Simple Summary
Western flower thrips (Frankliniella occidentalis) have a wide range of hosts. Therefore, they can colonize new host plants with each seasonal change. This study examined whether the superoxide dismutase (SOD) gene regulates the feeding adaptation of F. occidentalis after host shifting. The coding sequences for CCS1 and MnSOD2 in F. occidentalis were cloned and the corresponding amino acid sequence was predicted, and the mRNA expression levels of these two genes at different developmental stages were determined. Further, the mRNA expression levels of FoCCS1 and FoMnSOD2 in second-instar larvae and adult females transferred to kidney bean and broad bean plants for rearing were analyzed. Decreasing the mRNA levels of FoCCS1 and SOD activity by RNA interference significantly reduced the survival rate and fecundity of adult F. occidentalis females. These findings provide a reference for analyzing the adaptive mechanism of F. occidentalis after host shifting.
Abstract
Western flower thrips (Frankliniella occidentalis) pose a serious threat to the global vegetable and flower crop production. The regulatory mechanism for superoxide dismutase (SOD) in the feeding adaptation of F. occidentalis after host shifting remains unclear. In this study, the copper chaperone for SOD (CCS) and manganese SOD (MnSOD) genes in F. occidentalis were cloned, and their expression levels at different developmental stages was determined. The mRNA expression of FoCCS1 and FoMnSOD2 in F. occidentalis second-instar larvae and adult females of F1, F2, and F3 generations was analyzed after shifting the thrips to kidney bean and broad bean plants, respectively. The F2 and F3 second-instar larvae and F2 adult females showed significantly upregulated FoCCS1 mRNA expression after shifting to kidney bean plants. The F1 second-instar larvae and F2 adult females showed significantly upregulated FoCCS1 mRNA expression after shifting to broad bean plants. The RNA interference significantly downregulated the FoCCS1 mRNA expression levels and adult females showed significantly inhibited SOD activity after shifting to kidney bean and broad bean plants. F. occidentalis adult females subjected to RNA interference and released on kidney bean and broad bean leaves for rearing, respectively, significantly reduced the survival rate and fecundity. These findings suggest that FoCCS1 plays an active role in regulating the feeding adaptation ability of F. occidentalis after host shifting.
1. Introduction
The western flower thrips, Frankliniella occidentalis Pergande (Thysanoptera: Thripidae) is one of the most difficult-to-manage pests of vegetable and flower crops worldwide. F. occidentalis directly damages crop production by feeding on the leaves, flowers, and fruits of host plants and laying eggs on the plants; it can also indirectly damage crops by transmitting plant viruses [1]. This was first reported as an invasive thrip species in China in 2000; since then, it has rapidly spread to more than 10 provinces. F. occidentalis can reproduce parthenogenetically and sexually and has a short generation cycle [2,3]. The wide host range (>500 plant species), effective hiding ability, and adaptability of F. occidentalis greatly contributes to its management difficulty [4]. These biological features also enable F. occidentalis to invade and colonize seasonal crops of the current season. For example, kidney and broad beans are the main hosts of F. occidentalis [5]. Kidney beans are mainly cultivated in the summer and autumn seasons and are one of the main summer hosts of F. occidentalis. In contrast, broad beans are cultivated in winter and spring and are one of the main winter hosts of F. occidentalis. The seasonal growth characteristics of kidney beans and broad beans may provide a habitat for the colonization of F. occidentalis in different seasons.
It is a common ecological phenomenon for herbivorous insects to transfer to new hosts for feeding and colonization because of factors such as seasonal alterations in crop growth [6,7,8]. When herbivorous insects who colonize and adapt to one host for a long time shift to another host, they must first overcome the new host plant’s defense barrier to successfully colonize it [8,9]. An apoplastic burst of reactive oxygen species (ROS) occurs in the host plant, which is the first barrier against a continued attack by herbivorous insects [10]. ROS accumulation in host plants directly prevents insect feeding damage, and mediates the activation of defense genes and enzymes by regulating their transcription, participates in the interaction of signaling pathways in the plant defense system and establishes corresponding defense mechanisms [10,11,12]. Among all ROS, H2O2 is a highly stable and freely diffusible core element of plant-induced defense response [11,12]. In plants, H2O2 directly acts on herbivorous insects or enters the insect cells and changes the redox state, resulting in oxidative damage in the insect midgut [10,13]. For example, Schizaphis graminum infestation in wheat for 20 min leads to H2O2 accumulation, which effectively protects wheat from subsequent feeding damage [14]. An increase in H2O2 concentration is also an effective defense mechanism used by the peanut crop against cotton bollworm Helicoverpa armigera [15].
Superoxide dismutase (SOD) is a key enzyme in the first-line antioxidant defense system and is the only enzyme that directly metabolizes the superoxide class of ROS [16]. SOD catalyzes the disproportionation of ROS to form H2O and H2O2. H2O2 is subsequently converted to H2O by the action of catalase or peroxidase, which ultimately scavenges ROS and protects the organism from oxidative damage [16]. SOD in insects could be categorized into Cu/ZnSOD and MnSOD. Cu/ZnSOD is abundant in the serum and cytoplasm of the insect midgut, fat body, and body wall, whereas MnSOD mainly exists in the mitochondria [17]. Additionally, MnSOD is usually classified and annotated as SOD2. The Cu/ZnSOD family usually comprises five members: cytoplasmic Cu/ZnSOD, extracellular SOD (SOD3), copper chaperone (CCS), related to SOD (RSOD), and SODesque (SODq) [18]. Studies have shown that CCS is essential for the activation of mammalian Cu/ZnSOD [19]. Moreover, CCS is an essential factor for the regulation of copper homeostasis and promotion of all levels of SOD1 activity maturation [20,21,22]. Studies have indicated that insect SOD plays an important role in regulating the elimination of ROS induced by temperature [23], ultraviolet light, and heavy metal stress [24], and actively participates in the regulation of insect feeding adaptation during host shifting [25].
