Simple Summary
Currently, bee viruses are one of the threats to honey bee populations. The increase in invasive Vespid species causes a negative impact on honey bee colonies, weakening them and making them more vulnerable to the presence of pathogens. This bibliographic review shows the main research on bee viruses in the Vespa, Vespula and Polistes genera belonging to the Vespidae family. This taxon contains several of the most widespread invasive Vespid species worldwide. Many of these species are predators of honey bees and cause great impacts, as is the case with the yellow-legged hornet Vespa velutina. The presence of viruses in species of Vespids that interact with honey bees could represent an emerging risk in the transmission of pathogens, weakening the defense strategies of native species. Gathering this information is necessary to promote the research on the spread of bee viruses associated with invasive species as well as in the development of bee virus control strategies.
Abstract
The increase in invasive alien species is a concern for the environment. The establishment of some of these species may be changing the balance between pathogenicity and host factors, which could alter the defense strategies of native host species. Vespid species are among the most successful invasive animals, such as the genera Vespa, Vespula and Polistes. Bee viruses have been extensively studied as an important cause of honey bee population losses. However, knowledge about the transmission of honey bee viruses in Vespids is a relevant and under-researched aspect. The role of some mites such as Varroa in the transmission of honey bee viruses is clearer than in the case of Vespidae. This type of transmission by vectors has not yet been clarified in Vespidae, with interspecific relationships being the main hypotheses accepted for the transmission of bee viruses. A majority of studies describe the presence of viruses or their replicability, but aspects such as the symptomatology in Vespids or the ability to infect other hosts from Vespids are scarcely discussed. Highlighting the case of Vespa velutina as an invader, which is causing huge losses in European beekeeping, is of special interest. The pressure caused by V. velutina leads to weakened hives that become susceptible to pathogens. Gathering this information is necessary to promote further research on the spread of bee viruses in ecosystems invaded by invasive species of Vespids, as well as to prevent the decline of bee populations due to bee viruses.
1. Vespid Species as an Invasive Alien Species
Globalization is causing decisive changes in human habitats and ecosystems. The introduction of invasive species and the appearance of infectious diseases in certain populations are well known processes in this field. It was estimated that the economic cost of invasive species has been $1.288 trillion over the past 50 years [1], including many pathogens that have direct impacts on human activities and the health of species worldwide.
The success of invasive species depends on several factors, but the most recent findings suggest that the success of the invasion may depend on their ability to respond to natural selection, highlighting the importance of their genetic architecture, leading to adaptations to new environments [2]. Furthermore, the absence of enemies in an area where the exotic species are introduced may contribute to pest species settling in. This is known as “The enemy release hypothesis”. This could be explained by a decrease in enemy diversity in the invasive area compared to the native area, or by the fact that non-native species are less affected by enemies than native species in the invaded community [3]. Moreover, when species move to new environments, the probability of transporting the pathogens that interact with them is low, since they generally experience a bottleneck that affects survival [4]. Despite this, the increase in the expansion of exotic species could be influencing the global spread of pathogens, since these species and infectious diseases follow similar patterns, especially the greater connection and globalization [5].
The Vespidae family is vastly widespread and comprises almost all species of eusocial wasps, as well as many solitary species [6,7]. Although the proportion of species that successfully settle in new areas is low, the solitary and social wasps are a very favorable group to be an invasive species and, in many cases, they are considered pests. Many Vespids alter the composition of ecosystems since, being generalist predators, they change interactions or cause complex interactions with the host community [8]. The social wasps from the genera Vespula, Vespa, and Dolichovespula are pests in temperate and tropical regions of the world [9]. Two of the most abundant species are Vespula vulgaris (V. vulgaris) and Vespula germanica (V. germanica). These two species show a very plastic generalist foraging behavior, which favors the appearance of a great variety of impacts on biodiversity and changes in the function of the ecosystems invaded [7,10,11]. The case of Vespa is relevant since some species such as Vespa velutina (V. velutina) or Vespa orientalis (V. orientalis) are species of concern for ecosystems and human health, when introduced as invasive species [7]. V. velutina nigrithorax is the only one included in the List of Invasive Alien Species of Union concern (Regulation (EU) 1143/2014 of the European Parliament and of the Council of 22 October 2014 on the prevention and management of the introduction and spread of invasive alien species) and today it is a widespread pest in Europe.
V. velutina or Asian yellow-legged hornet is common in central to eastern Asia [12,13,14,15]. In their natural range, populations of V. velutina are kept in balance with the species they prey on (Asian honey bee Apis cerana (A. cerana)) and with the pathogens that are involved in their biological cycle [8]. The species was detected for the first time in France in 2004, and since then it has been slowly spreading throughout the rest of Europe [7,14,16]. One of the main impacts is due to its high capacity for predation of multiple species of insects, especially of the honey bee, altering the beekeeping sector at all levels [14,16]. In addition, this invasive insect is very flexible in its diet, since in addition to preying on an enormous variety of insect species, it also consumes other small animals, carrion from animal carcasses, and other resources such as sugary substances from nectar or fruits. This wide and diverse diet makes it a risk factor since it gives rise to a wide spectrum of interactions in the invaded ecosystems.
Knowledge about pathogens could be crucial to better understand the ecology of pest species and thus develop an integrated control strategy. Henceforth, this study aims to deepen the knowledge about the most common bee viruses found in different Vespid species. Additionally, this review makes a special mention of the V. velutina hornet, as it is a species that is expanding rapidly in all European ecosystems, thus causing a notable environmental and socioeconomic impact, especially in the beekeeping sector [14,16]. The role of this invasive species in the spillover of potential pathogens is still not well known. However, since a higher number of interrelations between species increases the risk, understanding the factors that facilitate the transmission is essential to establish control strategies.
2. Vespid Species as Hosts of Bee Viruses
Most invasive Vespid species interact extensively with other species, when preying on other insects such as honey bees, and during foraging and the search for resources to build nests. The interaction between these species allows the transmission of many pathogens, including bee viruses [17]. Thus, virus transfer between pest species and target species such as honey bees and vice versa is an expected scenario. Genomic approaches have greatly expanded understanding of the diversity of bee viruses, their routes of transmission, and the strategies that bees have developed to combat viral infections [18]. These infections are further exacerbated by major stressors such as parasites, poor nutrition, or the arrival of new predatory pest species, such as Vespids. The recent confirmation of replicability of bee viruses in some species of Vespids [19,20] could suggest that the interactions between pathogen and newly introduced host are being reconfigured.
In recent years, with the entry of the invasive species V. velutina in Europe, more research on viruses such as pathogens has been conducted, searching for the best control method to reduce hornet populations [20,21,22,23]. In parallel, the interest of researchers and beekeepers in the study of honey bee viruses is increasing, mainly due to their association with certain parasites such as Varroa [24]. Moreover, the enhancement of diagnostic techniques such as Next Generation Sequencing [19,20,21,25,26] makes it easier to detect these viruses in large sample size. However, so far there is still little knowledge about the presence of bee viruses and their transmission to other hosts such as the different species of hornets and their interrelationships.
In this sense, invasive polyphagous species are more successful in the establishment than monophagous or oligophagous species [20]. The transmission of pathogens is another consequence of the introduction of these invasive species [20]. Although the expansion of some species of invasive Vespids, such as Vespa or Vespula, to new areas could imply a reduction of its native pathogens, its rapid adaptability to new ecological niches gives rise to new interactions with existing pathogens in the new invaded areas. Other successful invasive insects are several Polistes species (Polistes dominula (P. dominula) or Polistes chinensis (P. chinensis)) that can also be considered as a pest in several countries [7,27]. Although species of the genus Polistes do not prey on bees, the spillover effect of the different bee viruses in ecosystems makes it a carrier and host for these viruses.
These species affect invaded local biodiversity either through direct predation or competition for food, nest settlements or space. Many invasive social wasp species are also increasingly recognized for their predation on honey bees [4,14,15,16]. It is noteworthy that most of the research dedicated to these invasions appears in the simultaneous phase of the invasion. From the point of view of the literature, much of this research focuses on studying the spread of pathogens between Vespid and honey bee species (hereinafter cited). Most of these apply genomic technologies, and their objectives are mainly focused on managing control strategies for these invasive species [28].
3. Types of Viral Transmission
Knowing how viruses spread and cause infections is essential. There are various ways of transmitting a virus. These are summarized in horizontal and vertical transmission, or both. In the horizontal transmission, the virus is usually transmitted between members of the same species that do not have a progenitors–descendants relationship, that is, between individuals of the same generation [29]. In turn, this type of transmission can be direct or indirect. The direct route involves an infection transmitted by environmental contamination, food, feces, or sexual infection. The indirect route involves a transmission vector, an intermediate biological host (such as mosquitoes or mites [24]) that acquires and transmits the virus from one host to another [29]. In general, infection in vertical transmission takes place through the host’s offspring. The virus is transmitted vertically from the parental generation to the brood through the egg. This can occur in two ways, by transmission on the surface of the egg (transovum transmission) or inside the egg (transovarian transmission) [29,30].
