Novel Insights into the circRNA-Modulated Developmental Mechanism of Western Honey Bee Larval Guts
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bee Larvae
2.2. RNA-seq Data Source
2.3. sRNA-seq Data Source
2.4. Identification and Expression Level Calculation of circRNAs
2.5. Screening of DEcircRNAs
2.6. Parental Gene Prediction and Annotation
2.7. Analysis of ceRNA Regulatory Network
2.8. PCR Validation of DEcircRNAs
2.9. RT-qPCR Detection of DEcircRNAs
3. Results
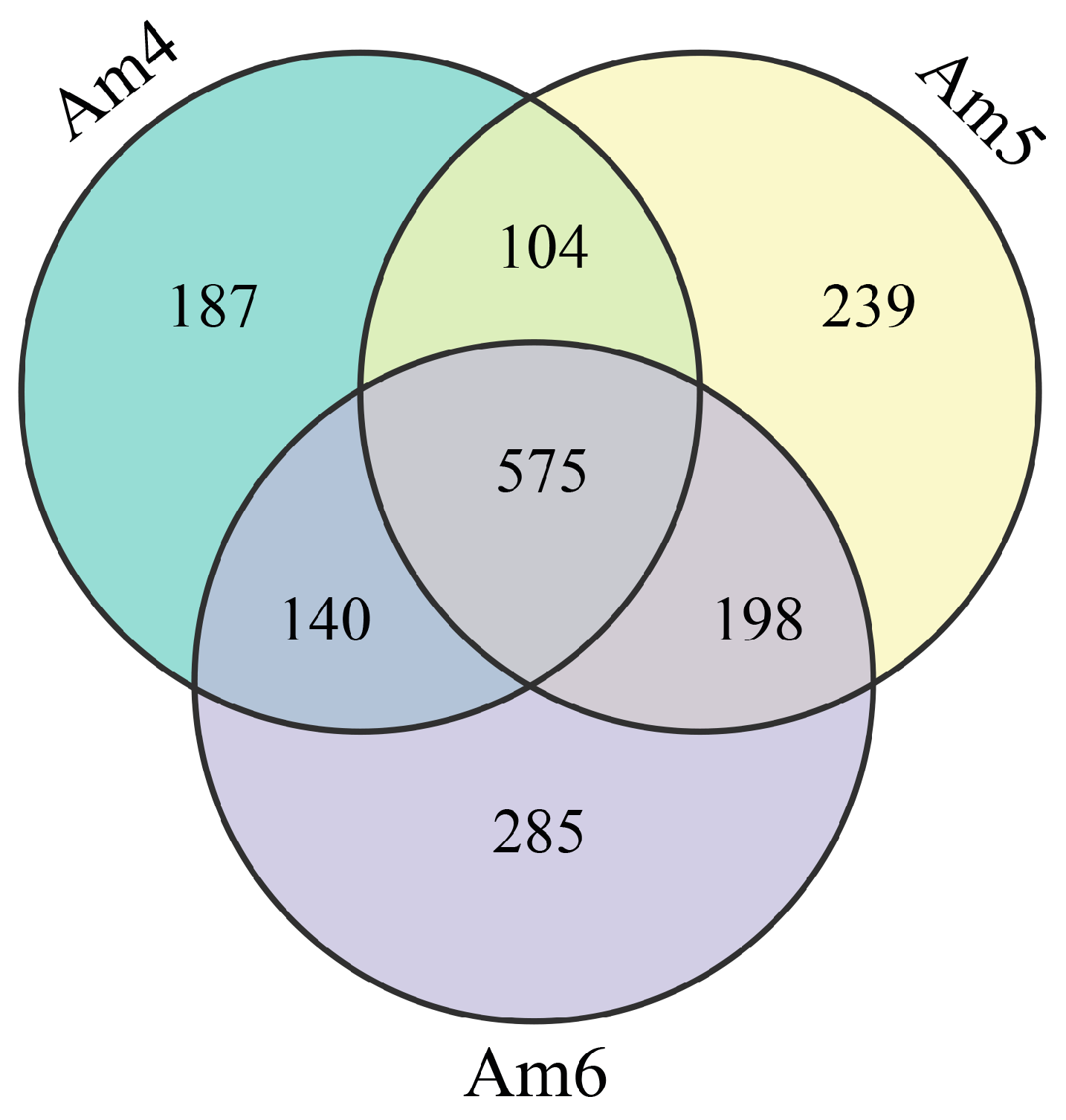
3.1. Highly Expressed circRNAs in the Gut Tissues of A. m. ligustica Worker Larvae
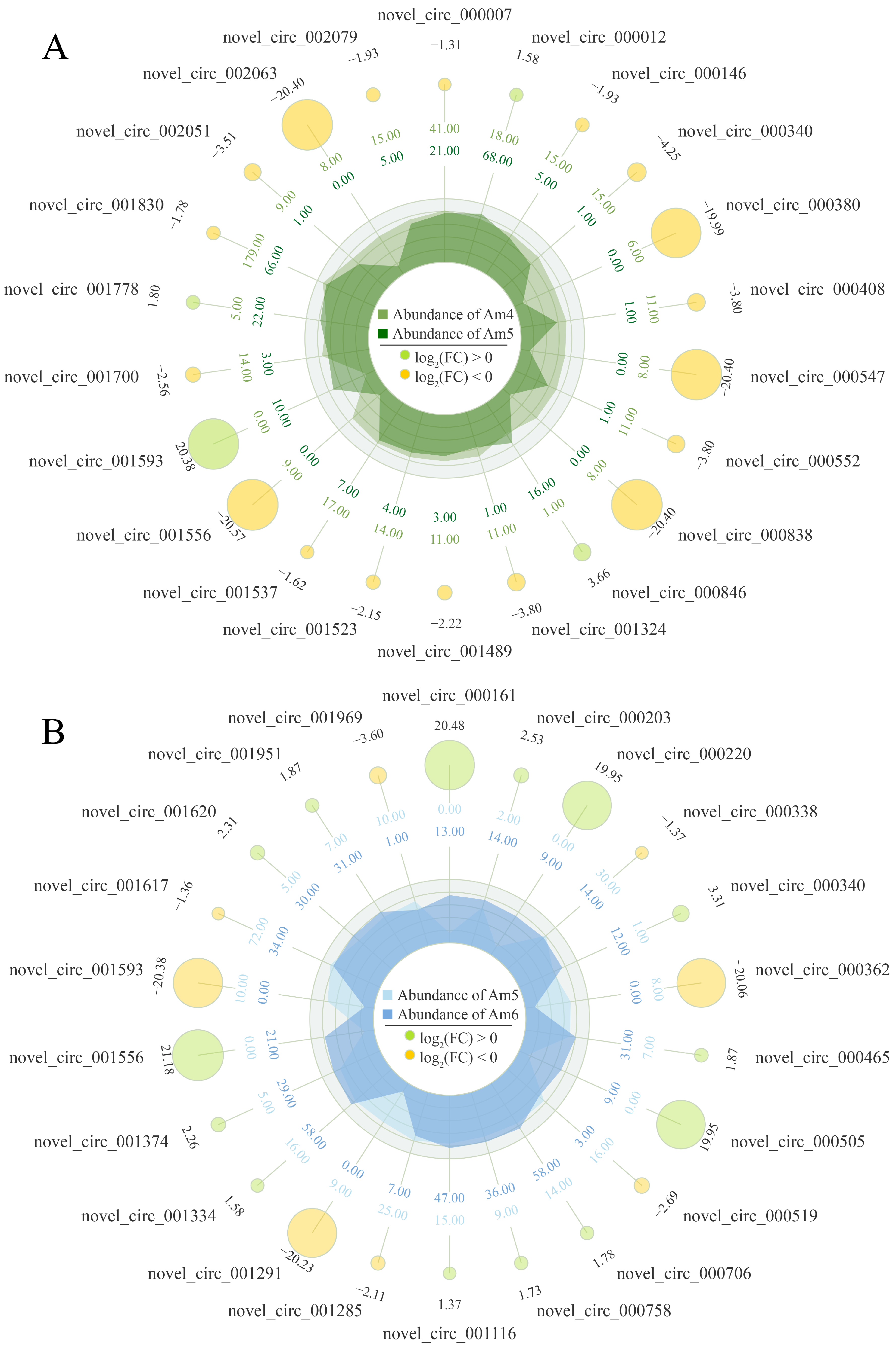
3.2. Dynamic Expression Profile of circRNAs in the Developmental Process of Larval Gut Tissues
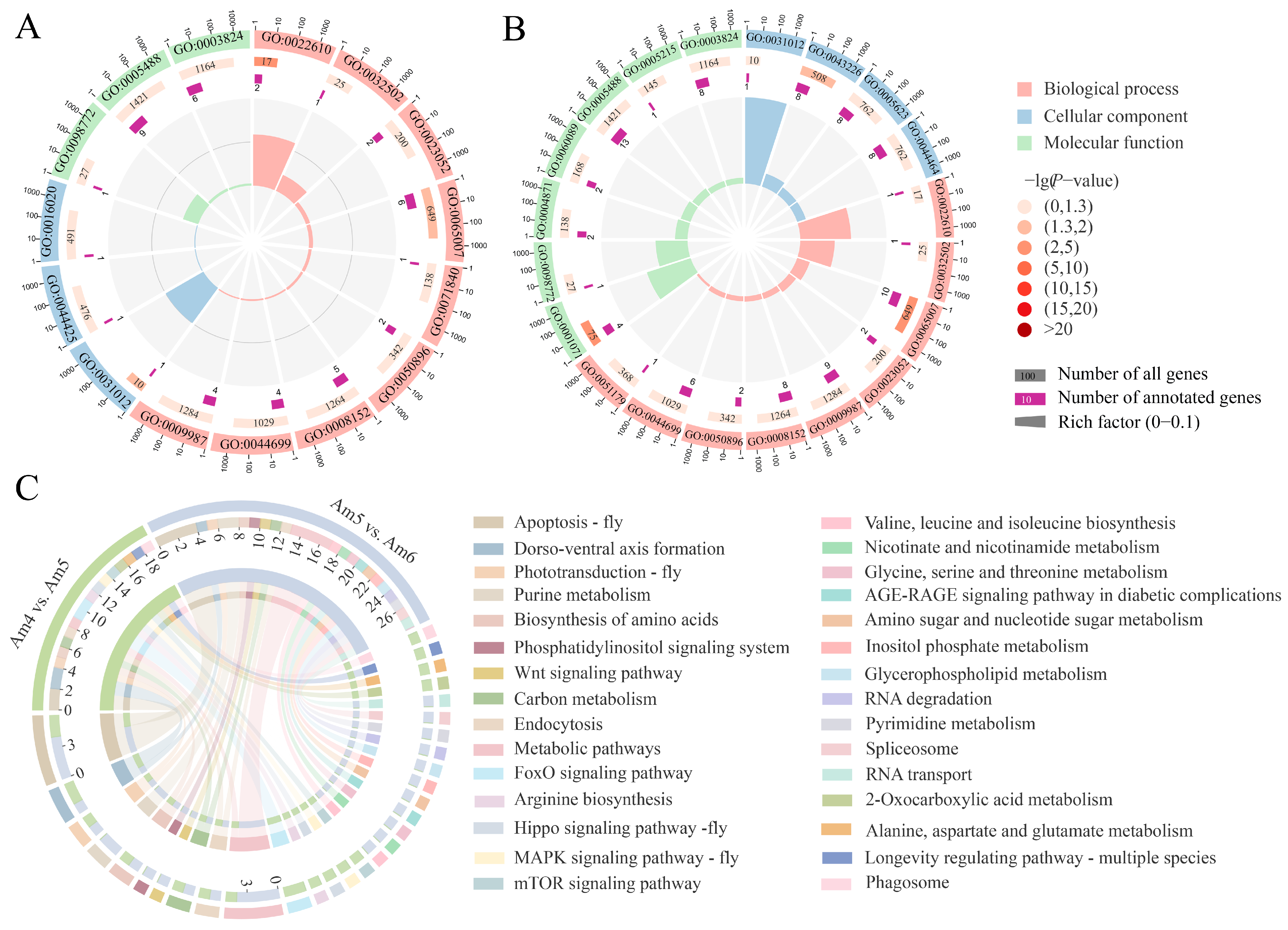
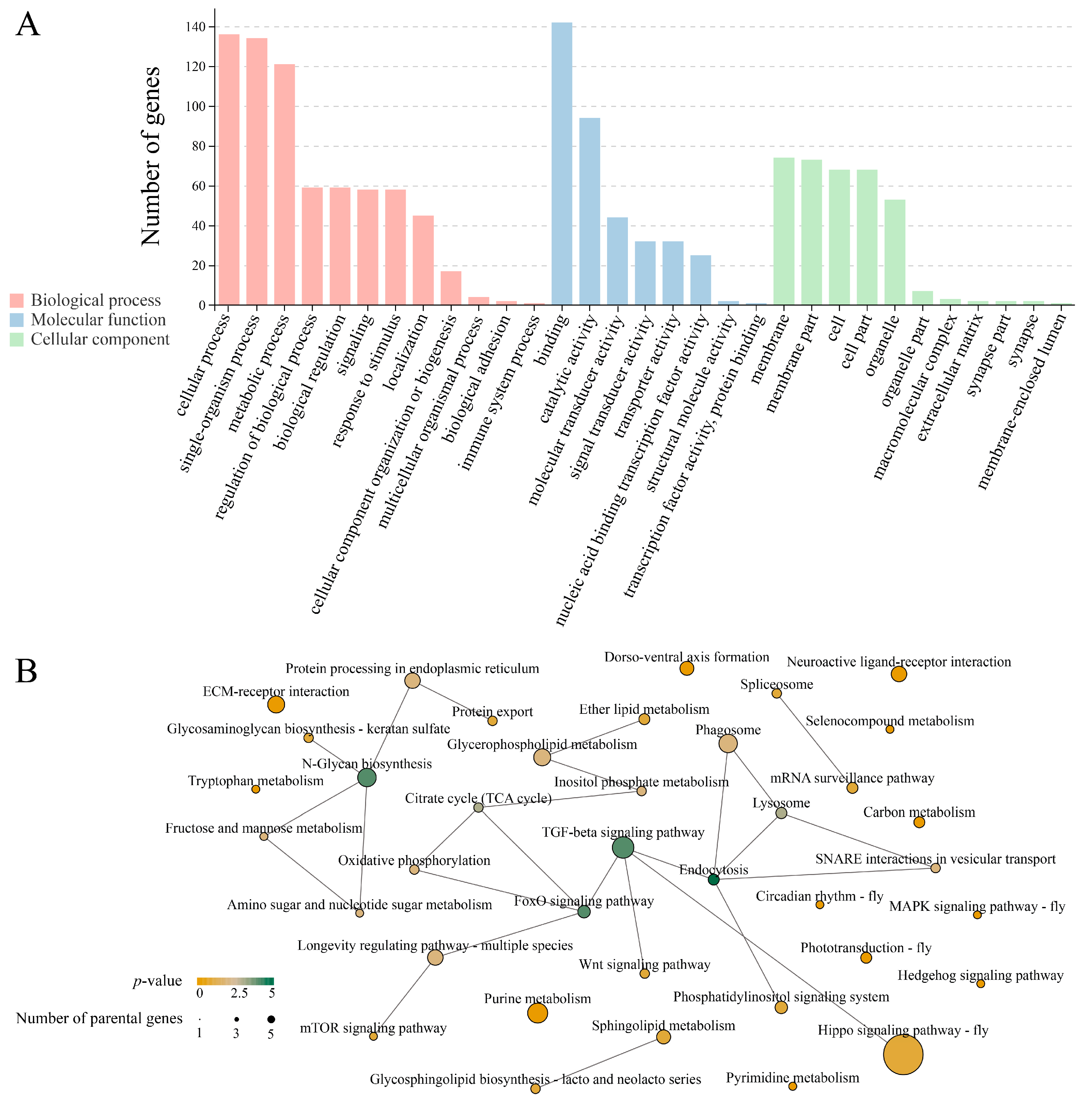
3.3. Analysis of DEcircRNAs’ Parental Genes
3.4. Investigation of DEcircRNA-Engaged Regulatory Networks
3.5. Analysis of DEcircRNA-Involved Sub-Networks Relative to Developmental Signaling Pathways
3.6. Investigation of DEcircRNA-Involved Sub-Networks Associated with Humoral and Cellular Immune Pathways
3.7. Validation of DEcircRNAs Based on PCR and RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Bunikis, I.; Pettersson, O.V.; Mosbech, M.B.; Childers, A.K.; Evans, J.D.; Mikheyev, A.S.; Robertson, H.M.; Robinson, G.E.; Webster, M.T. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genom. 2019, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Calin, G.A. Circular RNAs in cancer-lessons learned from microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2016, 34, e63. [Google Scholar] [CrossRef]
- Mackie, G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 1998, 395, 720–723. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Gan, H.; Feng, T.; Wu, Y.; Liu, C.; Xia, Q.; Cheng, T. Identification of circular RNA in the Bombyx mori silk gland. Insect Biochem. Mol. Biol. 2017, 89, 97–106. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef]