When insects shift to new hosts for feeding, a series of plant ROS-related defense responses are inevitably triggered [11,26,27]. Insect SOD plays an important role in protecting insects from oxidative damage and regulating their adaptability to host plants [13,17,25]. Previous studies have shown that F. occidentalis SODs are actively involved in the feeding response after host shifting [25]. However, the specific molecular mechanism underlying the regulation of the feeding adaptation of F. occidentalis after host shifting remains unclear. Therefore, in this study, the CCS and MnSOD gene subfamily of F. occidentalis were cloned and their mRNA expression levels in the second-instar larvae and adult females of the F1, F2, and F3 generations after host shifting were analyzed. Using RNAi technology, the effect of the downregulation of the highly expressed SOD genes on the feeding adaptation of F. occidentalis after host shifting was also investigated.
2. Materials and Methods
2.1. Insect Culture and Host Plant Cultivation
F. occidentalis were collected from Huaxi District, Guiyang City, China, and were raised in an RXZ-type multistage programming artificial climate box (Ningbo Jiangnan Instrument Factory, Ningbo, China) at the Institute of Entomology, Guizhou University. The breeding environment was maintained a 25 ± 1 °C, 70 ± 5% relative humidity, and 14:10 h light:dark photoperiod. Kidney bean pods were provided as food. F. occidentalis reared for more than 70 successive generations were used as the experimental population.
Kidney bean seeds (Phaseolus vulgaris L. (Fabales: Fabaceae), variety Jinshulu, were obtained from the Shengnong Seed Company in Xinji City, China. One seed was planted in each artificial climate chamber (environment conditions: 25 ± 1 °C, 65 ± 10% relative humidity, and 14:10 h light:dark photoperiod) in sterilized nutrient soil, and two seeds were planted in each vegetative bowl (11.5 cm diameter × 10.0 cm height). Approximately 16 days after germination, the second trifoliate leaves appeared, and healthy and undamaged kidney bean plants with similar growth characteristics were selected as the experimental hosts.
Broad bean seeds (Vicia faba L.), variety Lincan No. 5, were purchased from Kangle County Jinzhong Agricultural Products Industrial Development Co., Ltd., Linxi, China, and cultivated under the same conditions as that of kidney bean plants. The seedlings were grown for 20 days after germination, and healthy and undamaged broad bean plants with 8–10 leaves and approximately 15 cm in height showing similar growth characteristics were selected as the experimental hosts.
2.2. Reverse-Transcription PCR and Cloning by Rapid Amplification of the cDNA Ends of the F. occidentalis SOD Gene
Total RNA from the F. occidentalis adults from pod-feeding populations was extracted using Eastp® Super Total RNA Extraction Kit (Promega (Beijing) Biotech Co., Ltd., Beijing, China). cDNA was synthesized by reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Vilnius, Lithuania) as per the manufacturer’s instructions. SOD sequences were screened on the basis of the tested transcriptome data. The online tool primer designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome, accessed on 14 March 2019) was used to design cDNA cloning primers FoCCS1-F/FoCCS1-R and FoMnSOD2-F/FoMnSOD2-R (Table S1). PCR amplification was performed using Taq PCR Master Mix (2×, with blue dye) (Sangon Biotech, Shanghai, China). The PCR product was purified using the SanPrep Column PCR Product Purification Kit (Sangon Biotech), ligated using the pMD18T vector cloning kit (Takara Bio USA, Inc., San Jose, CA, USA), and transformed into competent DH5α cells. The transformed DH5α cells were cultured in Luria broth (LB) without antibiotics at 37 °C for 1.5 h and then streaked on LB agar plates containing 100 μg/mL ampicillin and incubated. Subsequently, the two intermediate gene fragments were obtained by sequencing after colony PCR and detected by electrophoresis. As per the obtained intermediate fragment, 3′-end primers (Table S1) were designed according to the instructions of SMARTer® RACE 5′/3′ Kit Components (Takara Bio USA, Inc.). The 3′-end cDNA sequence was obtained through PCR amplification, gel-cutting purification, ligation, transformation, and sequencing. Using SeqMan v.7.1.0 software (DNASTAR, Inc., Madison, WI, USA), the amplified intermediate fragment sequence of the target gene and the 3′-end cDNA sequence were spliced to obtain the complete coding sequence (CDS) of the target gene. The full-length verification primers cFoCCS1-F/cFoCCS1-R and cFoMnSOD2-F/cFoMnSOD2-R (Table S1) were set at both ends of the CDS region for full-length sequence verification by PCR amplification.
2.3. Bioinformatics Analysis of the SOD Gene Sequence
The cloned gene sequences were compared and analyzed using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 29 May 2019). The complete open reading frame (ORF) of the gene sequence was identified using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 29 May 2019). The amino acid sequences were predicted using the Translate tool (http://web.expasy.org/translate/, accessed on 29 May 2019), and phosphorylation sites were predicted with KinasePhos (http://kinasephos.mbc.nctu.edu.tw, accessed on 29 May 2019). Using ExPASy–PROSITE (https://prosite.expasy.org/, accessed on 29 May 2019), protein sequences were analyzed as PROSITE signatures. The ExPASy-ProtParam tool (https://web.expasy.org/protparam/, accessed on 29 May 2019) was used to analyze the physicochemical properties of the predicted proteins. NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 29 May 2019) was used to analyze catalytically active sites and conserved domains. SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html, accessed on 29 May 2019) was used to predict the secondary structure of proteins, SWISS-MODEL (https://swissmodel.expasy.org/interactive, accessed on 29 May 2019) was used to predict protein tertiary structures, and PyMOL 1.1 was used for three-dimensional modeling. A phylogenetic tree was constructed using the neighbor-joining method and bootstrapped 1000 times in MEGA 6.0 (Mega Limited, Auckland, New Zealand).