Virulence tends to increase in horizontal transmission, unlike in vertical transmission [29]. The higher the level of pathogen multiplication, the greater its virulence; thus, transmission and virulence will be positively correlated [29]. In vertical transmission, the reproduction of the host contributes to the reproduction of the parasite, so this type of transmission exerts a natural selection on the parasites to be less harmful to its hosts since the survival of the one implies the survival of the other [31]. This type of natural selection can eliminate the pathogen unless they develop special routes of transmission such as biparental transmission, in which the virus is transmitted by males (sperm) and females (ovules) to their offspring. Another method of increasing representation in future generations in vertical transmission, and thus balancing the effects of natural selection, is the sex-ratio distortion. This is the case of Pteromalus puparum negative-strand RNA virus 1 (PpNSRV-1), which is an RNA virus from a parasitoid wasp (Pteromalus puparum). This virus is transmitted vertically by infected wasps, biparentally (both female and male), but at the same time the virus reduces the number of female offspring by modifying the secondary sex ratio of the parasitoid host [32]. Although each type of transmission has certain characteristics to succeed in its reproduction and expansion, there are many viruses spreading through a combination of vertical and horizontal transmission [31].
Deformed Wing Virus (DWV), one of the most widely studied bee viruses, may be an example of both the vertical and horizontal transmission in the honey bee. Numerous studies describe the presence of DWV in honey bee queen ovaries and therefore a venereal–vertical infection [30,33,34,35]. Although it can also be transmitted horizontally [17], vertical transmission has been demonstrated to be the most generalized natural route of spread [30]. This class of viruses can be considered hereditary genetic agents that can cause potential impacts on the ecology and evolution of insects [36]. Apart from that, studies on DWV in Vespids suggest horizontal transmission mainly due to predation on honey bees and other insects [21]. In addition, the presence of DWV has been demonstrated in nectar and pollen of plants [17] that insects forage, which might be an added kind of horizontal transmission. However, this relationship between Vespids and DWV in nectar has not yet been proven.
The type of viral transmission depends largely on the type of relationship that exists between the interacting species. The relationships between pathogen and wasps can vary from pathogens to mutualists. An example is a relationship between Ascovirus DpAV4 and wasps of the genus Diadromus [37]. This relationship can be pathogenic, mutualistic or non-pathogenic, depending on the regulatory factors that control virus replication and on the type of relationship that has been established. Many viruses can be beneficial for their hosts, through a symbiotic interaction [38,39]. This is the case of parasitic wasp species of the genus Diadromus that harbors reovirus (family Reoviridae). It has been demonstrated that some of these reoviruses can suppress host defense, facilitating the development of parasitoid eggs [40]. In addition, some viral pathogens can influence other parameters such as the size of the nests. Thus, the Kashmir bee virus (KBV) affects the size of the nest in the common invasive wasp V. vulgaris [41]. The longevity of wasp colonies has also been influenced by the presence of diverse viral loads. This is the case for colonies of the Vespula pensylvanica (V. pensylvanica) wasp, whose longevity may be affected by the viral load of the Moku virus [42].
4. Transmission Vectors and Activators
4.1. Mites
Many of the bee viruses use transmission vectors such as mites. According to De Jong et al. [43], mites that affect honey bees or that are found in hives can be parasites, phoretic mites, and house hosts. Some of the parasitic mite species present in hives can cause serious diseases in honey bees. The main mites that cause problems are the Varroa genus, mainly the Varroa jacobsoni (V. jacobsoni) and Varroa destructor (V. destructor), Acarapis woodi, and Tropilaelaps mercedesae. Among them, the Varroa is the causative agent of varroosis one of the most serious diseases in honey bees [24]. In addition, it has the potential to contribute to the worldwide mortality of honey bee colonies through several RNA viral infections [44]. One of the most extended viruses associated with this mite is the DWV. Although this virus can appear without the presence of Varroa, it has been proven that its presence increases the frequency and quantity of this infection in honey bees [44]. This virus has been detected symptomatically and asymptomatically in some species of Vespidae and some predators of Apis mellifera (A mellifera), which suggests the possibility that the virus can be transmitted by ingesting infected bees [21,25,41,45,46]. Another virus associated with varroa (V. destructor) is the Acute bee Paralysis Virus-(ABPV). As in the previous case, it is a virus that affects the honey bee and that can also be transmitted using the varroa mite as a transmission vector to predatory species such as V. velutina [45]. Recently, a study carried out in northern Spain [47] has detected for the first time ectoparasitic mites of the genus Varroa attached to the lateral–ventral part of the abdomen of V. velutina specimens. In this study, a prevalence rate of the Varroa parasite in the V. velutina of 1.75% has been described. Despite this suggestion, there is no study in which the presence of Varroa has been detected in the life cycle of V. velutina. Such detection would confirm the relationship between the two species. However, this could still suggest a risk in the dispersion of mites that can transmit bee viruses. On the contrary, Varroa individuals (probably V. destructor, although initially published as V. jacobsoni) were detected in V. vulgaris larvae from Poland [48]. This finding could make this mite a pathogen affecting this larval stage of this wasp species. There are studies that also demonstrate the role of Varroa in the dynamics of hymenopteran pathogens in insect communities. Specifically, they reveal how the hidden effects of the Varroa mite can, by spillover, transform the composition of the pathogens in the interacting species [49]. Thus, the arrival of the V. destructor mite in Hawaiian populations of Apis modified the diversity of the DWV virus in the invasive population of the Vespula wasp, with DWV type A being the predominant variant [49].
There are transmission vectors other than Varroa. As Vespid species disperse to areas other than their native origins, the detection of new species linked to viral diseases is more evident. Pneumolaelaps niutirani (P. niutirani) is a species of mite that has recently been discovered in New Zealand [50], hence the name niutirani, which derives from the Maori “Niu Tīrani”, which means “New Zealand”. It is a species of mite identified for the first time in association with the nests of the wasp Vespula germanica. It has also been discovered in other Vespula species (V. germanica and V. vulgaris) in both New Zealander and Belgian individuals [50,51]. The study of pathogens associated with Vespula wasp nests has made it possible to detect viruses such as Moku and DWV in the P. niutirani mite [51]. This association between P. niutirani with specific pathogens could lead to invasive wasp infections. In addition, this mite can be transported by other species of Hymenoptera such as bumblebees and honey and solitary bees, making it a possible vector of viral transmission in the interaction between different species of Hymenoptera [50,51]. This finding creates new opportunities for the development of a biological control agent. However, this goal would require a better understanding of their feeding, biology, and the mechanism of transmission of pathogens in wasps.
4.2. Other Transmission Vectors
There are routes of transmission different to the mites such as the Nosema microsporidium [52]. Several species of the genus Nosema cause the disease known as nosemosis. Nosema apis (N. apis), Nosema ceranae (N. ceranae), Nosema neumanni (N. neumanni) are the most studied and have been found related to the honey bees [53]. In addition to causing nosemosis, this microsporidium (N. apis) could transmit and increase the spread of some viruses such as Black Queen Cell Virus (BQCV) [19]. Although it mainly affects honey bee pupae, this virus can infect the midgut of adult honey bees, probably transmitted by ingestion of Nosema microsporidia, with a correlation between peak infections of both pathogens [54]. As is the case with Varroa, Nosema can be transmitted to predatory Vespids such as V. crabro or V. velutina [19,22,23], Polybia scutellaris [55] and even to pest species for honey bees, such as the small hive beetle (Aethina tumida) [56]. Although both pathogens have been identified separately, further studies are needed to determine their possible infection and/or co-infection in Vespids.
5. The Incidence of Bee Viruses on Vespa, Vespula, and Polistes
Honey bee viruses have been demonstrated to have an ability to infect other hosts such as the Vespidae species. One of the most defended hypotheses that explains the presence of viruses in Vespids is the one that supports this infection as being caused by a predator–prey interaction [20,21,25,45,57]. The increase in host range could determine the long-term evolution and virulence of pathogens, as well as the emergence and spread of new diseases [58]. However, there are studies that currently provide information on infection due to other vector-type transmission factors.
The scientific literature focused on viruses found in different species of the family Vespidae is scarce. However, further knowledge is essential because social insects with large families play an important role in ecosystems. Moreover, some of them, such as V. velutina, have an intense predatory effect on bee colonies. This close relationship could favor the exchange of pathogens between the species and enables V. velutina to act as a vector for transmitting pathogens to other bee populations. The main investigations regarding the presence of the most common honey bee viruses in Vespid species are presented below. This study was carried out considering a total of 117 documents found in the time frame of 1968–2021. The search focused especially on those documents that identified bee viruses in the Vespid genera with the greatest capacity to invade new ecosystems. This search allowed for a characterization of bee viruses in the genera Vespa, Vespula as honey bee predators and Polistes as species that share ecosystems with bee viruses hosts.
