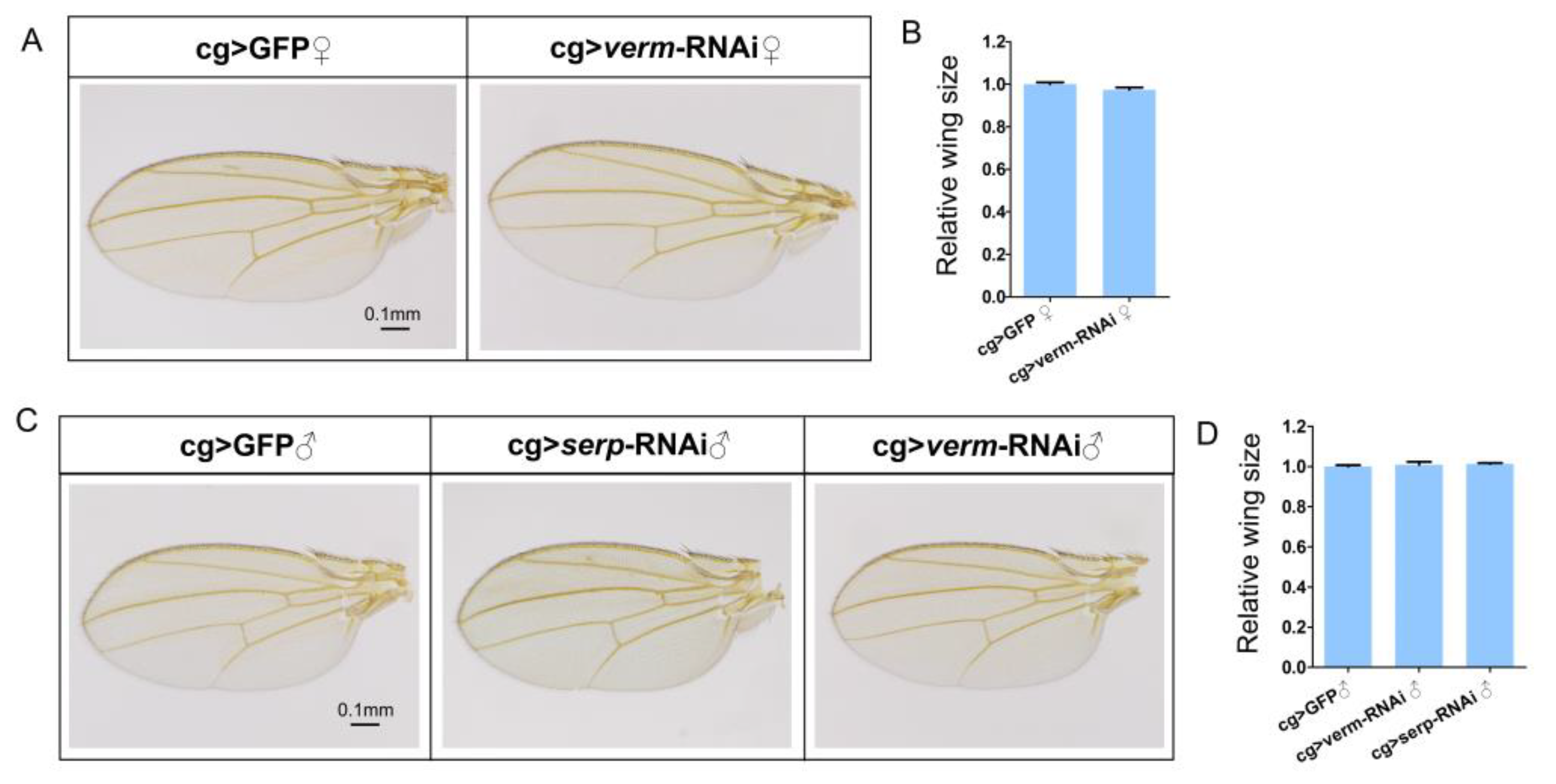
3.1. Knockdown of Serp and Verm in the Fat Body Does Not Cause Wing Deficiency
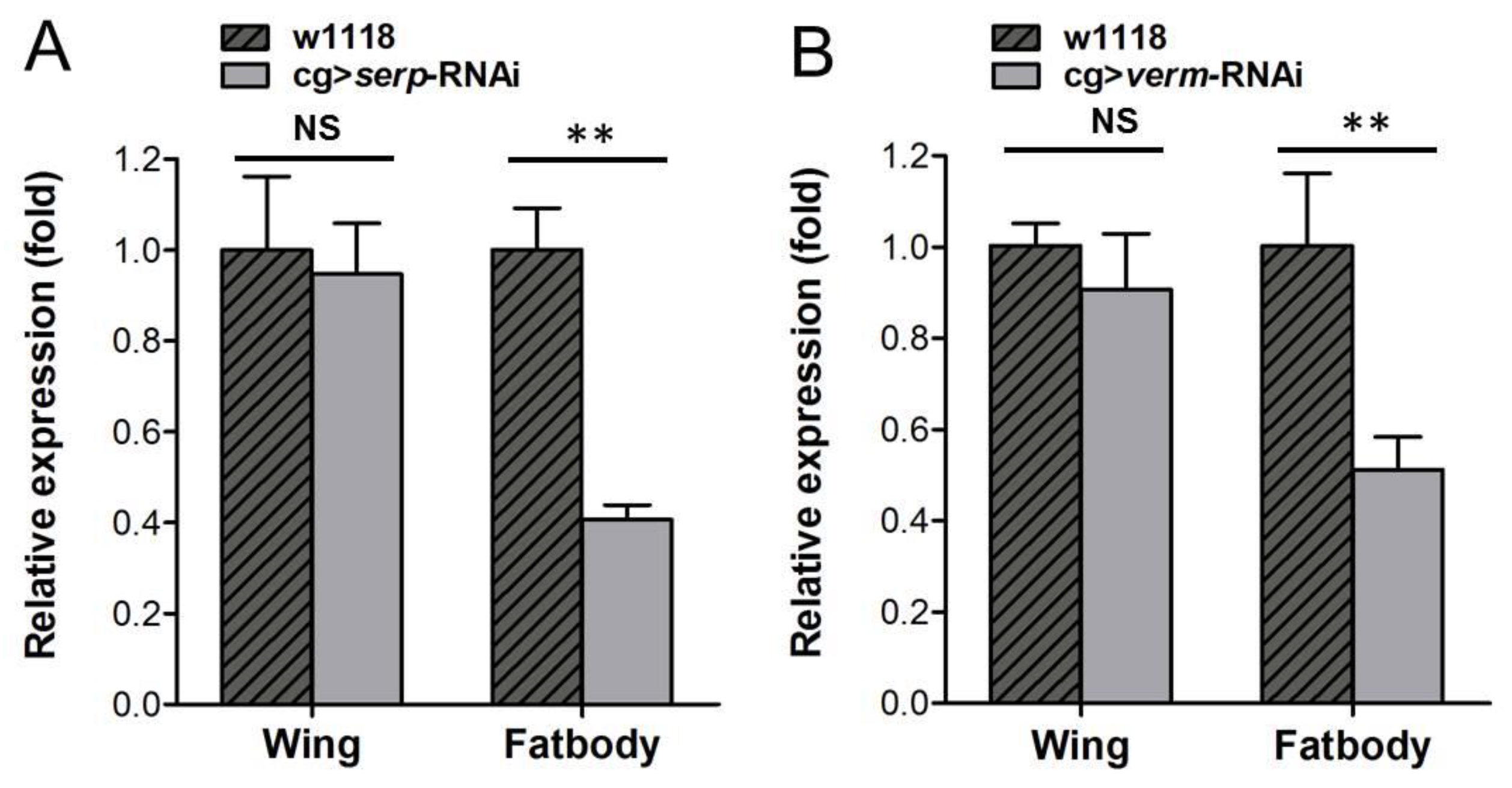
3.2. Knockdown of Serp and Verm in the Fat Body by cg-Gal4 Has no Effect on the Expression Level of Serp and Verm in the Wing
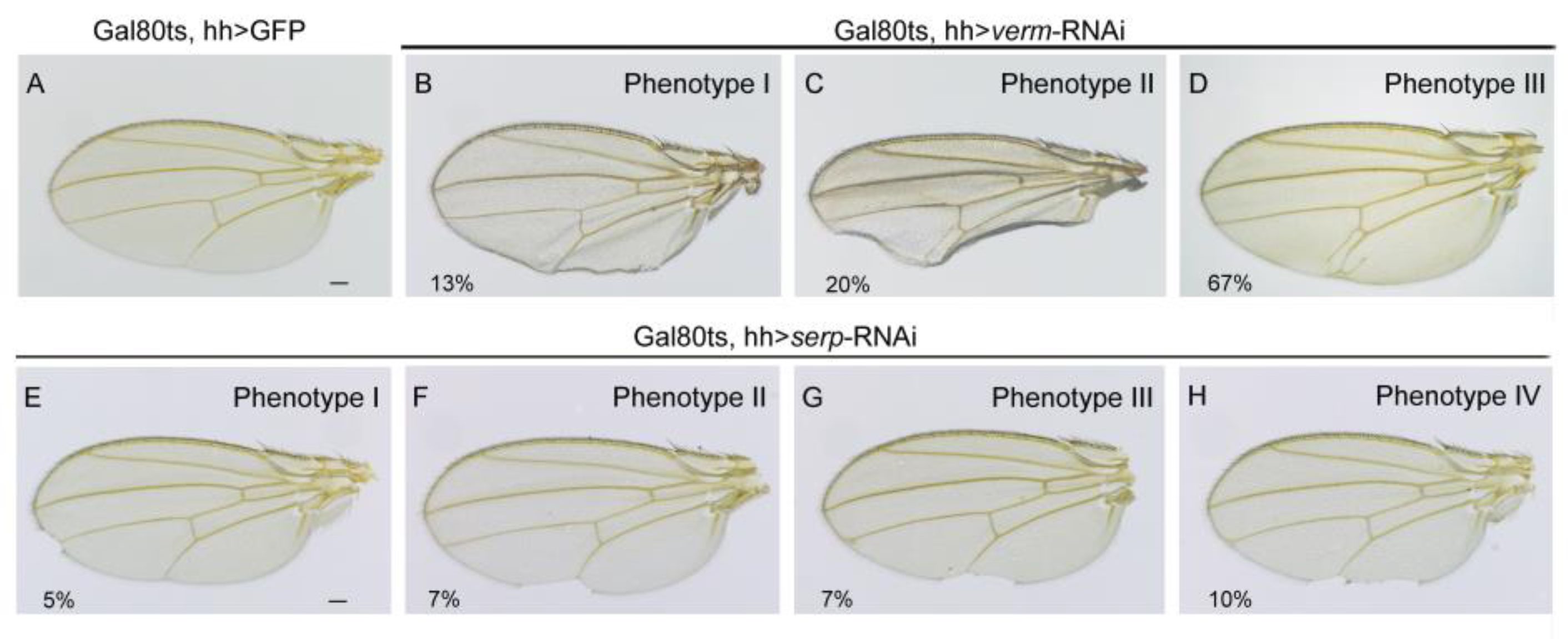
3.3. Knockdown of Serp and Verm in Posterior Areas Caused Wing Deficiency In Situ
3.4. Wing Deformations Caused by Serp or Verm Suppression Were Not Due to Cell Apoptosis of Wing Disc
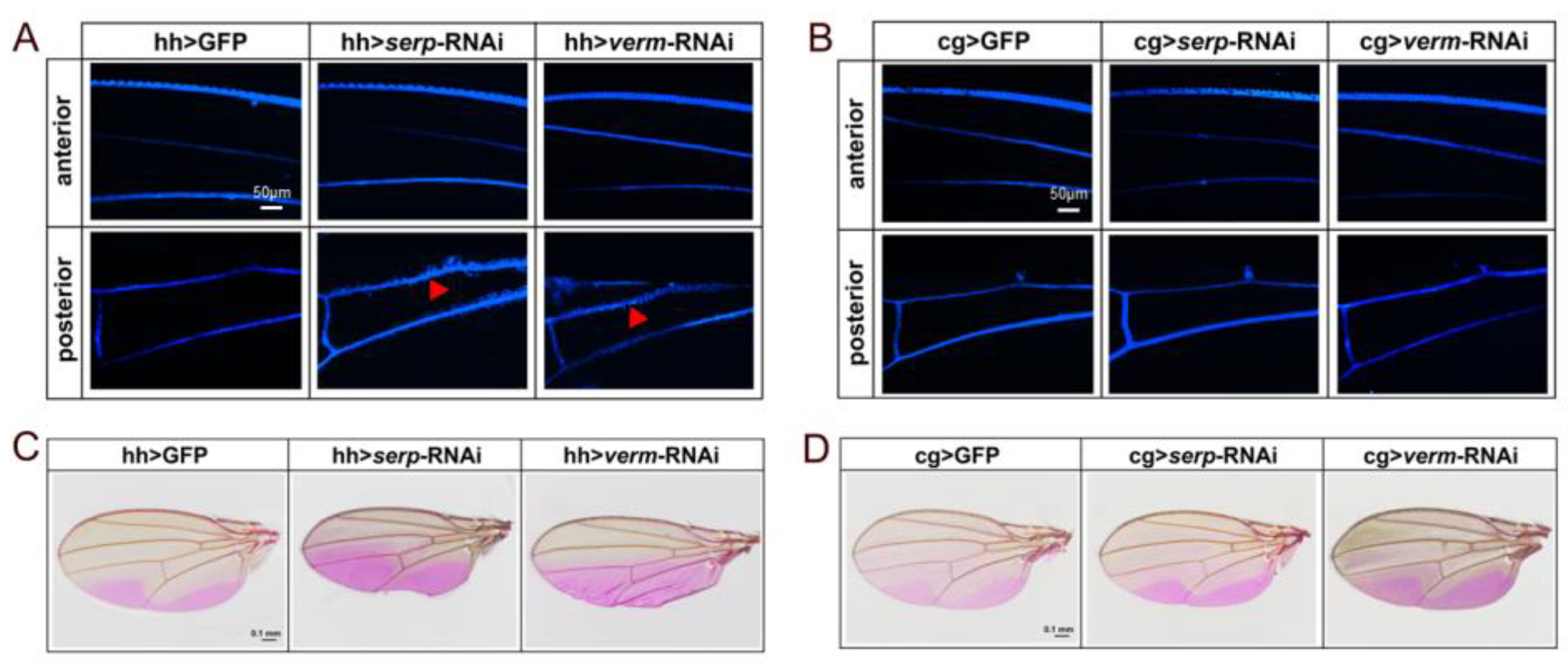
3.5. Repressing Serp and Verm in the Posterior Compartment Had no Non-Autonomous Effect on the Barrier Function of the Anterior Compartment
4. Discussion
4.1. Serp and Verm Play an Important Role in Wing Development of D. melanogaster
4.2. Serp or Verm Function in the Wing Is Independent of Fat Body–Hemolymph Transport System
4.3. Serp and Verm Were Produced Autonomously in the Wing
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, J.; Bhushan, B. Structure and mechanical properties of beetle wings: A review. R. Soc. Chem. 2019, 2, 12606–12623. [Google Scholar] [CrossRef]
- Brosson, D.; Kuhn, L.; Prensier, G.; Vivarès, C.P.; Texier, C. The putative chitin deacetylase of Encephalitozoon cuniculi: A surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 2005, 247, 81–90. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor rouxii. PNAS 1993, 90, 2564–2568. [Google Scholar] [CrossRef]