2.4. Expression Profiles of FoCCS1 and FoMnSOD2 at the Different Developmental Stages of F. occidentalis
The expression profiles of FoCCS1 and FoMnSOD2 at different developmental stages were determined using the F. occidentalis population reared on kidney bean pods. The larval stages (first instar, La-1; second instar on day 1, La2-1; and second instar on day 3, La2-3); pupal stage (Pu; on day 2 after pupation); and adult males and females on days 2 (Fe-2 and Ma-2), 5 (Fe-5 and Ma-5), 8 (Fe-8 and Ma-8), 11 (Fe-11 and Ma-11), and 14 (Fe-14 and Ma-14) after eclosion were collected. Three replicates per instar and 100 F. occidentalis individuals per replicate were used. The Primer Design tool was used to design primers for quantitative reverse-transcription PCR (RT-qPCR) of target genes (Table S2). RT-qPCR was performed on a CFX96TM real-time quantitative PCR system (BioRad, Hercules, CA, USA) using EF-1a as an internal reference gene (GenBank: AB277244.1) [28] and FastStart Essential DNA Green Master kit (Roche Diagnostics GmbH, Penzberg, Germany) according to manufacturer’s instructions. The reaction mixture (20 μL) contained 2 μL of cDNA (300 ng/μL), 1 μL each of the upstream and downstream primers, 6 μL of H2O, and 10 μL of DNA Green Master Mix. The following PCR program was used: 40 cycles of pre-denaturation at 95 °C for 10 min, denaturation at 95 °C for 30 s, annealing (Tm = 57 °C) for 30 s, and final extension at 72 °C for 30 s.
2.5. Effects of Host Shifting on the Expression Levels of FoCCS1 and FoMnSOD2 in F. occidentalis
F. occidentalis were reared after host shifting, and samples were collected as described by Liu et al. [25]. Adult females from the experimental F. occidentalis population were released into rearing boxes containing kidney bean and broad bean plants, respectively, 3 days after eclosion. There were 12 pots of kidney bean plants per rearing box, and approximately 100 female adult F. occidentalis were released per pot of kidney bean. After 24 h of egg laying, all F. occidentalis adults were removed. The developmental status of F. occidentalis on the host plant was recorded daily. When the hatched larvae developed into second-instar larvae, samples of these second-instar larvae of the F1 generation were collected. The remaining larvae continued to be reared to the adult stage. During this period, kidney bean and broad bean plants were replaced every 2 days to ensure continued feeding by F. occidentalis larvae. F1 generation adult females were collected 3 days after eclosion. The second-instar larvae and adult females reared on kidney bean plants were denoted as PLaF1 and PFeF1, respectively. The second-instar larvae and adult females reared on broad bean plants were denoted as VLaF1 and VFeF1, respectively. Samples from F2 and F3 generations were collected using the same rearing method. The F2 generation samples were denoted as PLaF2, PFeF2, VLaF2, and VFeF2, respectively, and the F3 generation samples were denoted as PLaF3, PFeF3, VLaF3, and VFeF3, respectively. In this experiment, the F. occidentalis populations reared on kidney bean pods were used as controls, and the corresponding samples were denoted as LaCK and FeCK, respectively. There were three biological replicates per treatment with 100 F. occidentalis individuals per replicate. The samples were flash-frozen in liquid nitrogen and stored at −80 °C. Total RNA extraction, cDNA synthesis, and RT-qPCR were performed as described above.
2.6. Synthesis of FoCCS1 Double-Stranded RNA
The FoCCS1 CDS was used (https://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl, accessed on 9 December 2020) to design primers for double-stranded (ds) RNA synthesis. The T7 promoter sequence (TAATACGACTCACTATAGGG), dsFoCCS1-F, and dsFoCCS1-R were added to the 5′ ends of the upstream and downstream primers, respectively. The TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific) was used to synthesize FoCCS1 dsRNA, and the GeneJET RNA Purification Kit (Thermo Scientific) was used to purify the synthesized dsRNA, which was stored at −80 °C. eGFP was used as a negative control (Table S1).
2.7. FoCCS1 RNA Interference and Determination of SOD Enzyme Activity
The F2 generation adult females of F. occidentalis reared on kidney bean and broad bean plants were used as experimental insects on day 3 after emergence. According to the method described by Zhang et al. [29], F. occidentalis were fed 300 µL of dsRNA-free honey solution, 300 ng/µL of dseGFP honey solution, and 300 ng/µL of dsFoCCS1 honey solution, respectively. The F. occidentalis reared on kidney bean plants and broad bean plants feeding on dsRNA-free honey solution were denoted as HK and HB, respectively. Three biological replicates per treatment and 40 adult females per replicate were used. The samples were collected after 24 h of feeding with the dsRNA solution, and the expression level of FoCCS1 after RNA interference was determined using RT-qPCR. The F2 generation F. occidentalis adult females that were transferred to kidney bean plants for rearing were subjected to RNAi, and samples were collected to determine the SOD activity using SOD activity assay kit (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China).
2.8. Effects of RNAi on the Survival Rate of Adult Females and the Number of Offspring Nymphs of F. occidentalis
Using the RNAi treatment method described in Section 2.7, adult females were fed dseGFP and dsFoCCS1 solutions for 24 h, respectively. Then, 35 RNAi-treated F. occidentalis adult females were released onto the kidney bean and broad bean leaves, respectively, to continue rearing. The rearing was performed in a transparent cylindrical plastic cup with a lid (5.5 cm in diameter and 4.0 cm in height, covered with a square hole with a length of 3.5 cm, and sealed with a 200-mesh gauze to ensure air permeability and prevent the thrips from escaping). Kidney bean and broad bean leaves were kept moist by wrapping the petioles with moist absorbent cotton. The survival rate was recorded every 2 days, and the surviving F. occidentalis were transferred to fresh kidney bean and broad bean leaves to continue rearing. The survival rates of F. occidentalis released back to the host plants and reared for 2, 4, 6, 8, and 10 days were recorded. The bean and broad bean leaves on which adult females had laid eggs were maintained in the artificial climate box, and the number of hatched nymphs was counted after 6 days. The number of larvae hatched on the replaced leaves on days 2, 4, 6, 8, and 10 were also recorded.