- Ren, Y.; Yue, H.; Li, L.; Xu, Y.; Wang, Z.; Xin, Z.; Lin, T. Identification and characterization of circRNAs involved in the regulation of low nitrogen-promoted root growth in hexaploid wheat. Biol. Res. 2018, 51, 43. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, D.; Chen, H.; Fu, Z.; Xiong, C.; Hou, C.; Zheng, Y.; Guo, Y.; Wang, H.; Du, Y.; et al. Systematic investigation of circular RNAs in Ascosphaera apis, a fungal pathogen of honeybee larvae. Gene 2018, 678, 17–22. [Google Scholar] [CrossRef]
- Guo, R.; Chen, D.; Chen, H.; Xiong, C.; Zheng, Y.; Hou, C.; Du, Y.; Geng, S.; Wang, H.; Dingding, Z.; et al. Genome-wide identification of circular RNAs in fungal parasite Nosema ceranae. Curr. Microbiol. 2018, 75, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C. Evolutionary trends of digestion and absorption in the major insect orders. Arthropod Struct. Dev. 2020, 56, 100931. [Google Scholar] [CrossRef]
- Holtof, M.; Lenaerts, C.; Cullen, D.; Vanden Broeck, J. Extracellular nutrient digestion and absorption in the insect gut. Cell Tissue Res. 2019, 377, 397–414. [Google Scholar] [CrossRef]
- Zeng, T.; Jaffar, S.; Xu, Y.; Qi, Y. The Intestinal Immune Defense System in Insects. Int. J. Mol. Sci. 2022, 23, 15132. [Google Scholar] [CrossRef] [PubMed]
- Foronda, D.; Weng, R.; Verma, P.; Chen, Y.W.; Cohen, S.M. Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes. Dev. 2014, 28, 2421–2431. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fu, Y.; Cheng, H.; Zhao, C.; Huang, Q.; Chang, M.; Qiu, W.; Shen, Y.; Li, D. lncR26319/miR-2834/EndophilinA axis regulates oogenesis of the silkworm, Bombyx mori. Insect Sci. 2023, 30, 65–80. [Google Scholar] [CrossRef]
