An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Species Identification
2.2. RNA Extraction and Sequencing
2.3. Virus Genome Identification and Small RNA Analysis
2.4. Virus Genome Annotation
2.5. Phylogenetic Analyses
3. Results
3.1. Discovery of RNA Virus-Related Sequences in A. sinensis
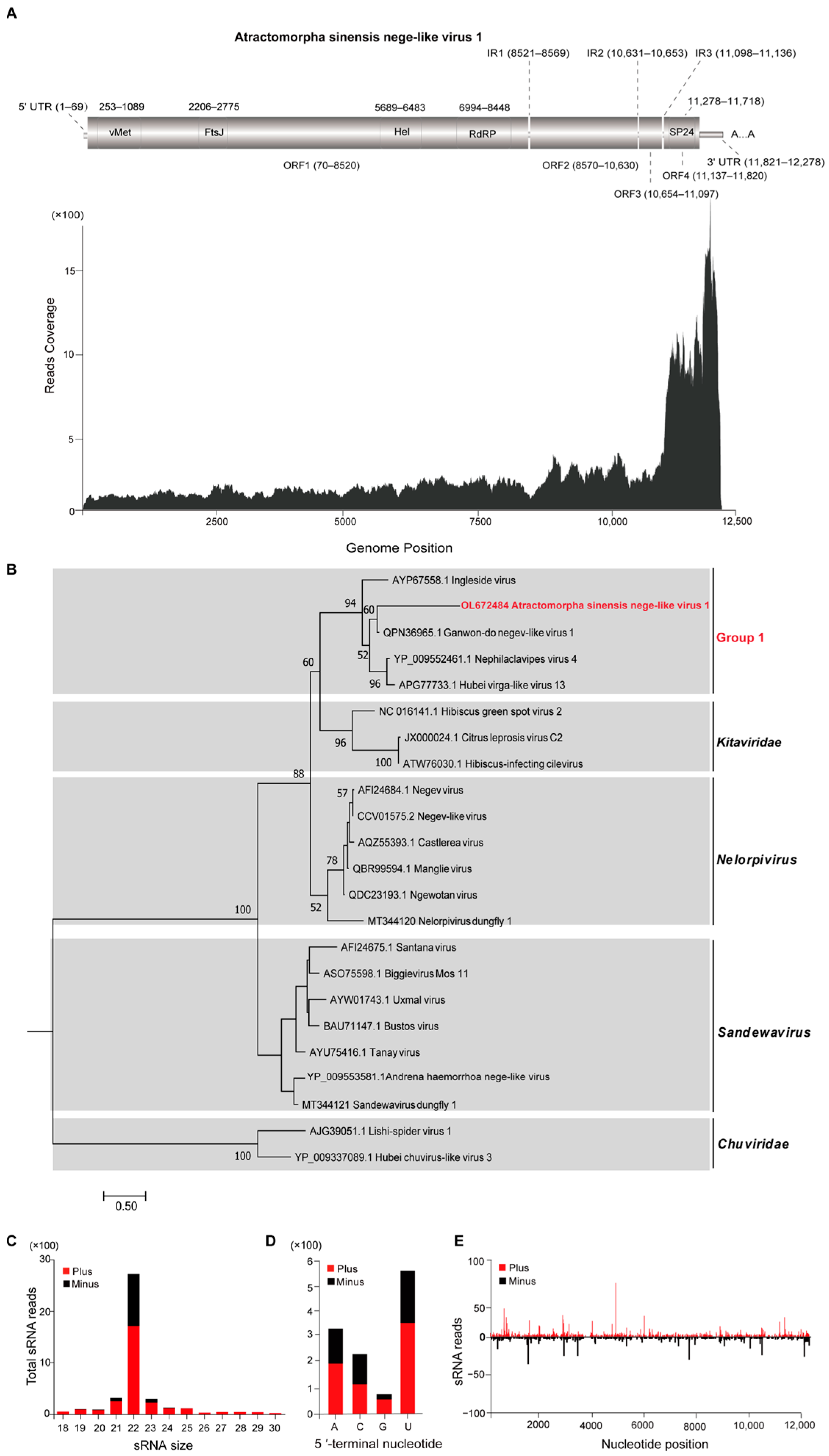
3.2. Characterization of Atractomorpha sinensis Nege-Like Virus 1
3.3. Characterization of Atractomorpha sinensis Iflavirus 1
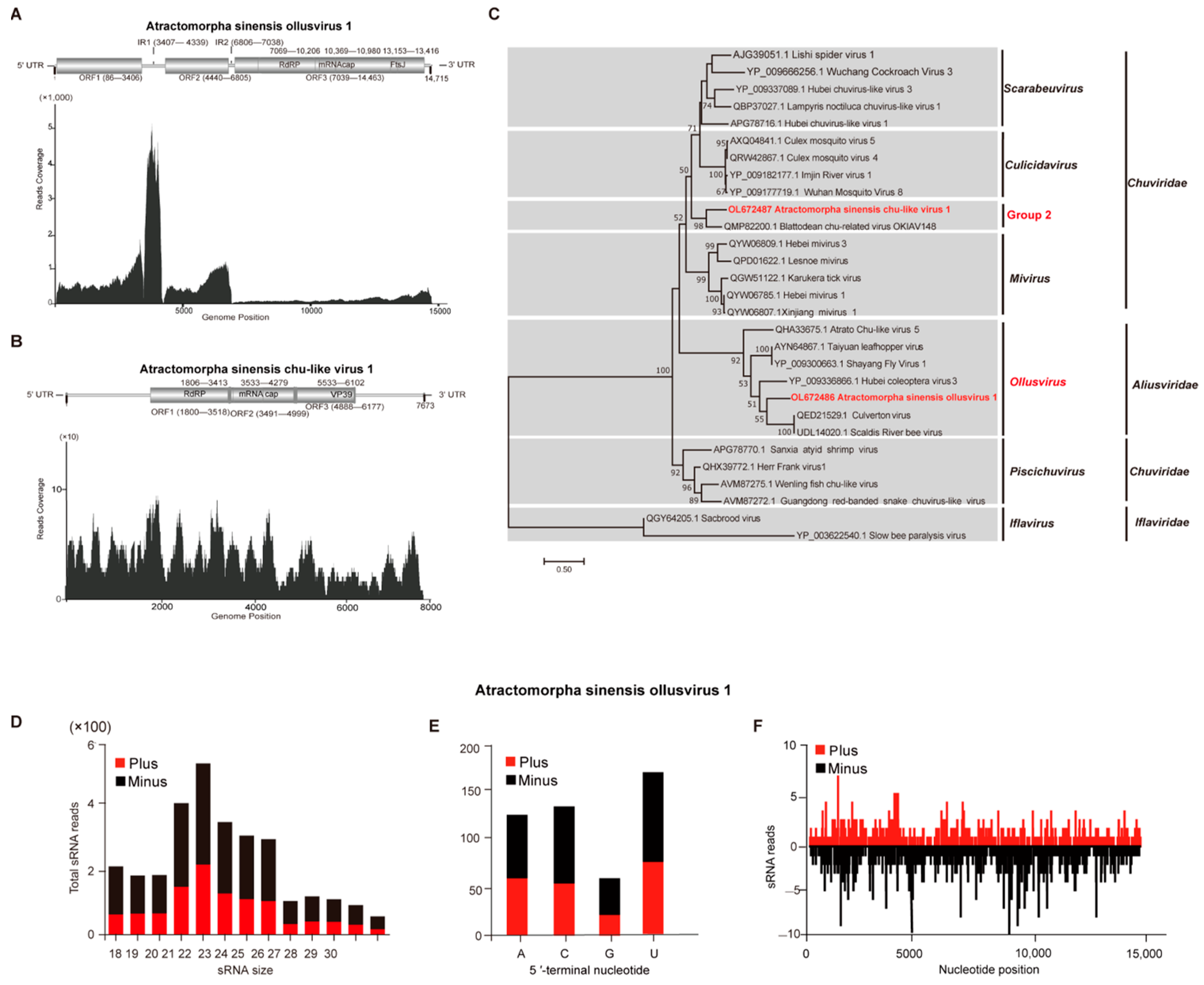
3.4. Characterization of Atractomorpha sinensis Ollusvirus 1 and Atractomorpha sinensis Chu-Like Virus 1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Li, C.-X.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Kang, Y.-J.; Chen, L.-J.; Qin, X.-C.; Xu, J.; Holmes, E.C.; Zhang, Y.-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4, e05378. [Google Scholar] [CrossRef]
- Vasilakis, N.; Forrester, N.L.; Palacios, G.; Nasar, F.; Savji, N.; Rossi, S.L.; Guzman, H.; Wood, T.G.; Popov, V.; Gorchakov, R.; et al. Negevirus: A Proposed New Taxon of Insect-Specific Viruses with Wide Geographic Distribution. J. Virol. 2013, 87, 2475–2488. [Google Scholar] [CrossRef]
- Nouri, S.; Matsumura, E.E.; Kuo, Y.-W.; Falk, B.W. Insect-specific viruses: From discovery to potential translational applications. Curr. Opin. Virol. 2018, 33, 33–41. [Google Scholar] [CrossRef]
- Carvalho, V.L.; Long, M.T. Insect-Specific Viruses: An overview and their relationship to arboviruses of concern to humans and animals. Virology 2021, 557, 34–43. [Google Scholar] [CrossRef]
- Goenaga, S.; Kenney, J.L.; Duggal, N.K.; Delorey, M.; Ebel, G.D.; Zhang, B.; Levis, S.C.; Enria, D.A.; Brault, A.C. Potential for Co-Infection of a Mosquito-Specific Flavivirus, Nhumirim Virus, to Block West Nile Virus Transmission in Mosquitoes. Viruses 2015, 7, 5801–5812. [Google Scholar] [CrossRef]