2.9. Statistical Analyses
Microsoft Excel 2019 was used for data analysis, and relative gene expression was calculated using the 2−ΔΔCt quantitative method [30]. SPSS v.21.0 (IBM, Armonk, NY, USA) was used for one-way analysis of variance, and Tukey’s method was used for multiple comparisons (α = 0.05). The survival rate of adult females and the number of offspring larvae of F. occidentalis fed with dsFoCCS1 and dseGFP was calculated by independent samples t-test. GraphPad Prism v.8.0 (GraphPad Software, San Diego, CA, USA) was used to plot the survival curve of F. occidentalis after RNAi treatment. SigmaPlot v.14.0 (Systat Software, SigmaPlot for Windows, San Jose, CA, USA) was used to construct graphs.
3. Results
3.1. Sequence Characterization of FoCCS1 and FoMnSOD2
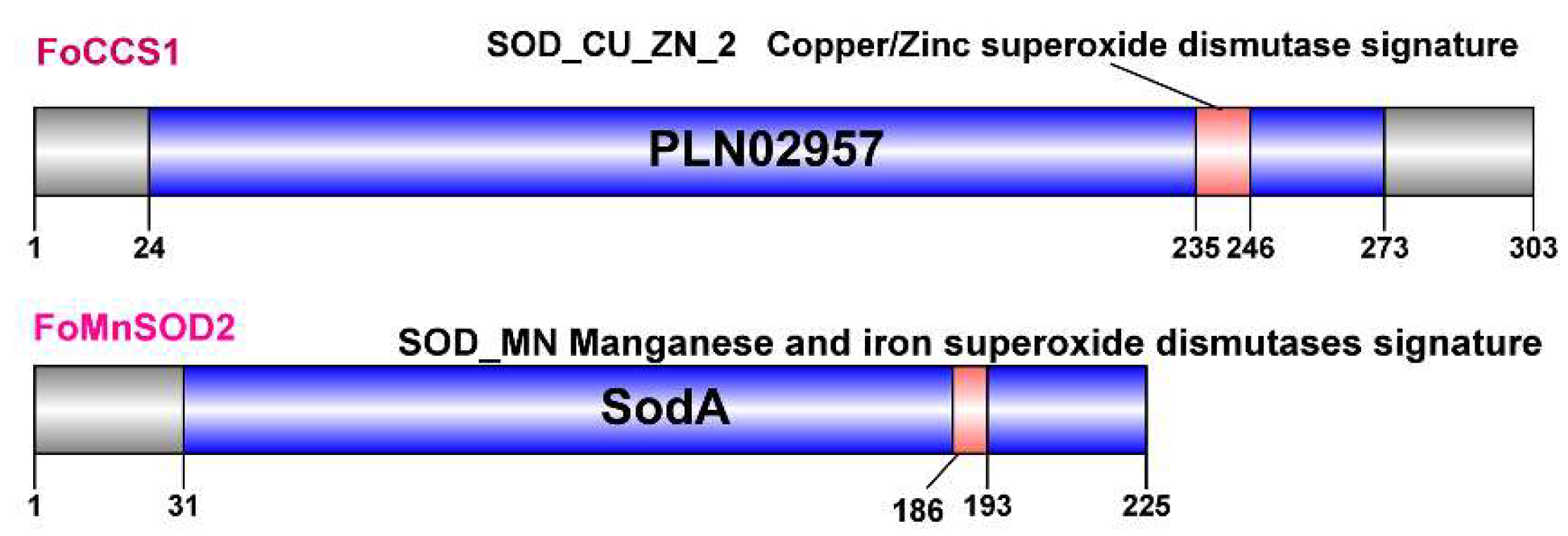
Two full-length CDS sequences of the SOD gene of F. occidentalis were cloned and identified as belonging to the CCS and MnSOD (SOD2) subfamily, respectively. They were labeled as FoCCS1 (GenBank accession number: MT571493) and FoMnSOD2 (GenBank accession number: MT460164), respectively. The bioinformatics features of FoCCS1 and FoMnSOD2 sequences were markedly different (Table 1). FoCCS1 contains a Cu/ZnSOD signature 2 sequence (GNSGhRlACgiI) and a CCS-specific conserved domain PLN02957, which is described as copper, zinc superoxide dismutase (Table 1 and Figure 1). FoMnSOD2 contains a manganese and iron SOD signature sequence (DvWEHAYY) and a specific conserved domain SodA (Table 1 and Figure 1). Secondary structure prediction showed that α-helices, extended strands, and random coils accounted for 0.66%, 36.30%, and 63.04%, respectively, in FoCCS1 and 56.44%, 3.11%, and 40.44%, respectively, in FoMnSOD2 (Figure S1). The results of protein tertiary structure prediction showed that FoCCS1 had 60.67% homology with the human copper chaperone for SOD1 and had a global model quality estimation (GMQE) value of 0.40. The homology between FoMnSOD2 and Caenorhabditis elegans mitochondrial MnSOD2 was 61.54%, and the GMQE value was 0.78. The PyMOL modeling results of FoMnSOD2 protein tertiary structures are shown in Figure S2.

Table 1.
Basic biological information for Frankliniella occidentalis CCS and MnSOD2 sequences.
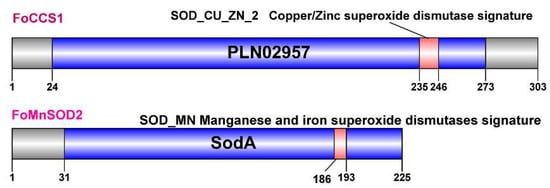
Figure 1.
Analysis of the conserved domains of CCS1 and MnSOD2 proteins of Frankliniella occidentalis using NCBI Conserved Domain Search (nih.gov). The blue box indicates the superoxide dismutase (SOD) domain, and the red box indicates the signature sequence.