- Chen, X.; Shi, W.; Chen, C. Differential circular RNAs expression in ovary during oviposition in honey bees. Genomics 2019, 111, 598–606. [Google Scholar] [CrossRef]
- Thölken, C.; Thamm, M.; Erbacher, C.; Lechner, M. Sequence and structural properties of circular RNAs in the brain of nurse and forager honeybees (Apis mellifera). BMC Genom. 2019, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Fan, X.; Cai, Z.; Wu, Y.; Zhang, W.; Zhao, H.; Guo, S.; Feng, P.; Li, Q.; Zou, P.; et al. Unveiling the circRNA-mediated immune responses of western honey bee larvae to Ascosphaera apis invasion. Int. J. Mol. Sci. 2022, 24, 613. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, J.; Fan, X.; Long, Q.; Chen, H.; Ye, Y.; Zhang, K.; Ren, Z.; Zhang, Y.; Niu, Q.; et al. CircRNA-regulated immune responses of asian honey bee workers to microsporidian infection. Front. Genet. 2022, 13, 1013239. [Google Scholar] [CrossRef]
- Chen, D.; Chen, H.; Du, Y.; Zhu, Z.; Wang, J.; Geng, S.; Xiong, C.; Zheng, Y.; Hou, C.; Diao, Q.; et al. Systematic identification of circular RNAs and corresponding regulatory networks unveil their potential roles in the midguts of eastern honeybee workers. Appl. Microbiol. Biotechnol. 2020, 104, 257–276. [Google Scholar] [CrossRef]
- Nation, J.L. Insect Physiology and Biochemistry, 4th ed.; CRC Press: Boca Raton, FL, USA, 2022; pp. 27–33. [Google Scholar] [CrossRef]
- Caccia, S.; Casartelli, M.; Tettamanti, G. The amazing complexity of insect midgut cells: Types, peculiarities, and functions. Cell Tissue Res. 2019, 377, 505–525. [Google Scholar] [CrossRef]
- Ye, Y.; Fan, X.; Long, Q.; Wang, J.; Zhang, W.; Cai, Z.; Sun, M.; Gu, X.; Zou, P.; Chen, D.; et al. Comprehensive investigation and regulatory function of lncRNAs engaged in western honey bee larval immune response to Ascosphaera apis invasion. Front. Physiol. 2022, 13, 1082522. [Google Scholar] [CrossRef] [PubMed]