- Tsigos, I.; Bouriotis, V. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef]
- Shao, Z.; Thomas, Y.; Hembach, L.; Xing, X.; Duan, D.; Moerschbacher, B.M.; Bulone, V.; Tirichine, L.; Bowler, C. Comparative characterization of putative chitin deacetylases from Phaeodactylum tricornutum and Thalassiosira pseudonana highlights the potential for distinct chitin-based metabolic processes in diatoms. New Phytol. 2019, 221, 1890–1905. [Google Scholar] [CrossRef]
- Wang, C.Y.; Cheng, J.; Lyu, Z.H.; Li, Z.X.; Chen, J.X.; Lin, T. Chitin deacetylase 1 and 2 are indispensable for larval–pupal and pupal–adult molts in Heortia vitessoides (Lepidoptera: Crambidae). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 237, 110325. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef]
- Cohen, E.; Moussian, B. Extracellular Composite Matrices in Arthropods. Camb. Int. Law J. 2016, 15, 44–57. [Google Scholar]
- Win, N.; Stevens, W. Shrimp chitin as substrate for fungal chitin deacetylase. Appl. Microbiol. Biotechnol. 2001, 57, 334–341. [Google Scholar] [PubMed]
- Sarmiento, K.P.; Panes, V.A.; Santos, M.D. Molecular cloning and expression of chitin deacetylase 1 gene from the gills of Penaeus monodon (black tiger shrimp). Fish Shellfish. Immunol. 2016, 55, 484–489. [Google Scholar] [CrossRef]