3.2. Homologous Alignment and Phylogenetic Analysis
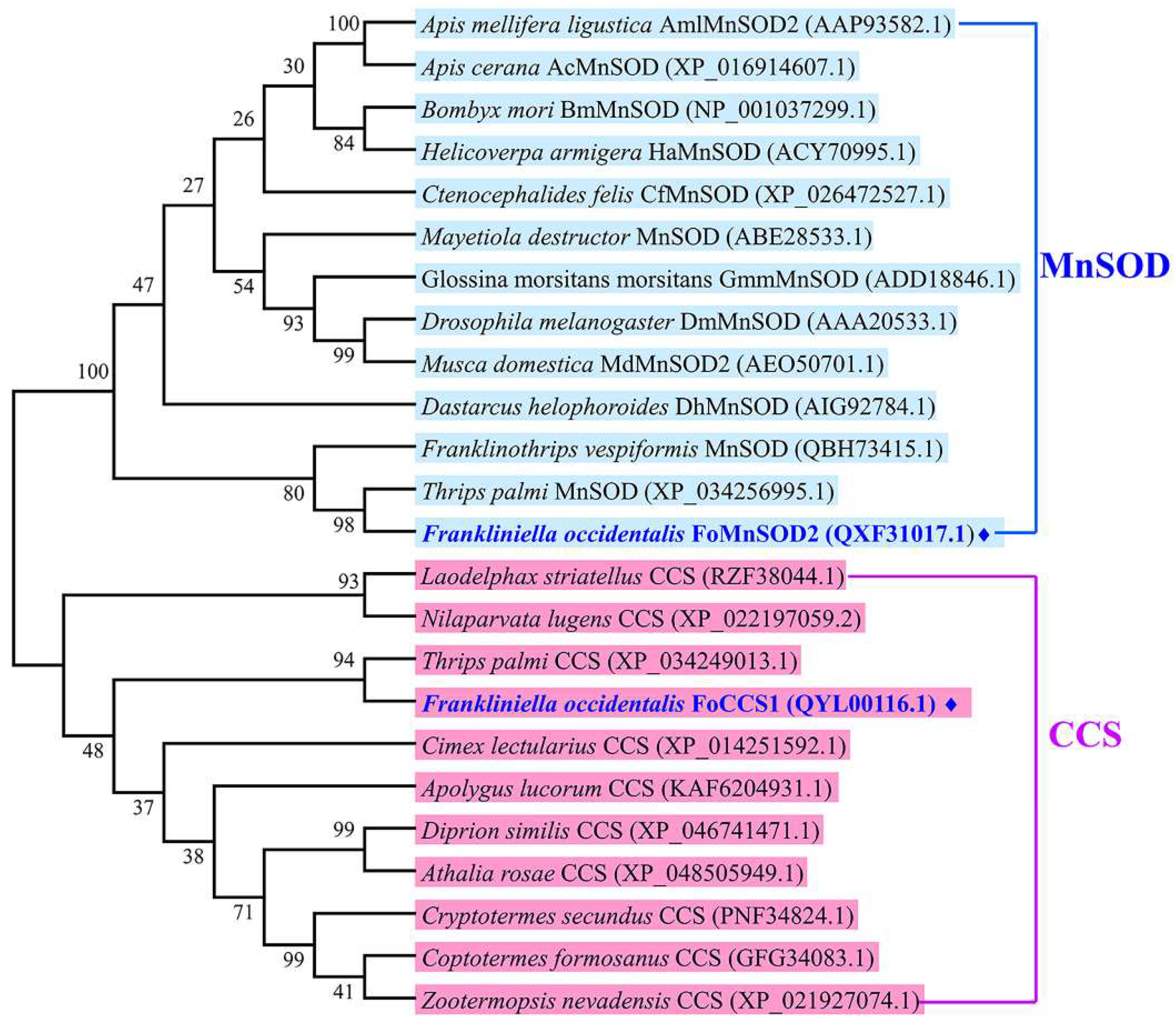
The amino acid sequences of FoCCS1 and FoMnSOD2 were used as query sequences for BLASTp alignment analysis. FoCCS1 showed the highest sequence homology (76%) with CCS of Thrips palmi (Thysanoptera; GenBank accession number: XP_034249013.1). It also showed high sequence homology at 60%, 60%, and 59.47%, respectively, with CCS of insects belonging to other orders, such as Nilaparvata lugens (GenBank accession number: AQW43017.1), Cimex lectularius (GenBank accession number: XP_014251592.1), and Diprion similis (GenBank accession number: XP_046741472.1). FoMnSOD2 showed high sequence homology of 96%, 90%, and 69%, respectively, with MnSOD of F. cephalica (GenBank accession number: QBH73414.1), T. palmi (GenBank accession number: XP_034256995.1), and Franklinothrips vespiformis (GenBank accession number: QBH73415.1). It also showed high sequence homology at 64%, 61%, and 67%, respectively, with MnSOD of insects belonging to other orders, such as Oxya chinensis (GenBank accession number: QFZ96018.1), Anoplophora glabripennis (GenBank accession number: XP_018577318.1), and Heortia vitessoides (GenBank accession number: AWX63642.1).
The amino acid sequences of SOD genes belonging to the CCS and MnSOD subfamilies in other insects were downloaded from the NCBI database, and a phylogenetic tree was constructed using the neighbor-joining method in MEGA 6.0. The results showed that FoCCS1 and FoMnSOD2 belonged to two branches of CCS and MnSOD. FoCCS1 is most closely related to T. palmi CCS, and with the CCS of Laodelphax striatellus, Apolygus lucorum, and Nilaparvata lugens, and other insects grouped together. FoMnSOD2 is also most closely related to MnSOD of T. palmi and F. vespiformis (Thysanoptera), and with the MnSOD of Dastarcus helophoroides, Mayetiola destructor, and Bombyx mori and other insects grouped together (Figure 2).
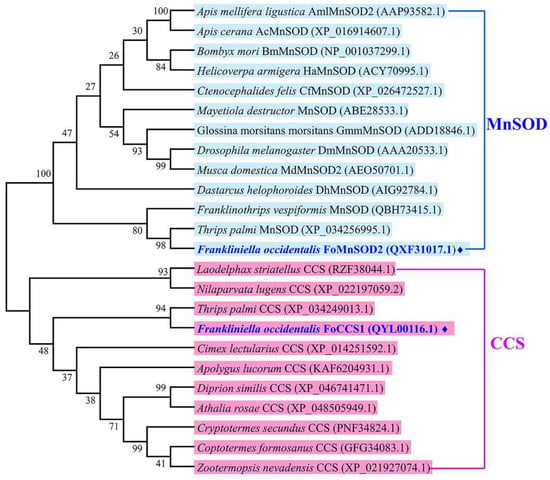
Figure 2.