- Baidaliuk, A.; Miot, E.F.; Lequime, S.; Moltini-Conclois, I.; Delaigue, F.; Dabo, S.; Dickson, L.B.; Aubry, F.; Merkling, S.H.; Cao-Lormeau, V.-M.; et al. Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo. J. Virol. 2019, 93, e00705-19. [Google Scholar] [CrossRef]
- Romo, H.; Kenney, J.L.; Blitvich, B.J.; Brault, A.C. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg. Microbes Infect. 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Patterson, E.I.; Kautz, T.F.; Contreras-Gutierrez, M.A.; Guzman, H.; Tesh, R.B.; Hughes, G.L.; Forrester, N.L. Negeviruses reduce replication of alphaviruses during coinfection. J. Virol. 2021, 95, e00433-21. [Google Scholar] [CrossRef]
- Huang, H.-J.; Ye, Z.-X.; Wang, X.; Yan, X.-T.; Zhang, Y.; He, Y.-J.; Qi, Y.-H.; Zhang, X.-D.; Zhuo, J.-C.; Lu, G.; et al. Diversity and infectivity of the RNA virome among different cryptic species of an agriculturally important insect vector: Whitefly Bemisia tabaci. NPJ Biofilms Microbiomes 2021, 7, 1–15. [Google Scholar] [CrossRef]
- Kondo, H.; Fujita, M.; Hisano, H.; Hyodo, K.; Andika, I.B.; Suzuki, N. Virome Analysis of Aphid Populations That Infest the Barley Field: The Discovery of Two Novel Groups of Nege/Kita-Like Viruses and Other Novel RNA Viruses. Front. Microbiol. 2020, 11, 509. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, P.; Liu, W.; Cao, M.; Massart, S.; Wang, X. Complete genome sequence and characterization of a new iflavirus from the small brown planthopper (Laodelphax striatellus). Virus Res. 2019, 272, 197651. [Google Scholar] [CrossRef]
- Fu, Y.; Cao, M.; Wang, H.; Du, Z.; Liu, Y.; Wang, X. Discovery and characterization of a novel insect-specific reovirus isolated from Psammotettix alienus. J. Gen. Virol. 2020, 101, 884–892. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Yuan, B.; Yang, L.; Yang, Y.; Fang, Q.; Kuhn, J.H.; Song, Q.; Ye, G. A novel cripavirus of an ectoparasitoid wasp increases pupal duration and fecundity of the wasp’s Drosophila melanogaster host. ISME J. 2021, 15, 3239–3257. [Google Scholar] [CrossRef]