- Forfert, N.; Natsopoulou, M.E.; Frey, E.; Rosenkranz, P.; Paxton, R.J.; Moritz, R.F. Parasites and Pathogens of the Honeybee (Apis mellifera) and Their Influence on Inter-Colonial Transmission. PLoS ONE 2015, 10, e0140337. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Guo, R.; Du, Y.; Xiong, C.L.; Zheng, Y.Z.; Fu, Z.M.; Xu, G.J.; Wang, H.P.; Chen, H.Z.; Geng, S.H.; Zhou, D.D.; et al. Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Sci. Agric. Sin. 2018, 51, 4197–4209. (In Chinese) [Google Scholar] [CrossRef]
- Burge, S.W.; Daub, J.; Eberhardt, R.; Tate, J.; Barquist, L.; Nawrocki, E.P.; Eddy, S.R.; Gardner, P.P.; Bateman, A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013, 41, D226–D232. [Google Scholar] [CrossRef]
- Qin, H.; Peng, J.; Liu, L.; Wu, J.; Pan, L.; Huang, X.; Huang, M.; Qiu, H.; Du, B.; China Critical Care Clinical Trials Group (CCCCTG). A Retrospective Paired Comparison between Untargeted Next Generation Sequencing and Conventional Microbiology Tests with Wisely Chosen Metagenomic Sequencing Positive Criteria. Front. Med. 2021, 8, 686247. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Li, S.Y.; Wang, C.Y.; Xiao, Y.X.; Tang, X.B.; Yuan, Z.W.; Bai, Y.Z. RNA-Seq Profiling of Circular RNAs During Development of Hindgut in Rat Embryos with Ethylenethiourea-Induced Anorectal Malformations. Front. Genet. 2021, 12, 605015. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mitteer, D.R.; Greer, B.D.; Fisher, W.W.; Cohrs, V.L. Teaching behavior technicians to create publication-quality, single-case design graphs in graphpad prism 7. J. Appl. Behav. Anal. 2018, 51, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; An, S.; Cheng, P.; Zhang, K.; Gong, M.; Zhang, Z.; Zhang, R. Whole-transcriptome profiling across different developmental stages of Aedes albopictus (Diptera: Culicidae) provides insights into chitin-related non-coding RNA and competing endogenous RNA networks. Parasit. Vectors 2023, 16, 33. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Dai, K.; Liang, Z.; Zhu, M.; Zhang, M.; Pan, J.; Hu, X.; Zhang, X.; Xue, R.; et al. circEgg regulates histone H3K9me3 by sponging bmo-miR-3391-5p and encoding circEgg-P122 protein in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2020, 124, 103430. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, J.; Zhang, J.; Zhang, K.; Gu, X.; Yao, Y.; Ren, Z.; Zhang, Y.; Chen, D.; Guo, R. Circular RNA ame_circ_000115 regulates expression of genes in larval gusts of Apis mellifera ligustica stressed by Ascosphaera apis. Chin. J. Biotechnol. 2023, 39, 217–230. (In Chinese) [Google Scholar] [CrossRef]
- Van der Graaf, K.; Jindrich, K.; Mitchell, R.; White-Cooper, H. Roles for RNA export factor, Nxt1, in ensuring muscle integrity and normal RNA expression in Drosophila. G3 2021, 11, jkaa046. [Google Scholar] [CrossRef]
- Weigelt, C.M.; Sehgal, R.; Tain, L.S.; Cheng, J.; Eßer, J.; Pahl, A.; Dieterich, C.; Grönke, S.; Partridge, L. An insulin-sensitive circular RNA that regulates lifespan in Drosophila. Mol. Cell 2020, 79, 268–279.e5. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Wu, W.; Dong, Y.; Wang, M.; Yi, D.; Zhou, Y.; Xu, Q. Systematic identification and functional analysis of circular RNAs during Rrice black-streaked dwarf virus infection in the Laodelphax striatellus (Fallén) midgut. Front. Microbiol. 2020, 11, 588009. [Google Scholar] [CrossRef]
- Fan, Y.X.; Andoh, V.; Chen, L. Multi-omics study and ncRNA regulation of anti-BmNPV in silkworms, Bombyx mori: An update. Front. Microbiol. 2023, 14, 1123448. [Google Scholar] [CrossRef] [PubMed]