Phylogenetic tree of copper chaperone for superoxide dismutase (CCS) and manganese superoxide dismutase (MnSOD) in Frankliniella occidentalis and other insect species using the neighbor-joining method with a bootstrap value of 1000. The branch name of the phylogenetic tree is the species name and GenBank accession number corresponding to each subfamily gene. ♦ represents the amino acid sequence of the gene cloned in this study.
3.3. Analysis of Expression Profiles in the Different Developmental Stages
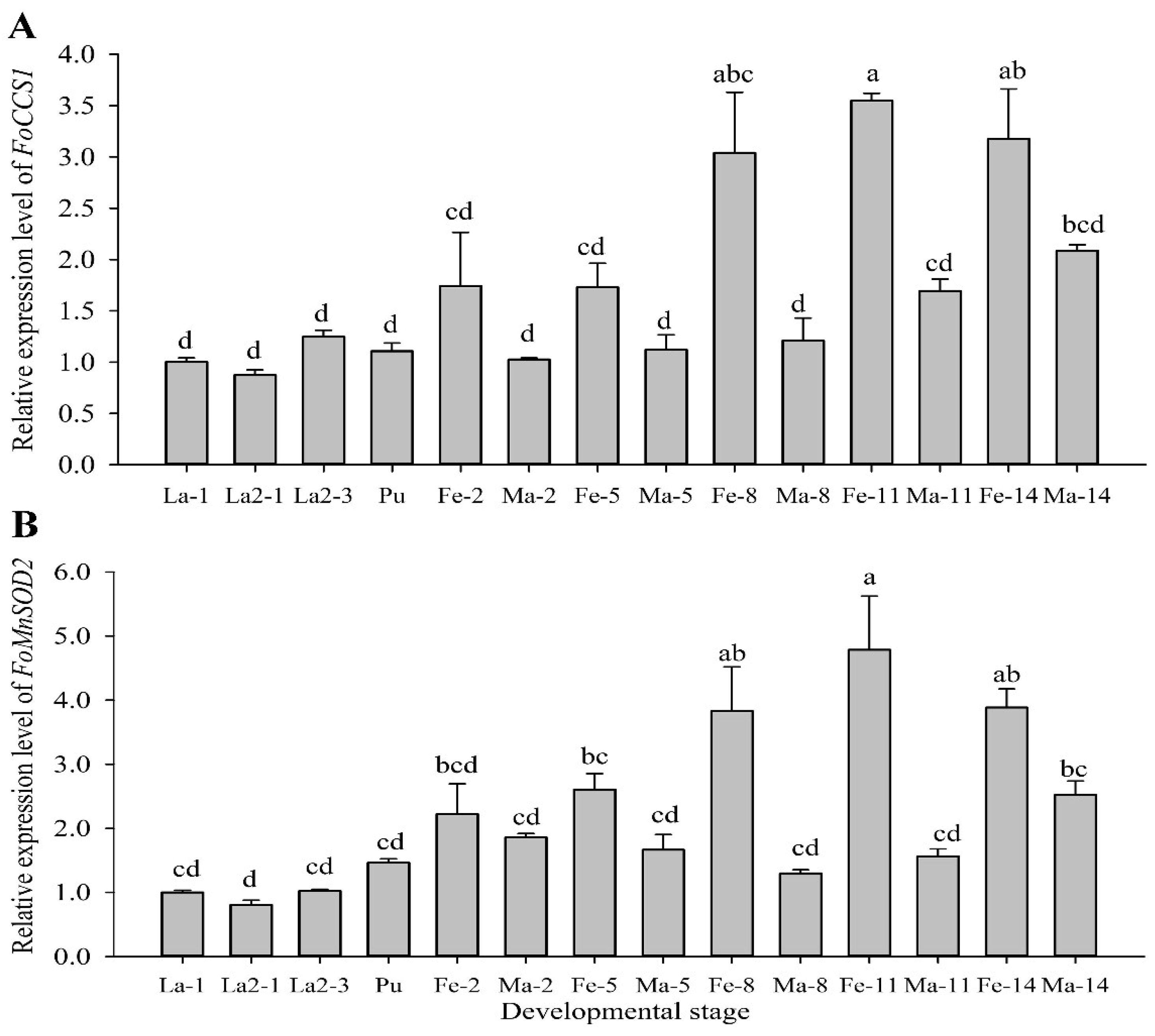
RT-qPCR assay at different developmental stages showed that FoCCS1 and FoMnSOD2 were expressed in the larval, pupal, and adult stages. The expression levels of the two genes were higher in adult females than in the larvae, pupae, and adult males (Figure 3). The expression levels of FoCCS1 in adult females on days 2 (Fe-2), 5 (Fe-5), 8 (Fe-8), 11 (Fe-11), and 14 (Fe-14) were 1.74, 1.73, 3.0, 3.5, and 3.2 times higher than those in the first-instar larvae (La-1), respectively. There was no significant difference in terms of FoCCS1 expression among the larvae, pupae, and adult males. The expression levels of FoMnSOD2 in adult females on days 2 (Fe-2), 5 (Fe-5), 8 (Fe-8), 11 (Fe-11), and 14 (Fe-14) were 2.2, 2.6, 3.8, 4.8, and 3.9 times higher than those in the first-instar larvae (La-1), respectively. The expression levels of FoMnSOD2 in adult males on days 2 (Ma-2) and 14 (Ma-14) were 2.2 and 2.5 times higher than those of La-1 on the, respectively, and the expression levels were similar among other stages.