A total of 22 viruses (Acute Bee Paralysis Virus (ABPV), Acypi-like Virus, Aphid Lethal Paralysis Virus (ALPV), Apis mellifera Filamentous Virus (AmFV), recently identified Macula-like Virus (BeeMLV), Black Queen Cell Virus (BQCV), Chronic Bee Paralysis Virus (CBPV), Deformed Wing Virus (DWV), Israeli Acute Paralysis Virus (IAPV), Ifla-like virus, Kashmir bee virus (KBV), La Jolla virus, Lake Sinai Virus (LSV), Luteo-like virus 1, Menton virus, Moku, Mott mill virus, Nora-like virus, Partiti-like virus, Permutotetra-like virus, and Triato-like Virus) have been found in Vespids. These viruses are summarized in Table 1, Table 2 and Table 3.

Table 1.
Occurrence of viruses in Vespa.

Table 2.
Occurrence of viruses in Vespula.

Table 3.
Occurrence of viruses in Polistes.
The tables specify where the replication was detected. The tables refer to the following 15 species. Six belonged to the genus Vespa (V. velutina, V. crabro, V. bicolor, V. affinis, Vespa sp., and V. orientalis), four to the genus Vespula (V. vulgaris, V. germanica, V. pensylvanica, and Vespula sp.), and five to the genus Polistes (P. chinensis, P. humilis, P. rothneyi, P. fuscatus, and P. metricus).
In addition, this review highlighted the description of bee viruses in V. velutina, as it is an invasive alien species whose rapid and impactful introduction could be altering the composition of pathogens in invaded ecosystems (Figure 1).
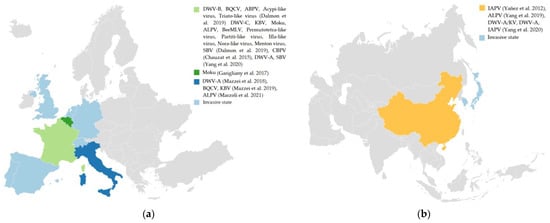
Figure 1.
Studies on bee viruses of V. velutina in the invasive area in Europe; (a) and in its native area; (b) In addition, figures also show its distribution as an invasive species in light blue (Invasive state). Created with Datawrapper. The references correspond to the following call-out numbers: Dalmon et al. [45], Chauzat et al. [23], Yang et al. [59,60], Garigliany et al. [61], Mazzei et al. [19,21], Marzoli et al. [20] and Yañez et al. [46]. Created with Datawrapper.
5.1. Deformed Wing Virus (DWV)
One of the most main viruses associated with the collapse of the colony is DWV [72]. This can be observed in the fact that all studies have demonstrated the presence of this virus in the different genera investigated. It is a positive-stranded RNA virus [72,73], which causes a deformation of the wings of honey bees. The disease is characterized by morphological abnormalities that include malformed appendages, a reduced abdomen, and erroneous coloration [74]. Being one of the most widespread viruses in Apis, the study of species that interact with this species, such as Vespids, is also a highly pursued objective. The transmission of DWV in honey bees is usually associated with the mite V. destructor [72,74,75,76]. However, it has also been associated with secondary infections by other pathogens [77]. In addition, a horizontal transmission can exist from the pollen in flowers, pollen loads and other bee products to insects [17,29,65,76,78]. High levels of infection in the hive may be the cause of DWV transmission to different predators belonging to the Vespidae family. As in honey bees, the main visible symptom in Vespids has been the presence of deformed wings. This symptom, such as the predation of honey bees, has been evaluated to genetically characterize the existing variants of DWV in Vespids. There are three DWV genetic variants defined as type A, B, and C [25,70,79], with type A being the most virulent [79]. In addition to these three variants, a subtype of this virus known as the Kakugo virus (KV) has been studied, which usually appears with variant A [60]. Table 1 and Table 2 show its presence in Vespa, Vespula, and Polistes individuals. In the case of V. crabro [25,60], the presence of the replicating virus in the abdomen and thorax of asymptomatic and symptomatic individuals has been found [25,62]. In V. velutina, DWV variants A, B and C have been detected [20,21,45,60]. The presence of variants A and B has also been detected in adult individuals and pupae of other species of the Vespa genus, such as V. bicolor, V. affinis, and Vespa sp. [60]. Power et al. [63] have identified this virus in the recently introduced V. orientalis in Italy. The DWV has been demonstrated to be present in more than 64 species of insects, including all the species of the genera Vespula and Polistes (with the exception of P. humilis and P. rothneyi) treated in this document [65,66,69,71]. Polistes spp. are common social wasps that hunt caterpillar prey and feed on nectar by occasionally visiting flowers [80]. It would then be a horizontal transmission that could explain the presence of DWV in this species. Nonetheless, studies exist which associate DWV infection in Polistes spp. with the presence of Varroa, since when Varroa is absent in the ecosystems shared with Polistes, DWV has not been detected either. This suggests that Varroa could promote increased horizontal transmission within a honey bee colony, therefore increasing transmission in the environment [80].
The replication of this virus has also been studied in the various species of Vespids. In V. velutina, the replicative form has been detected by Mazzei et al. [21] in samples collected in Italy and by Dalmon et al. [45] in France, demonstrating active viral replication. Of note is the finding of replicative forms in V. germanica from New Zealand and Europe [66,69], V. vulgaris in samples from New Zealand [66] and in Vespula sp. from the USA [71].
5.2. Black Queen Cell Virus (BQCV)
Another virus that has been extensively and frequently studied in Vespids due to its high incidence in honey bees is the BQCV [19,45,60,63,64,65,71]. This virus was also found in all the genera (Table 1 and Table 2). The action of the BQCV in the honey bees causes the death of queens in the larval or prepupal stages. The clinical symptomatology implies a dark coloration accompanied by decomposition, presenting the queen cells’ black spots on their walls. If the queen were to complete her development, she would darken, showing a dark-brown coloration [81]. This virus does not affect adult worker bees, because although they contain the virus, they do not show alteration or symptoms [82]. The most studied transmission mechanism in honey bees is due to contagion during larval feeding [82]. However, it is noteworthy that its presence correlates with the presence of N. apis, as mentioned previously [19,54,83]. Like DWV, the spillover effect of this virus is evident, causing infection in several species, including those of Vespidae [58]. BQCV has been studied globally, through its identification in Vespid species, where a high prevalence (from 2.2% to 67%) has been recorded [45]. In this sense, a pronounced genetic diversity of this virus is suggested from the phylogenetic point of view [60]. This virus has also been observed in V. bicolor, V. crabro, and V. velutina [19,45,60]. In addition to being found in a co-infection with N. apis, this virus has been observed in co-infection with the Kashmir Bee Virus (KBV) in V. velutina, where the absence of vertical transmission is demonstrated [19].
The replicative form has also been found in the gut and muscle samples of this species, as well as in honey bees collected at the same time, suggesting transmission by predator–prey [45]. As for other Vespids species, it has been found in V. vulgaris, P. rothneyi, P. metricus, and Vespula sp. [7,60,65,71]. It is of special attention in the study by Levitt et al. [71], since this virus has been found in several insects and arachnid species. This work mentions the infection potential of this virus, even though replication was not studied.
5.3. Viruses That Cause Paralysis
Some viruses cause paralysis in some of the stages of A. mellifera; these are the Slow Bee Paralysis Virus (SBPV or SPV), Aphid Lethal Paralysis Virus (ALPV), Israeli Acute Paralysis Virus (IAPV), Chronic Bee Paralysis Virus (CBPV), and Acute Bee Paralysis Virus (ABPV) (Table 1). SBPV is an inflavirus with phylogenetic proximity to the Moku virus [61], which was discovered during the bee virus X propagation experiments in England in 1974 [84]. It affects adult honey bees by paralyzing their anterior legs [84]. This virus affects both the larvae and pupae of honey bee, although symptoms were not reported [85]. Its transmission has been related to the V. destructor mite and occurs especially during the mites’ feeding activities [86]. Although it has been studied in Vespids (V. velutina) [19], it has not been detected in any specimen.
The ALPV, a pathogenic aphid RNA virus, was first identified and classified in South Africa; based on its biophysical and biochemical properties, it was classified as a picorna-like virus [87]. It is currently included in the Dicistroviridae family [88]. Several subgroups with the ability to infect bees have been determined, such as the ALPV-Am strain identified in Spain and in the United States [89], and the ALPV-Ac strain similar to ALPV-Vv isolated in China [59]. In the genera studied, it has only been detected in Vespa, and exactly in individuals of V. velutina. Major studies demonstrated that this virus has been identified in V. velutina hornets in China [59], France [45], and Italy [20]. They demonstrated that V. velutina can be an important viral reservoir between V. velutina and honey bees, resulting in potential impacts on hive health [59].