- Fessler, J.H.; Kramerova, I.; Kramerov, A.; Chen, Y.; Fessler, L.I. Papilin, a novel component of basement membranes, in relation to ADAMTS metalloproteases and ECM development. Int. J. Biochem. Cell Biol. 2004, 36, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Kramerova, I.A.; Kawaguchi, N.; Fessler, L.I.; Nelson, R.E.; Chen, Y.; Kramerov, A.A.; Kusche-Gullberg, M.; Kramer, J.M.; Ackley, B.D.; Sieron, A.L.; et al. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 2000, 127, 5475–5485. [Google Scholar] [CrossRef]
- Kramerova, I.A.; Kramerov, A.A.; Fessler, J.H. Alternative splicing of papilin and the diversity of Drosophila extracellular matrix during embryonic morphogenesis. Dev. Dyn. 2003, 226, 634–642. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, W.; Zhao, X.; Yu, Z.; Guo, H.; Yang, Y.; Zhang, J.; Moussian, B.; Zhang, J. Apolipophorin-II/I Contributes to Cuticular Hydrocarbon Transport and Cuticle Barrier Construction in Locusta migratoria. Front. Physiol. 2020, 11, 790. [Google Scholar] [CrossRef]
- Stączek, S.; Zdybicka-Barabas, A.; Mak, P.; Sowa-Jasiłek, A.; Kedracka-Krok, S.; Jankowska, U.; Suder, P.; Wydrych, J.; Grygorczuk, K.; Jakubowicz, T.; et al. Studies on localization and protein ligands of Galleria mellonella apolipophorin III during immune response against different pathogens. J. Insect Physiol. 2018, 105, 18–27. [Google Scholar] [CrossRef]
- Kamareddine, L.; Nakhleh, J.; Osta, M.A. Functional Interaction between Apolipophorins and Complement Regulate the Mosquito Immune Response to Systemic Infections. J. Innate Immun. 2016, 8, 314–326. [Google Scholar] [CrossRef]