- Wang, F.; Fang, Q.; Wang, B.; Yan, Z.; Hong, J.; Bao, Y.; Kuhn, J.H.; Werren, J.H.; Song, Q.; Ye, G. A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 2017, 13, e1006201. [Google Scholar] [CrossRef]
- Dolja, V.V.; Krupovic, M.; Koonin, E.V. Deep Roots and Splendid Boughs of the Global Plant Virome. Annu. Rev. Phytopathol. 2020, 58, 23–53. [Google Scholar] [CrossRef]
- Li, W.-B.; Gao, Y.; Cui, J.; Shi, S.-S. Effects of Temperature on the Development and Fecundity of Atractomorpha sinensis (Orthoptera: Pyrgomorphidae). J. Econ. Entomol. 2020, 113, 2530–2539. [Google Scholar] [CrossRef]
- Buleu, O.G.; Jetybayev, I.Y.; Bugrov, A. Comparative analysis of chromosomal localization of ribosomal and telomeric DNA markers in three species of Pyrgomorphidae grasshoppers. Comp. Cytogenet. 2017, 11, 601–611. [Google Scholar] [CrossRef]
- Nunamaker, R.A.; Lockwood, J.A.; Stith, C.E.; Campbell, C.L.; Schell, S.P.; Drolet, B.S.; Wilson, W.C.; White, D.M.; Letchworth, G.J. Grasshoppers (Orthoptera: Acrididae) could serve as reservoirs and vectors of vesicular stomatitis virus. J. Med. Entomol. 2003, 40, 957–963. [Google Scholar] [CrossRef]
- Drolet, B.S.; Stuart, M.A.; Derner, J.D. Infection of Melanoplus sanguinipes Grasshoppers following Ingestion of Rangeland Plant Species Harboring Vesicular Stomatitis Virus. Appl. Environ. Microbiol. 2009, 75, 3029–3033. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Amit, I. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Lu, G.; Ye, Z.-X.; He, Y.-J.; Zhang, Y.; Wang, X.; Huang, H.-J.; Zhuo, J.-C.; Sun, Z.-T.; Yan, F.; Chen, J.-P.; et al. Discovery of Two Novel Negeviruses in a Dungfly Collected from the Arctic. Viruses 2020, 12, 692. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Guo, L.; Lu, X.; Liu, X.; Li, P.; Wu, J.; Xing, F.; Peng, H.; Xiao, X.; Shi, M.; Liu, Z.; et al. Metatranscriptomic Analysis Reveals the Virome and Viral Genomic Evolution of Medically Important Mites. J. Virol. 2021, 95, e01686-20. [Google Scholar] [CrossRef] [PubMed]