IAPV is a virus recently incorporated into the Dicistroviridae family. It has been discovered as a consequence of the spread of the virus in the pupae of white-eyed honey bees [90]. IAPV is a virus that displays high genetic diversity [60] and it is well established in Apis colonies, hence its multiple forms of transmission, both horizontal and vertical [91]. Varroa can be a vector for the transmission of this virus and its association also promotes the replication of IAPV, causing more damaging effects on honey bee colonies [24,46]. This virus has also been detected in sperm through artificial insemination, causing infections in the tissues of the honey bee queen [91]. This has confirmed the possibility of sexual transmission. Like DWV, its establishment in Apis populations allows a spillover effect towards other species that interact with Apis, in both predators and non-predators. Therefore, IAPV uses a wide variety of carriers and additional hosts [46,65]. In Vespids, the literature suggests that IAPV infection is generally related to bee ingestion [19,46]. Thus, this virus has been detected in wasps whenever the virus is also present in nearby bee populations [65]. As main results, IAPV has been detected in V. velutina [30,46], V. bicolor, V. affinis, and Vespa sp. [30] (Table 1), V. vulgaris [65], V. germanica in its replicative form [69] and P. rothneyi [60] (Table 3). It is important to add that this virus has been detected in pupae samples of some Vespids such as V. bicolor, P. rothneyi, V. affinis, and V. velutina [60]. For infection in pupae, feeding by adult individuals suggests the possibility of virus transmission. However, Yang et al. [60] suggests that it would be necessary to go deeper into the knowledge of infection in pupae, since it could be a more susceptible stage for the replication of this virus since they come from larvae fed on large quantities of masticated bees.
Chronic bee paralysis virus (CBPV) was isolated in Apis [92] and described in France as “maladie noire”; it causes a Type 2 syndrome [73,92,93] characterized with clusters of flightless trembling and crawling bees. In addition, some black individuals standing at the entrance of the hive are described in infected hives [92,94]. Furthermore, this virus can cause another version of infection known as Type 1 syndrome, first described in the UK. This is characterized by trembling and crawling bees and by less frequent or null black individuals and occurs at any time of the year [77,93]. These syndromes correspond to those described by Ribière et al. [94], who consider that it is composed mainly of two segments, RNA 1 and RNA 2. One of the syndromes causes the appearance of dysentery that cannot be excreted and is known as “Bloated abdomen”; the other syndrome is known as “black thieves” or “hairless black syndrome”. CBPV causes a very contagious infection with high multiplication levels causing very significant losses in bees that can be aggravated when accompanied by nutritional deficiencies or bad weather conditions in the summer [94,95]. The high replication of this virus promotes its spread to other species such as ants [96] but also Vespids [45]. In Europe, CBPV has been studied in V. velutina [19,23] for its high contagion when predating honey bees. However, only the study carried out by Chauzat et al. [23] demonstrated the presence in V. velutina from France. The presence of the virus, in coinfection with SBV (Sacbrood virus) and BQCV viruses, in diseased larvae with pathogens such as A. woodi and Nosema spp., was highlighted in the latest study. Symptomatology involved the non-conversion of pupae and a change in larvae color from pearly white to pale yellow and turning dark brown [23].
Parallel to the discovery of this latter virus (CBPV), the acute bee paralysis virus (ABPV) was discovered [77,93,94,97]. ABPV causes tremors and paralysis in a few days after infection; however, individuals die earlier than with chronic paralysis. High levels of the virus, at around 100 particles of ABPV, generate symptoms of paralysis, abnormal wings and body tremor in 2–4 days and the death in 1 or 2 days more [93]. Regarding the Vespid species, to date no symptoms have been detected. ABPV has been studied in Vespids, such as V. velutina [19,45,46,60] or V. germanica [69]. Regarding V. velutina, infection with this virus was detected in muscles and/or cuticle, legs, and mandibles, as well as in the guts of the hornet [45]. The presence of the virus in the body of V. velutina could be related to the infection cycle; however, further experimentation is necessary to characterize the virus and its virulence in the species [45]. In V. germanica, the replicative form has been detected [69], as shown in Table 2. It is important to note that the sequences analyzed in V. velutina [45] demonstrated 99% identity with the acute bee paralysis virus (ABPV) isolated Hungary 1 (GenBank AF486072.2), but with differences with other ABPV sequences obtained from bees of the same experiment. Furthermore, phylogenetic analysis in V. germanica demonstrated that the ABPV sequences were more closely related to each other than to the global GenBank sequences. These different sequences to others could support the hypothesis that Vespid species could be, in addition to carriers, hosts for virus infection.
ABPV is frequently studied with the Kashmir bee virus (KBV) and Israeli acute bee paralysis virus (IAPV), being a part of the AKI complex. These viruses are closely related virus species from the Dicistroviridae family [97,98]. ”AKI” is a complex formed by three viruses (acute bee paralysis virus ABPV, Kashmir bee virus KBV, Israeli acute bee paralysis IAPV) that have a worldwide distribution [45]. The presence of this complex is related to the high loss of bee colonies, especially when the colonies are infested with the parasitic mite V. destructor [97]. KBV is an RNA virus belonging to the Dicistroviridae family [99] and is one of the most virulent viruses that affect Apis [81,100]. This virus has been detected in the feces of both worker and queen bees and feeding is the most probable cause of transmission [65]. In addition, a vertical transmission has been detected; this would be in the case of the infection of bee queens that could transmit the virus through the eggs (transovarian transmission) [97,100,101]. The V. destructor could be involved in the initiation of KBV replication, which can also transmit KBV between adult bees and pupae [85,99,102]. It is genetically related to the acute bee paralysis virus (ABPV) and both can co-infect the same hive and the same honey bee. Some factors, such as stress in the hive, could propitiate to increased infection of this virus in the hive [19]. KBV has been detected in the genera Vespa and Vespula. In the case of V. velutina [19,46], an active viral replication in individuals has been demonstrated even at the end of the season. It is worth mentioning that the virus has been detected in parts other than the gut, such as in the brain of the same species [45]. This fact and the finding of replication in this species indicate that this virus could be adapted to V. velutina and, therefore, it is not specific to the honey bee. This virus has also been observed in other Vespids of the Vespula genus such as V. vulgaris [41,51,66,67] and V. germanica [51,66,69]. In both species, the replicative form has been detected, as well as coinfection with DWV. The replication of this virus in Vespid species and the high viral load detected in the sampled specimens could indicate that KBV is a potential pathogen of these species. Coinfection with other viruses also opens a range of different hypotheses about the infection mechanisms of this virus in Vespids, such as the weakening of the host defense mechanisms [19].
5.4. Sacbrood Virus (SBV)
The Sacbrood virus or Saciform Brood Virus (SBV) is also an RNA virus with a picornavirus in the genus Iflavirus and belongs to the family Iflaviridae [100,103,104]. SBV can present causes failure to pupate and subsequent death in A. mellifera, affecting both larvae and adult honey bees [103]. SBV in the early stages can result in the larvae dying shortly after being capped and before pupation. Therefore, the larva is unable to detach from the last larval cuticle and does not reach the operculation stage. The following days, eccidial fluids continue to accumulate between the cuticle and the tissues, causing the characteristic color of this infection and forming the sac [100]. The spread of this virus has been detected in both Apis predator and non-Apis predator hymenopteran [65,71]. A high prevalence has been also detected in studies that have tested this virus in species other than honey bees [45,60]. Regarding the main Vespid species in the literature, this virus has been observed in V. velutina [45,60]. Other species in which this virus has been detected were V. vulgaris [64,65], P. metricus, North American native wasp [65] and Vespula sp. [71] (Table 2 and Table 3). In the case of V. velutina, it has been detected in the part of the gut. This could lead to the suspicion that the transmission of this species occurs by preying on honey bees. However, as mentioned, it is a virus with a high prevalence, which has even been detected in pollen pellets and in non-predatory species of honey bee. In addition, finding this species in a stage such as the pupa of V. vulgaris (without symptoms of infection), it can be thought that it is a well-established virus and a pathogen for a high diversity of pollinating hosts.
5.5. Moku Virus (MV)
The inflavirus Moku virus (MV) was first discovered in the predatory social wasp Vespula pensylvanica, probably its native host, on Big Island, Hawaii [70]. It is an RNA virus, genetically different from any other virus at the nucleotide level, hence its nomenclature as “Moku”, which means island in Hawaiian. A close relationship has been found between this virus and the slow bee paralysis virus in studies on Belgian individuals of V. velutina [61]. However, no symptoms have been described for this virus. The Moku virus has also been detected in honey bee and Varroa samples. Although it was detected for the first time in V. pensylvanica [42,70,105], it has also been identified in other Vespids such V. crabro in UK [57] or V. velutina in Belgium [61] and France [45]. It has also been detected in P. chinensis and P. humilis, V. germanica, and V. vulgaris [66,68]. It is worth mentioning that in the specific case of V. velutina, as it is a predator that exerts great pressure on honey bees, a transmission in the direction of bees to Vespids could be suggested. Furthermore, the sequencing data and the higher abundance of this virus in Vespids versus honey bees could reinforce this suggestion and pose a future risk in the transmission of the virus from hornets to honey bees during predation events in hives [57,61,70]. However, it would be necessary to carry out more studies on the transmission of this virus that could demonstrate it. This virus has also been detected in co-occurrence with other pathogens; thus, in individuals of V. vulgaris from both New Zealand and Belgium, it has been detected in co-infection with high viral diversity. The majority belong to the Picorna family, but other families such as Luteo-Sobemo, Partiti and Tombus were also detected [68]. Of note is the detection of Vespula vulgaris Luteo-like virus 1, whose high presence in all life stages suggests a persistent infection in V. vulgaris colonies [68]. It is important to point out that the replicative form has also been detected, as it has been found in individuals of V. crabro [25], V. pensylvanica [42,57], P. chinensis, V. germanica, and V. vulgaris [66], as shown in Table 2 and Table 3. Although the Moku virus can infect honey bees, it is better established in Vespid species. In fact, it has been proven that in populations of V. vulgaris, the viral load of the Moku virus can affect vital traits such as the longevity of the colony [42].