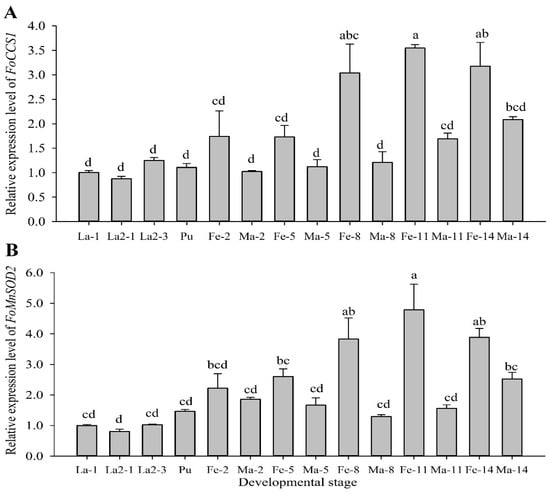
Figure 3.
Expression profiles of Frankliniella occidentalis FoCCS1 and MnSOD2 genes at different developmental stages. Data are presented as mean ± standard error of the mean (n = 3). Significant differences in comparison with the levels in first-instar larvae are indicated by different letters (Tukey’s test, p < 0.05).
3.4. Expression of FoCCS1 in F. occidentalis Feeding on Different Hosts
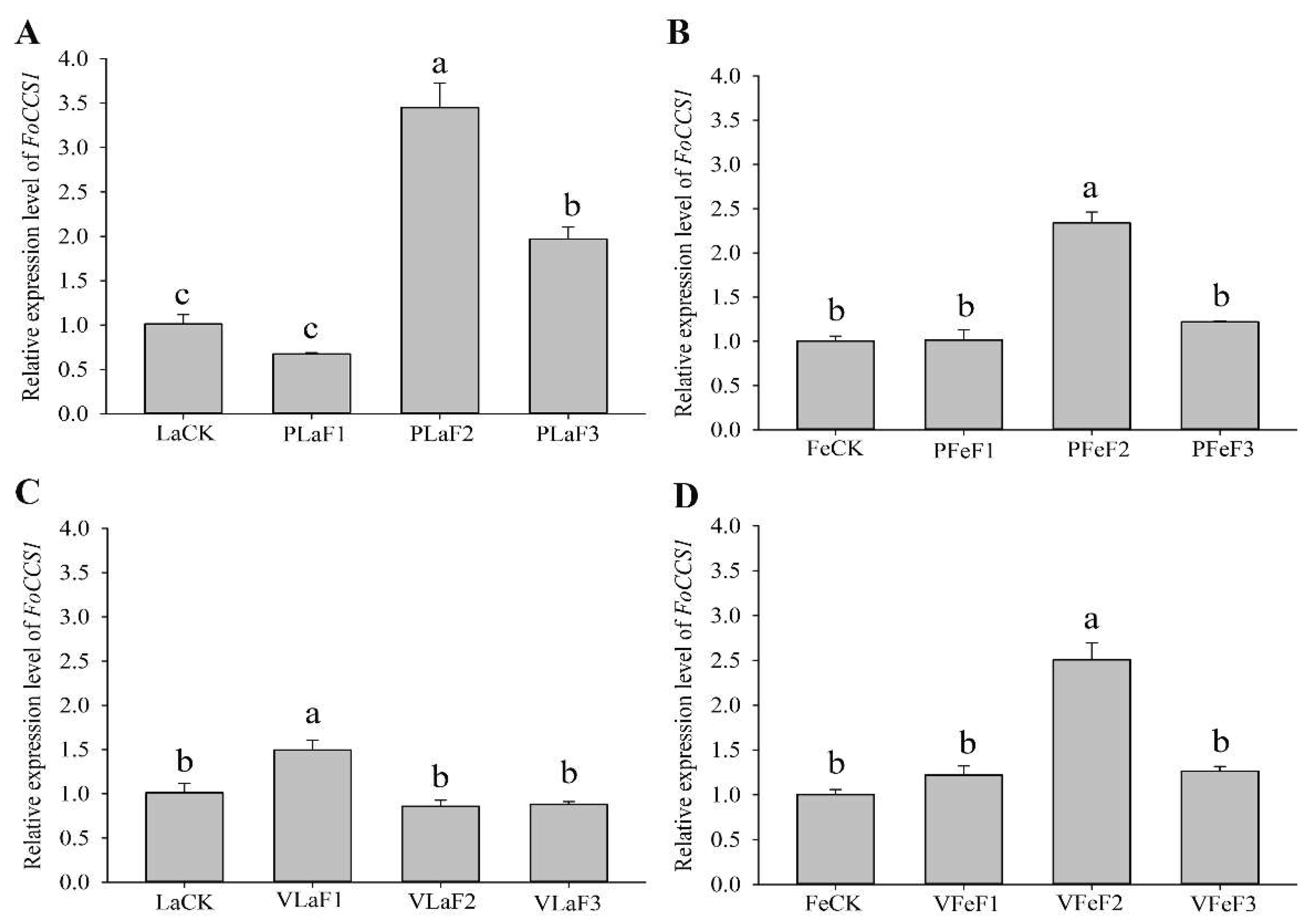
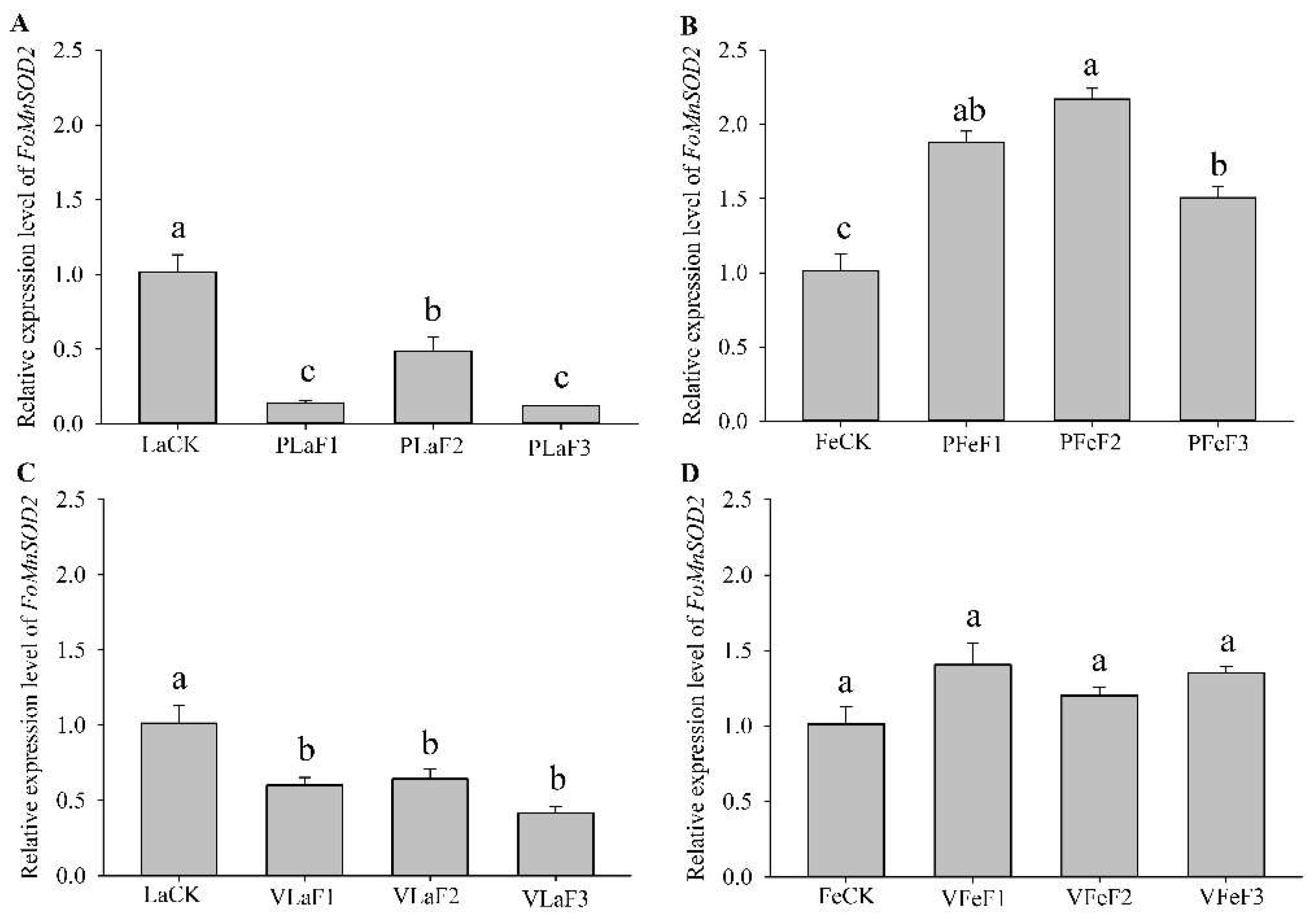
The expression levels of FoCCS1 in F. occidentalis larvae feeding on kidney bean and broad bean plants differed after host shifting. The expression level of FoCCS1 significantly increased in adult females in the F2 generation (Figure 4). The expressions of FoCCS1 in the second-instar larvae of F2 and F3 generations fed on kidney bean plants were significantly upregulated (3.42-fold and 1.95-fold, respectively) compared with the control (LaCK). However, the expression of FoCCS1 in the second-instar larvae shifted to broad bean plants for rearing was significantly upregulated only in the F1 generation. The expression levels of FoCCS1 in F1 and F3 generation adult females shifted to the two host plants for rearing were not significantly different from those in the control. However, the expression levels of FoCCS1 in F2 adult females feeding on kidney bean and broad bean plants were significantly upregulated (2.3-fold and 2.5-fold, respectively).
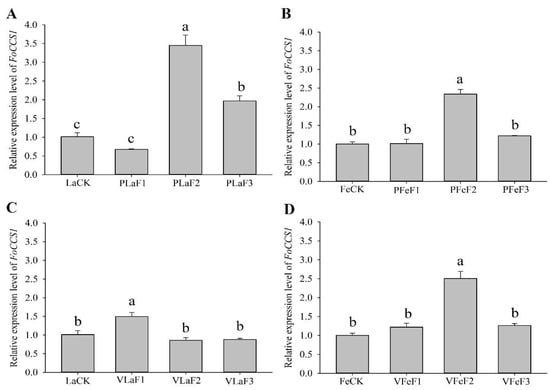
Figure 4.