- Heustis, R.J.; Ng, H.K.; Brand, K.J.; Rogers, M.C.; Le, L.T.; Specht, C.A.; Fuhrman, J.A. Pharyngeal polysaccharide deacetylases affect development in the nematode C. elegans and deacetylate chitin in vitro. PLoS ONE 2012, 7, e40426. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Liu, W.; Li, D.; Zhao, X.; Ding, G.; Zhang, M.; Ma, E.; Zhu, K.; Li, S.; Moussian, B.; et al. Helicoidal Organization of Chitin in the Cuticle of the Migratory Locust Requires the Function of the Chitin Deacetylase2 Enzyme (LmCDA2). J. Biol. Chem. 2016, 291, 24352–24363. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.A.; Qiang, Z.; Shu, Y.D.; Xiao, H.; Bu, C. Comparative characterization of putative chitin deacetylases from Tetranychus cinnabarinus. Biosci. Biotechnol. Biochem. 2019, 83, 1306–1309. [Google Scholar]
- Zhang, Z.; Yan, J.; Liu, Q.; Zhang, Y.; Gong, J.; Hou, Y. Genome-Wide Analysis and Hormone Regulation of Chitin Deacetylases in Silkworm. Int. J. Mol. Sci. 2019, 20, 1679. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, G.; Pang, Y.; Wang, P. A novel chitin-binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 2005, 35, 1224–1234. [Google Scholar] [CrossRef]