5.6. Lake Sinai Virus (LSV)
LSV, belonging to the genus Sinaivirus, was discovered in Western honey bee (A. mellifera) samples near Lake Sinai, South Dakota, USA [106]. Initially, only two variants known as LSV1 and LSV2 were discovered that can infect continuously over time. Thus, LSV2 could reach maximum peaks in January and April while LSV1 in July [106,107]. These two viruses can give symptoms especially when the bee colony is weakened by other factors such as colony collapse disorder (CCD) [108] or Nosema spp. [109]. More variants were later detected and included in this virus—LSV3 [108], LSV-Navarra [108], LSV4, LSV5 and several LSV discovered in Belgium [110,111]. LSV1 and LSV2 are characterized by not presenting obvious symptoms and by their predominance in adults [107]. Contradictorily, the identification of this virus in Vespidae species has only been referred to its presence in a V. bicolor pupa from China [60]. In this study, an occurrence has also been observed with other viruses such as Apis mellifera Filamentous Virus (AmFV), IAPV, DWV, CSBV, and BQCV. However, given that no virus replication was found in these samples, the appearance of the virus is probably due to contamination by feeding.
5.7. Apis Mellifera Filamentous Virus (AmFV)
AmFV was originally described as a rickettsial disease of honey bees (A. mellifera) [112]; however, it was later characterized as a large, enveloped DNA virus [107,112]. One of its main diagnostic features is that it renders the hemolymph of adult bees milky white due to cell degradation and the large number of rod-shaped virions present, when examined by electron microscopy [73,107,112,113]. Despite being a widely distributed virus, studies on other possible hosts, such as Vespids, are scarce. In fact, only the study by Yang et al. [60] has identified AmFv in hornets from China, in V. bicolor pupae and in both pupae and adults in P. rothneyi [60]. In these detections, no symptoms have been detected. Coexistence with other pathogens such as IAPV, DWV, CSBV, BQCV, and LSV has also been detected in V. bicolor, while in P. rothneyi, AmFV has been detected together with BQCV or together with IAPV [60]. In honey bees, it can occur together with Nosema [75] as a prerequisite; however, in Vespids this association is not mentioned. The scant information on its transmission in Vespids only explains that its presence is due to consumption by bees, hence its presence in larvae. Furthermore, the fact that there is no evidence yet for Vespid-specific sequences suggests that Vespids are only carriers of this virus.
5.8. Bee Viruses Detected Exclusively in V. Velutina
Recently identified Macula-like virus (BeeMLV) is a polyadenylated RNA virus in the family Tymoviridae in the order Tymovirales and close to the order of Maculavirus [75,114]. The presence of BeeMLV in bees has been related to V. destructor, in which the replicative form of the virus has been detected, suggesting its participation as a possible transmission vector [75,114]. In honey bees, this virus can infect both pupae and adults. However, in Vespids it has only been detected in the gut of adult individuals of V. velutina from France; thus, it is suggested that the presence of this virus is probably related to bee predation [45].
It is also worth mentioning the La Jolla virus (LJV) belonging to the Iflaviridae family, discovered for the first time in Drosophila and detected in honey bees in Australian samples [115]. This virus was detected for the first time in Italian samples of V. velutina [20]. In that study, the presence of La Jolla virus in bee samples was also confirmed, suggesting transmission through environmental contamination. Although no symptoms have been reported in V. velutina, melanization followed by rapid death has been observed in Drosophila within 3 days after injection of the LJV strain. This contribution has focused on LJV as a good biopesticide candidate for this species [20]. However, in the case of V. velutina, a better characterization of this virus would be necessary.
Finally, other viruses associated with the analysis of V. velutina from France were also detected, such as the Acypi-like virus, Triato-like virus, Permutotetra-like virus, Partiti-like virus, Ifla-like virus, Nora-like virus, Menton virus, and Mott mill virus [45].
The rapid spread of V. velutina in invaded ecosystems, coupled with its ability to carry a wide range of pathogens, is indicative of the potential risk it may pose to the species with which it interacts, such as honey bees. Most of these viruses have been detected as the target of a search for control of this pest species [20,21,22,23]. The adaptation and accommodation of V. velutina in the invaded ecosystems brings about the presence of new interactions between viral pathogens and insects. A potential indicator of this event could be represented by the ratio of the replicative virus/non-replicative virus [20]. In V. velutina, the presence of replicative forms of several of the viruses mentioned above has already been confirmed. A total of nine viruses have replicated in samples of this hornet (Table 1).
The literature found regarding the distribution of V. velutina in areas other than the native corresponds to three countries in Europe (Figure 1a), in which this pathogenic diversity has been detected. France is one of the countries invaded by V. velutina that has carried out the largest number of investigations into the bee virus. Figure 1 shows the main bee viruses identified in this country, highlighting the replicative forms of the viruses DWV-B, BQCV, SBV, ABPV, Acypi-like virus, and Triato-like virus that were identified [45]. France is followed by the Italian and Belgian studies, where replication has only been found in Italy (DWV-A; DWV-B; DWV-C, BQCV, KBV, ALPV, and Triato-like virus [19,21]). V. velutina is native to Southeast Asia, being common in southern China [14,15]. From these areas, it has been able to invade along two routes, through northeast Asia and Europe. In northeast Asia, it spread through South Korea to Japan [15], where it has been introduced as an invasive species (Figure 1b). The main studies on bee viruses in Asia have been carried out with individuals from China. From these studies, the replicative forms for the APV and ALPV viruses were found [46,60]. These studies suggest and reinforce the existence of colonization in these environments through the establishment of a balance between pathogenicity factors and V. velutina.
6. High Rate of Evolution of Bee Viruses as a Future Risk in Invaded Ecosystems
Bee viruses represent a threat to the loss of honey bee populations. For this reason, special attention should be paid to their propensity to jump between host species and, therefore, threaten other populations of wild pollinators with important ecological and economic roles [116]. This spread also affects the introduction of non-native species, such as Vespids. The research discussed in this review demonstrates that there is a virus transmission from honeybees to Vespids, and that virus replication does occur on certain occasions. This highlights the need to reformulate the specificity of bee viruses for honey bees and to deepen the scarce knowledge on their spread and infection of new hosts.
In contrast, there are no studies demonstrating reverse directionality, that is, from the Vespids to honey bees. However, the literature highlights the spillover of bee viruses into ecosystems invaded by invasive Vespids. This poses a risk for the spread of bee viruses and considers a gap regarding the future of bee virus control in apiaries.
The viruses presented above are often single-stranded RNA positive (+ss) viruses. They have a high replication rate and high error rates, resulting in them being genetically very heterogeneous. Some of the bee viruses discussed in this review, such as DWV, have been sequenced and structurally characterized. DWV can present three different variants with different virulence (variant types A, B, and C). Some molecular studies estimate a mean evolutionary rate of 1.346 × 10−3 substitutions/site/year [117], allowing the rapid adaptation to new host environments, with recombination representing an additional key source of genetic variation.
The main infections caused are usually transmitted by the invasive mite V. destructor. This may have favored the evolution of the virulence of the bee viruses it transmits and driven the emergence of viruses in other species in areas where it has been introduced. In parallel, the occurrence of these viruses in Vespid species has revealed more specific replication dynamics in Vespids. However, the lack of knowledge about this type of occurrence in Vespids may suggest that there is a future risk of changes in infection dynamics due to the high rate of evolution of these viruses. This review therefore highlights the need to study bee viruses in invasive species from the time of their early introduction.
7. Conclusions
An increasing number of shared viral genomes have been found between the family Apidae and the family Vespidae. These viruses are RNA viruses with a high rate of evolution, allowing a high frequency of virus exchange within and between the families of Hymenoptera. This could lead to the reformulation of the definition of species-specific viruses and specifically for the bee. Knowledge about bee viruses in Vespid species is scarce. Most of the investigations describe the presence and/or replication of the bee virus in Vespids. However, very little is known about the transmission mechanisms of these viruses. V. velutina, which has an increasing spatial distribution outside its native range, is having a major impact on beekeeping. This has promoted a growing number of investigations on bee viruses in this species. No less impactful are other invasive species that prey on bees or share their habitat, whose introduction has changed the dynamics of virus replication. These species could be a potential factor in the transmission of bee viruses, affecting the health and strength of the colony. The information collected in this review on honey bee viruses in invasive Vespids is relevant to promote the further research on honey bee virus transmission in Vespids and improve the management of infectious diseases caused by bee viruses in apiaries.
Author Contributions
A.F. and M.M. conceptualized the research. M.S.R.-F. and A.F. prepared the original draft; M.S.R.-F., A.F., M.M., A.D.-A. and M.C.S. wrote and reviewed the manuscript. M.S.R.-F. and M.C.S. edited the final manuscript. M.S.R.-F. acquired funds. All authors have read and agreed to the published version of the manuscript.