- Abbas, M.N.; Kausar, S.; Gul, I.; Li, J.; Yu, H.; Dong, M.; Cui, H. The potential biological roles of circular RNAs in the immune systems of insects to pathogen invasion. Genes 2023, 14, 895. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, M.; Liu, B.; Liang, Z.; Huang, L.; Xu, J.; Yu, L.; Li, K.; Jiang, M.; Xue, R.; et al. Circular RNA alterations in the Bombyx mori midgut following B. mori nucleopolyhedrovirus infection. Mol. Immunol. 2018, 101, 461–470. [Google Scholar] [CrossRef]
- Pandey, A.; Galeone, A.; Han, S.Y.; Story, B.A.; Consonni, G.; Mueller, W.F.; Steinmetz, L.M.; Vaccari, T.; Jafar-Nejad, H. Gut barrier defects, intestinal immune hyperactivation and enhanced lipid catabolism drive lethality in NGLY1-deficient Drosophila. Nat. Commun. 2023, 14, 5667. [Google Scholar] [CrossRef]
- Eleftherianos, I.; Heryanto, C.; Bassal, T.; Zhang, W.; Tettamanti, G.; Mohamed, A. Haemocyte-mediated immunity in insects: Cells, processes and associated components in the fight against pathogens and parasites. Immunology 2021, 164, 401–432. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Jing, X.; Douglas, A.E. The multi-tasking gut epithelium of insects. Insect Biochem. Mol. Biol. 2015, 67, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef]
- Militello, G.; Weirick, T.; John, D.; Döring, C.; Dimmeler, S.; Uchida, S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 2017, 18, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, L.; Chen, Y.; Liu, P.; Zhou, Y.; Chen, X.; Gu, J. Aal-circRNA-407 regulates ovarian development of Aedes albopictus, a major arbovirus vector, via the miR-9a-5p/Foxl axis. PLoS Pathog. 2023, 19, e1011374. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, Y.; Fan, X.; Zhao, H.; Zhang, Y.; Guo, S.; Jing, X.; Liu, Z.; Feng, P.; Liu, X.; et al. ame-miR-34 Modulates the larval body weight and immune response of Apis mellifera workers to Ascosphara apis invasion. Int. J. Mol. Sci. 2023, 24, 1214. [Google Scholar] [CrossRef]