- Yu, R.R.; Liu, W.M.; Zhao, X.M.; Zhang, M.; Li, D.Q.; Zuber, R.; Ma, E.B.; Zhu, K.Y.; Moussian, B.; Zhang, J.Z. LmCDA1 organizes the cuticle by chitin deacetylation in Locusta migratoria. Insect Mol. Biol. 2019, 28, 301–312. [Google Scholar] [CrossRef]
- Zhang, M.; Ji, Y.; Zhang, X.; Ma, P.; Wang, Y.; Moussian, B.; Zhang, J. The putative chitin deacetylases Serpentine and Vermiform have non-redundant functions during Drosophila wing development. Insect Biochem. Mol. Biol. 2019, 110, 128–135. [Google Scholar] [CrossRef]
- Luschnig, S.; Batz, T.; Armbruster, K.; Krasnow, M.A. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef]
- Wang, S.; Jayaram, S.A.; Hemphälä, J.; Senti, K.-A.; Tsarouhas, V.; Jin, H.; Samakovlis, C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006, 16, 180–185. [Google Scholar] [CrossRef]
- Liu, L.; Qu, M.; Liu, T.; Chen, Q.; Guo, X.; Yang, J.; Yang, Q. Biochemical characterization of three midgut chitin deacetylases of the Lepidopteran insect Bombyx mori. J. Insect Physiol. 2019, 113, 42–48. [Google Scholar] [CrossRef]
- Arakane, Y.; Dixit, R.; Begum, K.; Park, Y.; Specht, C.A.; Merzendorfer, H.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2009, 39, 355–365. [Google Scholar] [CrossRef]
- Dong, B.; Miao, G.; Hayashi, S. A fat body-derived apical extracellular matrix enzyme is transported to the tracheal lumen and is required for tube morphogenesis in Drosophila. Development 2014, 141, 4104–4109. [Google Scholar] [CrossRef]