- Harvey, E.; Rose, K.; Eden, J.-S.; Lawrence, A.; Doggett, S.L.; Holmes, E.C. Identification of diverse arthropod associated viruses in native Australian fleas. Virology 2019, 535, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Käfer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jiang, J.; Lin, X.; Shi, W.; Cao, C. Identification of diverse viruses associated with grasshoppers unveils the parallel relationship between host phylogeny and virome composition. Virus Evol. 2022, 8, veac057. [Google Scholar] [CrossRef]
- Nunes, M.R.; Contreras-Gutierrez, M.A.; Guzman, H.; Martins, L.C.; Barbirato, M.F.; Savit, C.; Balta, V.; Uribe, S.; Vivero, R.; Suaza, J.D.; et al. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specific viruses in the taxon Negevirus. Virology 2017, 504, 152–167. [Google Scholar] [CrossRef]
- Geng, P.; Li, W.; Lin, L.; de Miranda, J.R.; Emrich, S.; An, L.; Terenius, O. Genetic Characterization of a Novel Iflavirus Associated with Vomiting Disease in the Chinese Oak Silkmoth Antheraea pernyi. PLoS ONE 2014, 9, e92107. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Liu, W.; Cao, M.; Wang, X. Sequence analysis and genomic organization of a novel chuvirus, Tàiyuán leafhopper virus. Arch. Virol. 2018, 164, 617–620. [Google Scholar] [CrossRef]
- Di Paola, N.; Dheilly, N.M.; Junglen, S.; Paraskevopoulou, S.; Postler, T.S.; Shi, M.; Kuhn, J.H. Jingchuvirales: A new taxonomical framework for a rapidly expanding order of unusual monjiviricete viruses broadly distributed among arthropod subphyla. Appl. Environ. Microbiol. 2022, 88, e01954-21. [Google Scholar] [CrossRef]
- Han, X.; Wang, H.; Wu, N.; Liu, W.; Cao, M.; Wang, X. Leafhopper Psammotettix alienus hosts chuviruses with different genomic structures. Virus Res. 2020, 285, 197992. [Google Scholar] [CrossRef]
- Fujita, R.; Kato, F.; Kobayashi, D.; Murota, K.; Takasaki, T.; Tajima, S.; Lim, C.-K.; Saijo, M.; Isawa, H.; Sawabe, K. Persistent viruses in mosquito cultured cell line suppress multiplication of flaviviruses. Heliyon 2018, 4, e00736. [Google Scholar] [CrossRef]