Funding
Rodríguez-Flores, M.S. express his gratitude for the support of Postdoctoral Financial Aid of Xunta de Galicia and the University of Vigo, grant number ED481D-2022-021, founded by CONSELLERÍA DE CULTURA, EDUCACIÓN E ORDENACIÓN UNIVERSITARIA, and VICEPRESIDENCIA SEGUNDA E CONSELLERÍA DEECONOMÍA, EMPRESA E INNOVACIÓN (Government of Galicia, Spain). In addition, this research was funded by INTERREG ATLANTIC AREA PROGRAM (European Regional Development Fund—ERDF, European Union): EAPA_800/2018—Atlantic-POSitive.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful for the acquisition of funds to carry out a research stay at the Department of Veterinary Sciences of University of Pisa, with the support of a Post-doctoral Grant from the Xunta de Galicia and the University of Vigo.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zenni, R.D.; Essl, F.; García-Berthou, E.; McDermott, S.M. The economic costs of biological invasions around the world. NeoBiota 2021, 67, 1–9. [Google Scholar] [CrossRef]
- Lee, C.E. Evolutionary genetics of invasive species. Trends Ecol. Evol. 2002, 17, 386–391. [Google Scholar] [CrossRef]
- Colautti, R.I.; Ricciardi, A.; Grigorovich, I.A.; MacIsaac, H.J. Is invasion success explained by the enemy release hypothesis? Ecol. Lett. 2004, 7, 721–733. [Google Scholar] [CrossRef]
- Lester, P.J.; Bosch, P.J.; Gruber, M.A.M.; Kapp, E.A.; Peng, L.; Brenton-Rule, E.C.; Buchanan, J.; Stanislawek, W.L.; Archer, M.; Corley, J.C.; et al. No Evidence of Enemy Release in Pathogen and Microbial Communities of Common Wasps (Vespula vulgaris) in Their Native and Introduced Range. PLoS ONE 2015, 10, e0121358. [Google Scholar] [CrossRef] [PubMed]
- Crowl, T.A.; Crist, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef]
- Pickett, K.M.; Carpenter, J.M. Simultaneous analysis and the origin of eusociality in the Vespidae (Insecta: Hymenoptera). Arthropod Syst. Phylogeny 2010, 68, 3–33. [Google Scholar]
- Beggs, J.R.; Brockerhoff, E.G.; Corley, J.C.; Kenis, M.; Masciocchi, M.; Muller, F.; Rome, Q.; Villemant, C. Ecological effects and management of invasive alien Vespidae. BioControl 2011, 56, 505–526. [Google Scholar] [CrossRef]
- Cappa, F.; Cini, A.; Bortolotti, L.; Poidatz, J.; Cervo, R. Hornets and honey bees: A coevolutionary arms race between ancient adaptations and new invasive threats. Insects 2021, 12, 1037. [Google Scholar] [CrossRef]
- Rose, E.A.F.; Harris, R.J.; Glare, T.R.; Rose, E.A.F. Possible pathogens of social wasps (hymenoptera: Vespidae) and their potential as biological control agents. New Zeal. J. Zool. 1999, 26, 179–190. [Google Scholar] [CrossRef]
- Hammer, S.; Jensen, J.-K. The invasion of two species of social wasps (Hymenoptera, Vespidae) to the Faroe Islands. BioInvasions Rec. 2019, 8, 558–567. [Google Scholar] [CrossRef]
- D’Adamo, P.; Lozada, M. Flexible foraging behavior in the invasive social wasp Vespula germanica (Hymenoptera: Vespidae). Ann. Entomol. Soc. Am. 2009, 102, 1109–1115. [Google Scholar] [CrossRef]
- Nguyen, L.T.P.; Saito, F.; Kojima, J.; Carpenter, J.M. Vespidae of Viet Nam (Insecta: Hymenoptera) 2. Taxonomic Notes on Vespinae. Zool. Sci. 2006, 23, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Sonthichai, S. Nesting habits of some hornet species (Hymenoptera, Vespidae) in Northern Thailand. Agric. Nat. Resour. 2004, 38, 196–206. [Google Scholar]
- Monceau, K.; Bonnard, O.; Thiéry, D. Vespa velutina: A new invasive predator of honeybees in Europe. J. Pest Sci. 2014, 87, 1–16. [Google Scholar] [CrossRef]
- Choi, M.B. Foraging behavior of an invasive alien hornet (Vespa velutina) at Apis mellifera hives in Korea: Foraging duration and success rate. Entomol. Res. 2021, 51, 143–148. [Google Scholar] [CrossRef]
- Requier, F.; Rome, Q.; Chiron, G.; Decante, D.; Marion, S.; Menard, M.; Muller, F.; Villemant, C.; Henry, M. Predation of the invasive Asian hornet affects foraging activity and survival probability of honey bees in Western Europe. J. Pest Sci. 2019, 92, 567–578. [Google Scholar] [CrossRef]
- Mazzei, M.; Carrozza, M.L.; Luisi, E.; Forzan, M.; Giusti, M.; Sagona, S.; Tolari, F.; Felicioli, A. Infectivity of DWV associated to flower pollen: Experimental evidence of a horizontal transmission route. PLoS ONE 2014, 9, e113448. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee Viruses: Ecology, Pathogenicity, and Impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef]
- Mazzei, M.; Cilia, G.; Forzan, M.; Lavazza, A.; Mutinelli, F.; Felicioli, A. Detection of replicative Kashmir Bee Virus and Black Queen Cell Virus in Asian hornet Vespa velutina (Lepelieter 1836) in Italy. Sci. Rep. 2019, 9, 10091. [Google Scholar] [CrossRef]
- Marzoli, F.; Forzan, M.; Bortolotti, L.; Pacini, M.I.; Rodríguez-Flores, M.S.; Felicioli, A.; Mazzei, M. Next generation sequencing study on RNA viruses of Vespa velutina and Apis mellifera sharing the same foraging area. Transbound. Emerg. Dis. 2021, 68, 2261–2273. [Google Scholar] [CrossRef]
- Mazzei, M.; Forzan, M.; Cilia, G.; Sagona, S.; Bortolotti, L.; Felicioli, A. First detection of replicative deformed wing virus (DWV) in Vespa velutina nigrithorax. Bull. Insectol. 2018, 71, 211–216. [Google Scholar]
- Gabín-García, L.B.; Bartolomé, C.; Guerra-Tort, C.; Rojas-Nossa, S.V.; Llovo, J.; Maside, X. Identification of pathogens in the invasive hornet Vespa velutina and in native Hymenoptera (Apidae, Vespidae) from SW-Europe. Sci. Rep. 2021, 11, 11233. [Google Scholar] [CrossRef] [PubMed]
- Chauzat, M.-P.; Schurr, F.; Faucon, J.-P.; Blanchard, P.; Drajnudel, P. First detections of honey bee pathogens in nest of the Asian hornet (Vespa velutina) collected in France Magali Ribière-Chabert. CIHEAM Watch Lett. 2015, 33, 1–5. [Google Scholar]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y. Communication Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Forzan, M.; Sagona, S.; Mazzei, M.; Felicioli, A. Detection of deformed wing virus in Vespa crabro. Bull. Insectol. 2017, 70, 261–265. [Google Scholar]
- Liu, S.; Vijayendran, D.; Bonning, B.C. Next generation sequencing technologies for insect virus discovery. Viruses 2011, 3, 1849–1869. [Google Scholar] [CrossRef]
- Armstrong, T.R.; Stamp, N.E. Colony productivity and foundress behaviour of a native wasp versus an invasive social wasp. Ecol. Entomol. 2003, 28, 635–644. [Google Scholar] [CrossRef]
- Rankin, E.E.W. ScienceDirect Emerging patterns in social wasp invasions. Curr. Opin. Insect Sci. 2021, 46, 72–77. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Yañez, O.; Piot, N.; Dalmon, A.; de Miranda, J.R.; Chantawannakul, P.; Panziera, D.; Amiri, E.; Smagghe, G.; Schroeder, D.; Chejanovsky, N. Bee Viruses: Routes of Infection in Hymenoptera. Front. Microbiol. 2020, 11, 943. [Google Scholar] [CrossRef]
- Lipsitch, M.; Siller, S.; Nowak, M.A. Horizontal transmission. Evolution 1996, 50, 1729–1741. [Google Scholar] [PubMed]
- Wang, F.; Fang, Q.; Wang, B.; Yan, Z.; Hong, J.; Bao, Y.; Kuhn, J.H.; Werren, J.H.; Song, Q.; Ye, G. A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 2017, 13, e1006201. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Fries, I. Venereal and vertical transmission of deformed wing virus in honeybees (Apis mellifera L.). J. Invertebr. Pathol. 2008, 98, 184–189. [Google Scholar] [CrossRef]
- Fievet, J.; Tentcheva, D.; Gauthier, L.; De Miranda, J.; Cousserans, F.; Colin, M.E.; Bergoin, M. Localization of deformed wing virus infection in queen and drone Apis mellifera L. Virol. J. 2006, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Amiri, E.; Meixner, M.D.; Kryger, P. Deformed wing virus can be transmitted during natural mating in honey bees and infect the queens. Sci. Rep. 2016, 6, 33065. [Google Scholar] [CrossRef]
- Martinez, J.; Lepetit, D.; Ravallec, M.; Fleury, F.; Varaldi, J. Additional heritable virus in the parasitic wasp Leptopilina boulardi: Prevalence, transmission and phenotypic effects. J. Gen. Virol. 2016, 97, 523–535. [Google Scholar] [CrossRef]