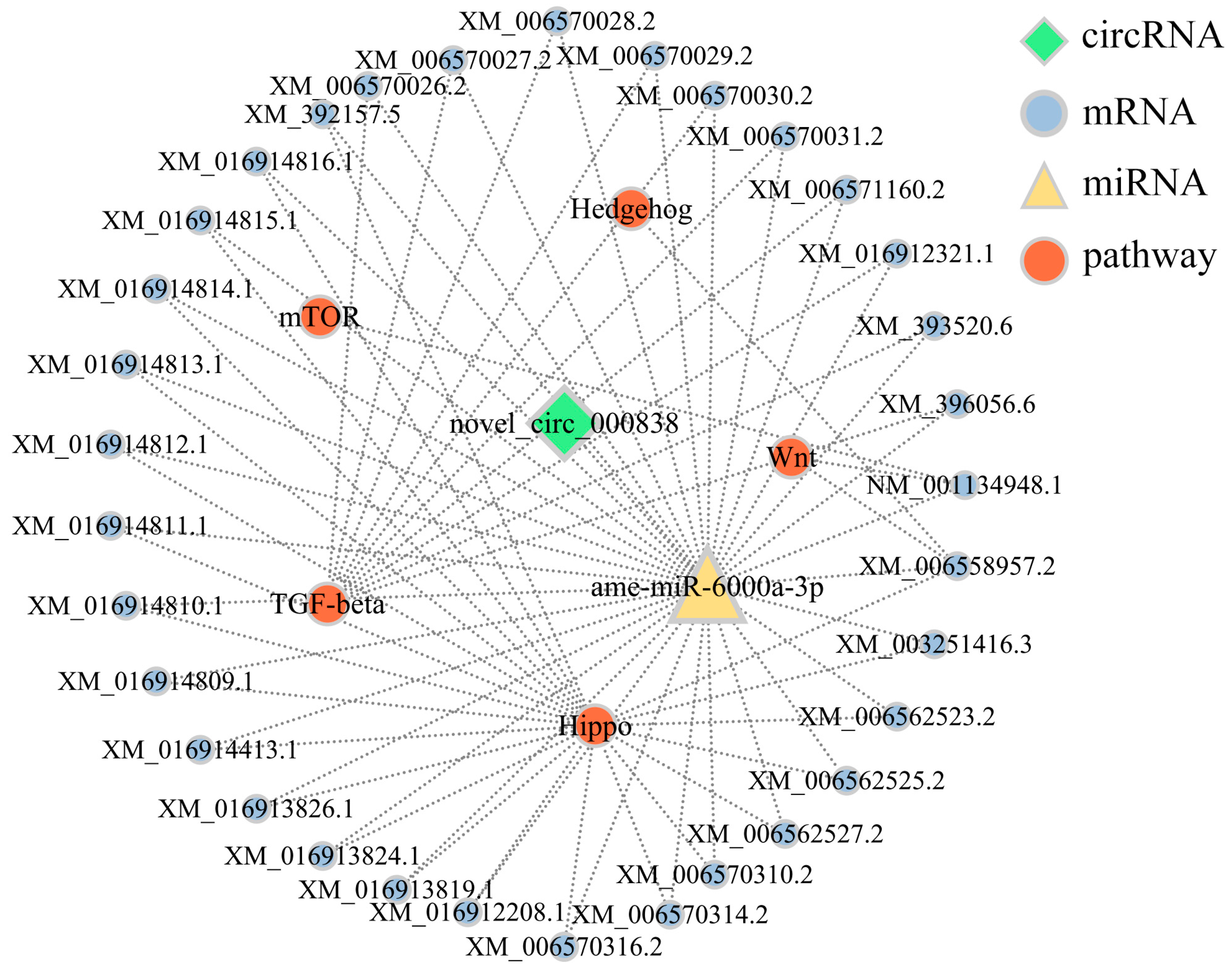



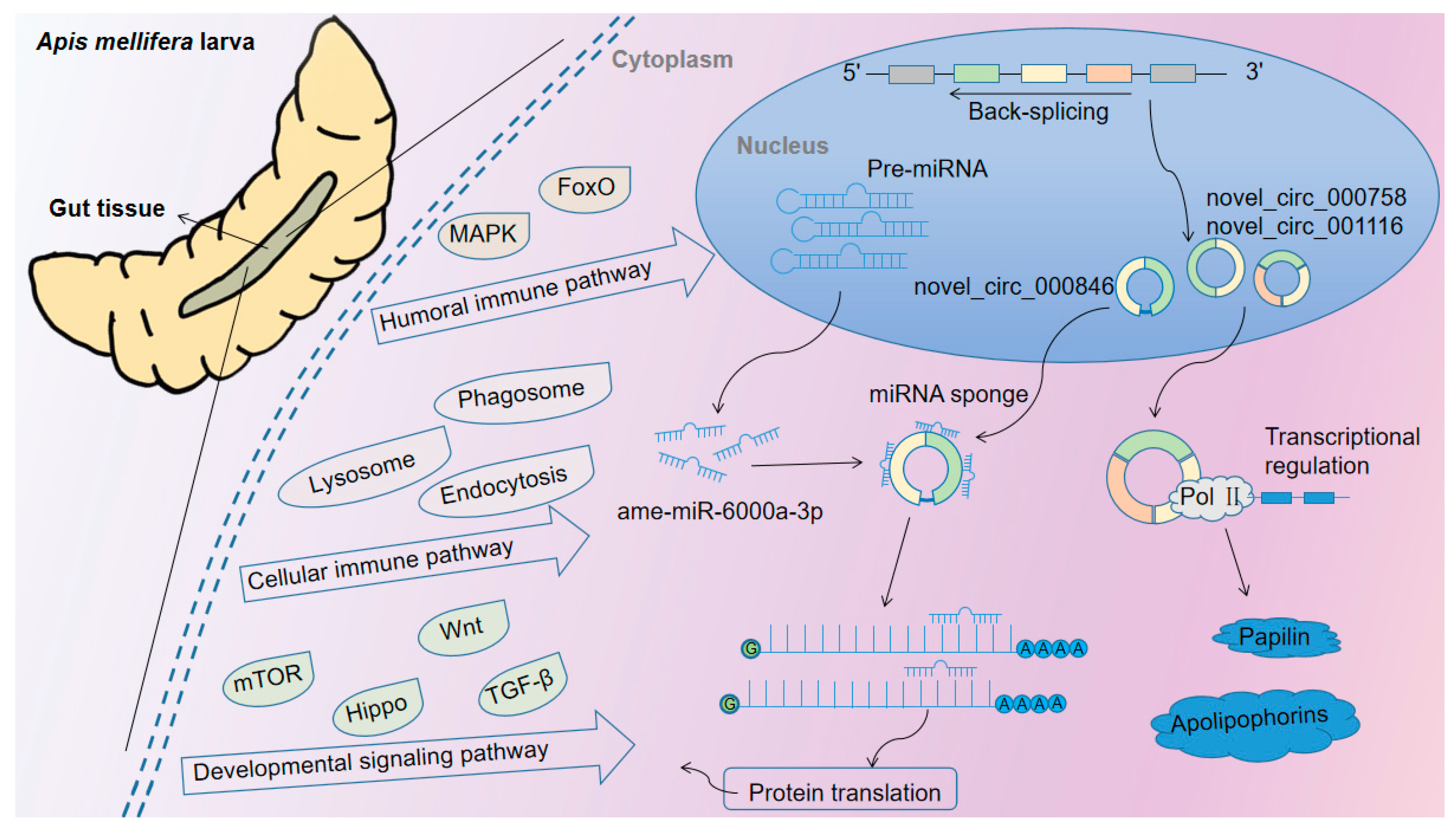
| CircRNA ID | Am4 Group RPM | Am5 Group RPM | Am6 Group RPM |
|---|---|---|---|
| novel_circ_000069 | 54,161.61966 | 67,741.05436 | 58,471.61079 |
| novel_circ_000027 | 46,028.72469 | 26,222.34362 | 21,108.47725 |
| novel_circ_000438 | 29,589.89445 | 50,942.36547 | 56,778.41743 |
| novel_circ_000115 | 28,378.61222 | 36,328.87189 | 28,897.16672 |
| novel_circ_000131 | 19,207.47534 | 19,393.6083 | 15,125.86071 |
| novel_circ_000630 | 16,957.9512 | 12,564.87299 | 14,900.10159 |
| novel_circ_000231 | 13,497.14483 | 17,071.8383 | 12,416.75133 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Fan, X.; Zang, H.; Liu, X.; Feng, P.; Ye, D.; Zhu, L.; Wu, Y.; Jiang, H.; Chen, D.; et al. Novel Insights into the circRNA-Modulated Developmental Mechanism of Western Honey Bee Larval Guts. Insects 2023, 14, 897. https://doi.org/10.3390/insects14110897
Zhang Y, Fan X, Zang H, Liu X, Feng P, Ye D, Zhu L, Wu Y, Jiang H, Chen D, et al. Novel Insights into the circRNA-Modulated Developmental Mechanism of Western Honey Bee Larval Guts. Insects. 2023; 14(11):897. https://doi.org/10.3390/insects14110897
Chicago/Turabian StyleZhang, Yiqiong, Xiaoxue Fan, He Zang, Xiaoyu Liu, Peilin Feng, Daoyou Ye, Leran Zhu, Ying Wu, Haibin Jiang, Dafu Chen, and et al. 2023. "Novel Insights into the circRNA-Modulated Developmental Mechanism of Western Honey Bee Larval Guts" Insects 14, no. 11: 897. https://doi.org/10.3390/insects14110897
APA StyleZhang, Y., Fan, X., Zang, H., Liu, X., Feng, P., Ye, D., Zhu, L., Wu, Y., Jiang, H., Chen, D., & Guo, R. (2023). Novel Insights into the circRNA-Modulated Developmental Mechanism of Western Honey Bee Larval Guts. Insects, 14(11), 897. https://doi.org/10.3390/insects14110897