- Patterson, E.I.; Villinger, J.; Muthoni, J.N.; Dobel-Ober, L.; Hughes, G.L. Exploiting insect-specific viruses as a novel strategy to control vector-borne disease. Curr. Opin. Insect Sci. 2020, 39, 50–56. [Google Scholar] [CrossRef] [PubMed]

| Tentative Virus Names | NCBI Accession | Length (nt) | Classification | Abundance | E-Value | RdRP Protein Identities | Top BlastP Hit Virus | Virus Reference |
|---|---|---|---|---|---|---|---|---|
| Atractomorpha sinensis nege-like virus 1 (ASNLV1) | OL672484 | 12,278 | Nege-like virus | 112.2 | 6 × 10−110 | 42.13% | Ganwon-do negev-like virus 1 | [31] |
| Atractomorpha sinensis iflavirus 1 (ASIV1) | OL672485 | 9412 | Iflavirus | 1689.9 | 2 × 10−129 | 45.63% | Hubei coleoptera virus 1 | [22] |
| Atractomorpha sinensis ollusvirus 1 (ASOV1) | OL672486 | 14,715 | Ollusvirus | 251.5 | 0.0 | 50.84% | Culverton virus | [32] |
| Atractomorpha sinensis chu-like virus 1 (ASCLV1) | OL672487 | 7673 | Unclassified chuviridae | 4.8 | 3 × 10−165 | 63.96% | Blattodean chu-related virus OKIAV148 | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.-J.; Ye, Z.-X.; Zhang, C.-X.; Li, J.-M.; Chen, J.-P.; Lu, G. An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis. Insects 2023, 14, 9. https://doi.org/10.3390/insects14010009
He Y-J, Ye Z-X, Zhang C-X, Li J-M, Chen J-P, Lu G. An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis. Insects. 2023; 14(1):9. https://doi.org/10.3390/insects14010009
Chicago/Turabian StyleHe, Yu-Juan, Zhuang-Xin Ye, Chuan-Xi Zhang, Jun-Min Li, Jian-Ping Chen, and Gang Lu. 2023. "An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis" Insects 14, no. 1: 9. https://doi.org/10.3390/insects14010009
APA StyleHe, Y.-J., Ye, Z.-X., Zhang, C.-X., Li, J.-M., Chen, J.-P., & Lu, G. (2023). An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis. Insects, 14(1), 9. https://doi.org/10.3390/insects14010009