- Stasiak, K.; Renault, S.; Federici, B.A.; Bigot, Y. Characteristics of pathogenic and mutualistic relationships of ascoviruses in field populations of parasitoid wasps. J. Insect Physiol. 2005, 51, 103–115. [Google Scholar] [CrossRef]
- Roossinck, M.J. Changes in population dynamics in mutualistic versus pathogenic viruses. Viruses 2011, 3, 12–19. [Google Scholar] [CrossRef]
- Roossinck, M.J. The good viruses: Viral mutualistic symbioses. Nat. Rev. Microbiol. 2011, 9, 99–108. [Google Scholar] [CrossRef]
- Renault, S.; Stasiak, K.; Federici, B.; Bigot, Y. Commensal and mutualistic relationships of reoviruses with their parasitoid wasp hosts. J. Insect Physiol. 2005, 51, 137–148. [Google Scholar] [CrossRef]
- Dobelmann, J.; Loope, K.J.; Wilson-Rankin, E.; Quinn, O.; Baty, J.W.; Gruber, M.A.M.; Lester, P.J. Fitness in invasive social wasps: The role of variation in viral load, immune response and paternity in predicting nest size and reproductive output. Oikos 2017, 126, 1208–1218. [Google Scholar] [CrossRef]
- Loope, K.J.; Wilson Rankin, E.E. Viral load, not food availability or temperature, predicts colony longevity in an invasive eusocial wasp with plastic life history. Sci. Rep. 2021, 11, 10087. [Google Scholar] [CrossRef] [PubMed]
- De Jong, D.; Morse, R.A.; Eickwort, G.C. Mite pests of honey bees. Annu. Rev. Entomol. 1982, 27, 229–252. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.M.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Dalmon, A.; Gayral, P.; Decante, D.; Klopp, C.; Bigot, D.; Thomasson, M.; AHerniou, E.; Alaux, C.; Conte, Y. Le Viruses in the invasive hornet Vespa velutina. Viruses 2019, 11, 1041. [Google Scholar] [CrossRef] [PubMed]
- Yañez, O.; Zheng, H.Q.; Hu, F.L.; Neumann, P.; Dietemann, V. A scientific note on Israeli acute paralysis virus infection of Eastern honeybee Apis cerana and vespine predator Vespa velutina. Apidologie 2012, 43, 587–589. [Google Scholar] [CrossRef]
- Sánchez, O.; Arias, A. All that glitters is not gold: The other insects that fall into the asian yellow-legged hornet Vespa velutina ‘specific’ traps. Biology 2021, 10, 448. [Google Scholar] [CrossRef]
- Jeliński, M. The mite Varroa jacobsoni Oudemans, 1904 on larvae of common wasp Vespa (Paravespula) vulgaris L. Wiad. Parazytol. 1990, 36, 55–58. [Google Scholar]
- Loope, K.J.; Baty, J.W.; Lester, P.J.; Wilson Rankin, E.E. Pathogen shifts in a honeybee predator following the arrival of the Varroa mite. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182499. [Google Scholar] [CrossRef]
- Fan, Q.H.; Zhang, Z.Q.; Brown, R.; France, S.; Bennett, S. New Zealand Pneumolaelaps Berlese (Acari: Laelapidae): Description of a new species, key to species and notes on biology. Syst. Appl. Acarol. 2016, 21, 119–138. [Google Scholar] [CrossRef][Green Version]
- Felden, A.; Baty, J.W.; Bulgarella, M.; Brown, R.L.; Dobelmann, J.; Gruber, M.A.M.; Quinn, O.; Lester, P.J. Viral and fungal pathogens associated with Pneumolaelaps niutirani (Acari: Laelapidae): A mite found in diseased nests of Vespula wasps. Insectes Soc. 2020, 67, 83–93. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Sagona, S.; Giusti, M.; Jarmela dos Santos, P.E.; Nanetti, A.; Felicioli, A. Nosema ceranae infection in honeybee samples from Tuscanian Archipelago (Central Italy) investigated by two qPCR methods. Saudi J. Biol. Sci. 2019, 26, 1553–1556. [Google Scholar] [CrossRef]
- Leat, N.; Ball, B.; Govan, V.; Davison, S. Analysis of the complete genome sequence of black queen-cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 2000, 81, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Porrini, M.P.; Porrini, L.P.; Garrido, P.M.; de Melo e Silva Neto, C.; Porrini, D.P.; Muller, F.; Nuñez, L.A.; Alvarez, L.; Iriarte, P.F.; Eguaras, M.J. Nosema ceranae in South American Native Stingless Bees and Social Wasp. Microb. Ecol. 2017, 74, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Cardaio, I.; dos Santos, P.E.J.; Ellis, J.D.; Nanetti, A. The first detection of Nosema ceranae (Microsporidia) in the small hive beetle, Aethina tumida Murray (Coleoptera: Nitidulidae). Apidologie 2018, 49, 619–624. [Google Scholar] [CrossRef]
- Highfield, A.; Kevill, J.; Kevill, J.; Mordecai, G.; Mordecai, G.; Hunt, J.; Henderson, S.; Sauvard, D.; Feltwell, J.; Martin, S.J.; et al. Detection and replication of moku virus in honey bees and socialwasps. Viruses 2020, 12, 607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, S.Y.; Evans, J.D.; Pettis, J.S.; Yin, G.F.; Chen, Y.P. New evidence that deformed wing virus and black queen cell virus are multi-host pathogens. J. Invertebr. Pathol. 2012, 109, 156–159. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, H.; Shi, J.; Xu, X.; Wu, Y.; Guo, R.; Chen, D.; Wang, X.; Deng, S.; Yang, S.; et al. Discovery of aphid lethal paralysis virus in Vespa velutina and Apis cerana in China. Insects 2019, 10, 157. [Google Scholar] [CrossRef]
- Yang, S.; Gayral, P.; Zhao, H.; Wu, Y.; Jiang, X.; Wu, Y.; Bigot, D.; Wang, X.; Yang, D.; Herniou, E.A.; et al. Occurrence and molecular phylogeny of honey bee viruses in Vespids. Viruses 2020, 12, 6. [Google Scholar] [CrossRef]
- Garigliany, M.; Taminiau, B.; El Agrebi, N.; Cadar, D.; Gilliaux, G.; Hue, M.; Desmecht, D.; Daube, G.; Linden, A.; Farnir, F.; et al. Moku virus in invasive Asian Hornets, Belgium, 2016. Emerg. Infect. Dis. 2017, 23, 2109–2112. [Google Scholar] [CrossRef] [PubMed]
- Forzan, M.; Felicioli, A.; Sagona, S.; Bandecchi, P.; Mazzei, M. Complete genome sequence of deformed wing virus isolated from Vespa crabro in Italy. Genome Announc. 2017, 5, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Power, K.; Altamura, G.; Martano, M.; Maiolino, P. Detection of Honeybee Viruses in Vespa orientalis. Front. Cell. Infect. Microbiol. 2022, 12, 896932. [Google Scholar] [CrossRef]
- Evison, S.E.; Roberts, K.E.; Laurenson, L.; Pietravalle, S.; Hui, J.; Biesmeijer, J.C. Pervasiveness of parasites in pollinators. PLoS ONE 2012, 7, e30641. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; Vanengelsdorp, D.; Lipkin, W.I.; Depamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef] [PubMed]
- Dobelmann, J.; Felden, A.; Lester, P.J. Genetic strain diversity of multi-host RNA viruses that infect a wide range of pollinators and associates is shaped by geographic origins. Viruses 2020, 12, 358. [Google Scholar] [CrossRef]
- Quinn, O.; Gruber, M.A.M.; Brown, R.L.; Baty, J.W.; Bulgarella, M.; Lester, P.J. A metatranscriptomic analysis of diseased social wasps (Vespula vulgaris) for pathogens, with an experimental infection of larvae and nests. PLoS ONE 2018, 13, e0209589. [Google Scholar] [CrossRef]
- Remnant, E.J.; Baty, J.W.; Bulgarella, M.; Dobelmann, J.; Quinn, O.; Gruber, M.A.M.; Lester, P.J. A diverse viral community from predatory wasps in their native and invaded range, with a new virus infectious to honey bees. Viruses 2021, 13, 1431. [Google Scholar] [CrossRef]
- Brenton-Rule, E.C.; Dobelmann, J.; Baty, J.W.; Brown, R.L.; Dvorak, L.; Grangier, J.; Masciocchi, M.; McGrannachan, C.; Shortall, C.R.; Schmack, J.; et al. The origins of global invasions of the German wasp (Vespula germanica) and its infection with four honey bee viruses. Biol. Invasions 2018, 20, 3445–3460. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Brettell, L.E.; Pachori, P.; Villalobos, E.M.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Moku virus; A new Iflavirus found in wasps, honey bees and Varroa. Sci. Rep. 2016, 6, 4–9. [Google Scholar] [CrossRef]
- Levitt, A.L.; Singh, R.; Cox-Foster, D.L.; Rajotte, E.; Hoover, K.; Ostiguy, N.; Holmes, E.C. Cross-species transmission of honey bee viruses in associated arthropods. Virus Res. 2013, 176, 232–240. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L. Honey bee pathology. Annu. Rev. Entomol. 1968, 13, 191–212. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 86, 3419–3424. [Google Scholar] [CrossRef]