- Chapman, R.F.; Chapman, R.F. The Insects: Structure and function; Cambridge university press: Cambridge, UK, 1998. [Google Scholar]
- Wang, Y.; Yu, Z.; Zhang, J.; Moussian, B. Regionalization of surface lipids in insects. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152994. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Carballo, R.G.; Moussian, B. Double cuticle barrier in two global pests, the whitefly Trialeurodes vaporariorum and the bedbug Cimex lectularius. J. Exp. Biol. 2017, 220, 1396–1399. [Google Scholar]
- Flaven-Pouchon, J.; Moussian, B. Fluorescent microscopy-based detection of chitin in intact Drosophila melanogaster. Front. Physiol. 2022, 694, 856369. [Google Scholar] [CrossRef]
- Otaki, J.M.; Nakazato, Y. Butterfly Wing Color Pattern Modification Inducers May Act on Chitin in the Apical Extracellular Site: Implications in Morphogenic Signals for Color Pattern Determination. Biology 2022, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Toprak, U.; Baldwin, D.; Erlandson, M.; Gillott, C.; Hou, X.; Coutu, C.; Hegedus, D.D. A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol. Biol. 2008, 17, 573–585. [Google Scholar] [CrossRef]
- Noh, M.Y.; Muthukrishnan, S.; Kramer, K.J.; Arakane, Y. Group I chitin deacetylases are essential for higher order organization of chitin fibers in beetle cuticle. J. Biol. Chem. 2018, 293, 6985–6995. [Google Scholar] [CrossRef]
- Zheng, H.; Yang, X.; Xi, Y. Fat body remodeling and homeostasis control in Drosophila. Life Sci. 2016, 167, 22–31. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ji, Y.; Moussian, B.; Yang, S.; Zhang, J.; Zhang, T.; Zhang, M. Serpentine and Vermiform Are Produced Autonomously to Fulfill Their Function in Drosophila Wings. Insects 2023, 14, 406. https://doi.org/10.3390/insects14050406
Zhang X, Ji Y, Moussian B, Yang S, Zhang J, Zhang T, Zhang M. Serpentine and Vermiform Are Produced Autonomously to Fulfill Their Function in Drosophila Wings. Insects. 2023; 14(5):406. https://doi.org/10.3390/insects14050406
Chicago/Turabian StyleZhang, Xubo, Yanan Ji, Bernard Moussian, Shumin Yang, Jianzhen Zhang, Tingting Zhang, and Min Zhang. 2023. "Serpentine and Vermiform Are Produced Autonomously to Fulfill Their Function in Drosophila Wings" Insects 14, no. 5: 406. https://doi.org/10.3390/insects14050406
APA StyleZhang, X., Ji, Y., Moussian, B., Yang, S., Zhang, J., Zhang, T., & Zhang, M. (2023). Serpentine and Vermiform Are Produced Autonomously to Fulfill Their Function in Drosophila Wings. Insects, 14(5), 406. https://doi.org/10.3390/insects14050406