- Gisder, S.; Genersch, E. Honey bee viruses. Viruses 2015, 7, 5603–5608. [Google Scholar] [CrossRef]
- Yue, C.; Schröder, M.; Gisder, S.; Genersch, E. Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 2007, 88, 2329–2336. [Google Scholar] [CrossRef]
- Ball, B.V.; Allen, M.F. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 1988, 113, 237–244. [Google Scholar] [CrossRef]
- Moeckel, N.; Gisder, S.; Genersch, E. Horizontal transmission of deformed wing virus: Pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J. Gen. Virol. 2011, 92, 370–377. [Google Scholar] [CrossRef]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.F.; Gogol-Döring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160811. [Google Scholar] [CrossRef]
- Santamaria, J.; Villalobos, E.M.; Brettell, L.E.; Nikaido, S.; Graham, J.R.; Martin, S. Evidence of Varroa-mediated deformed wing virus spillover in Hawaii. J. Invertebr. Pathol. 2018, 151, 126–130. [Google Scholar] [CrossRef]
- Bailey, L.; Woods, R.D. Two more small RNA viruses from honey bees and further observations on sacbrood and acute bee-paralysis viruses. J. Gen. Virol. 1977, 37, 175–182. [Google Scholar] [CrossRef]
- Calvo, M.Á.; Balart, M.; Baz, A.; Creus, S.; Dueso, M.; Farré, C. Principales enfermedades de etiología vírica de interés en apicultura. An. R. Acad. Dr. 2013, 17, 1138–2414. [Google Scholar]
- Benjeddou, M.; Leat, N.; Allsopp, M.; Davison, S. Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase PCR. Appl. Environ. Microbiol. 2001, 67, 2384–2387. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Woods, R.D. Three previously undescribed viruses from the honey bee. J. Gen. Virol. 1974, 25, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Ribière, M.; Ball, B.; Aubert, M. Natural history and geographical distribution of honey bee viruses. In Virology and the Honey Bee; Ball, B., Aubert, M., Fries, I., Eds.; Publications Office: Brussels, Belgium, 2008; pp. 15–84. [Google Scholar]
- Santillán-Galicia, M.T.; Ball, B.V.; Clark, S.J.; Alderson, P.G. Transmission of deformed wing virus and slow paralysis virus to adult bees (Apis mellifera L.) by Varroa destructor. J. Apic. Res. 2010, 49, 141–148. [Google Scholar] [CrossRef]
- Williamson, C.; Rybicki, E.P.; Kasdorf, G.G.F.; Von Wechmar, M.B. Characterization of a new picorna-like virus isolated from aphids. J. Gen. Virol. 1988, 69, 787–795. [Google Scholar] [CrossRef]
- Van Munster, M.; Dullemans, A.M.; Verbeek, M.; Van Den Heuvel, J.; Clerivet, A.; Van Der Wilk, F. Sequence analysis and genomic organization of Aphid lethal paralysis virus: A new member of the family Dicistroviridae. J. Gen. Virol. 2002, 83, 3131–3138. [Google Scholar] [CrossRef]
- Granberg, F.; Vicente-Rubiano, M.; Rubio-Guerri, C.; Karlsson, O.E.; Kukielka, D.; Belák, S.; Sánchez-Vizcaíno, J.M. Metagenomic detection of viral pathogens in Spanish honeybees: Co-infection by aphid lethal paralysis, Israel acute paralysis and Lake Sinai viruses. PLoS ONE 2013, 8, e57459. [Google Scholar] [CrossRef]
- Maori, E.; Lavi, S.; Mozes-Koch, R.; Gantman, Y.; Peretz, Y.; Edelbaum, O.; Tanne, E.; Sela, I. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: Evidence for diversity due to intra-and inter-species recombination. J. Gen. Virol. 2007, 88, 3428–3438. [Google Scholar] [CrossRef]
- Chen, Y.P.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Ribière, M.; Triboulot, C.; Mathieu, L.; Aurières, C.; Faucon, J.-P.; Pépin, M. Molecular diagnosis of chronic bee paralysis virus infection. Apidologie 2002, 33, 339–351. [Google Scholar] [CrossRef]
- de Miranda, J.R. Diagnostic techniques for virus detection in honey bees. In Virology and the Honey Bee; Ball, B., Aubert, M., Fries, I., Eds.; Publications Office: Brussels, Belgium, 2008; pp. 125–133. [Google Scholar]
- Ribière, M.; Olivier, V.; Blanchard, P. Chronic bee paralysis: A disease and a virus like no other? J. Invertebr. Pathol. 2010, 103, S120–S131. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Ball, B. The incidence and world distribution of honey bee viruses. Bee World 1996, 77, 141–162. [Google Scholar] [CrossRef]
- Celle, O.; Blanchard, P.; Olivier, V.; Schurr, F.; Cougoule, N.; Faucon, J.-P.; Ribière, M. Detection of Chronic bee paralysis virus (CBPV) genome and its replicative RNA form in various hosts and possible ways of spread. Virus Res. 2008, 133, 280–284. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Cordoni, G.; Budge, G. The Acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J. Invertebr. Pathol. 2010, 103, S30–S47. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Patterns of viral infection in honey bee queens. J. Gen. Virol. 2013, 94, 668. [Google Scholar] [CrossRef]
- Chen, Y.; Pettis, J.S.; Evans, J.D.; Kramer, M.; Feldlaufer, M.F. Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 2004, 35, 441–448. [Google Scholar] [CrossRef]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic Varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef]
- Hung, A.C.F. PCR detection of Kashmir bee virus in honey bee excreta. J. Apic. Res. 2000, 39, 103–106. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Drebot, M.; Tyler, S.; Shen, M.; Cameron, C.E.; Stoltz, D.B.; Camazine, S.M. Complete nucleotide sequence of Kashmir bee virus and comparison with acute bee paralysis virus. J. Gen. Virol. 2004, 85, 2263–2270. [Google Scholar] [CrossRef]
- Jin, L.; Mehmood, S.; Zhang, G.; Song, Y.; Su, S.; Huang, S.; Huang, H.; Zhang, Y.; Geng, H.; Huang, W.-F. Visualizing sacbrood virus of honey bees via transformation and coupling with enhanced green fluorescent protein. Viruses 2020, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, T.; Evans, J.D.; Rose, R.; Zhao, Y.; Li, Z.; Li, J.; Huang, S.; Heerman, M.; Rodríguez-García, C.; et al. The phylogeny and pathogenesis of sacbrood virus (SBV) infection in European honey bees, Apis mellifera. Viruses 2019, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Rothman, J.A.; Loope, K.J.; McFrederick, Q.S.; Wilson Rankin, E.E. Microbiome of the wasp Vespula pensylvanica in native and invasive populations, and associations with Moku virus. PLoS ONE 2021, 16, e0255463. [Google Scholar] [CrossRef]
- Runckel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; DeRisi, J.L. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS ONE 2011, 6, e20656. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.P.; Gauthier, L.; Genersch, E.; De Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; Vanengelsdorp, D.; Evans, J.D. Pathogen webs in collapsing honey bee colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; De Graaf, D.C. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS ONE 2013, 8, e72443. [Google Scholar] [CrossRef]
- Ravoet, J.; De Smet, L.; Wenseleers, T.; De Graaf, D.C. Genome sequence heterogeneity of Lake Sinai Virus found in honey bees and Orf1/RdRP-based polymorphisms in a single host. Virus Res. 2015, 201, 67–72. [Google Scholar] [CrossRef]
- Gauthier, L.; Cornman, S.; Hartmann, U.; Cousserans, F.; Evans, J.D.; De Miranda, J.R.; Neumann, P. The Apis mellifera filamentous virus genome. Viruses 2015, 7, 3798–3815. [Google Scholar] [CrossRef]
- Clark, T.B. A filamentous virus of the honey bee. J. Invertebr. Pathol. 1978, 32, 332–340. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Cornman, R.S.; Evans, J.D.; Semberg, E.; Haddad, N.; Neumann, P.; Gauthier, L. Genome characterization, prevalence and distribution of a macula-like virus from Apis mellifera and Varroa destructor. Viruses 2015, 7, 3586–3602. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.M.K.; Anderson, D.L.; Durr, P.A. Metagenomic analysis of Varroa-free Australian honey bees (Apis mellifera) shows a diverse Picornavirales virome. J. Gen. Virol. 2018, 99, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Nanetti, A.; Bortolotti, L.; Cilia, G. Pathogens Spillover from Honey Bees to Other Arthropods. Pathogens 2021, 10, 1044. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).