Expression of FoCCS1 in Frankliniella occidentalis feeding on kidney bean and broad bean plants. (A), second-instar larvae feeding on kidney bean plants; (B), adult females feeding on kidney bean plants; (C), second-instar larvae feeding on broad bean plants; (D), adult females feeding on broad bean plants. LaCK, second-instar larvae reared on kidney bean pods; FeCk, adult females reared on kidney bean pods; PLaF1–PLaF3, second larvae of F1–F3 generation reared on kidney bean plants; PFeF1–PfeF3, adult females of F1–F3 generation reared on kidney bean plants; VLaF1–VlaF3, second larvae of F1–F3 generation reared on broad bean plants; VFeF1–VFeF3, adult females of F1–F3 generation reared on broad bean plants. Data are expressed as mean ± standard error of the mean (n = 3). Different lowercase letters indicate significant difference among all treatments (Tukey’s test, p < 0.05).
3.5. Expression of FoMnSOD2 in F. occidentalis Feeding on Different Hosts
RT-qPCR revealed that the expression levels of FoMnSOD2 in F. occidentalis were different between those shifted to kidney bean and broad bean plants for rearing (Figure 5). The expression levels of FoMnSOD2 in the second-instar larvae of the F1, F2, and F3 generations of F. occidentalis reared on the kidney bean and broad bean plants were significantly lower than those in the control (LaCK). The expression levels of FoMnSOD2 in F1, F2, and F3 generation adult females shifted to kidney bean plants were significantly upregulated by 1.9-, 2.2-, and 1.5-fold, respectively, compared with the control (FeCK). However, the expression levels of FoMnSOD2 in adult females shifted to broad bean plants for rearing did not significantly differ from those in the control.
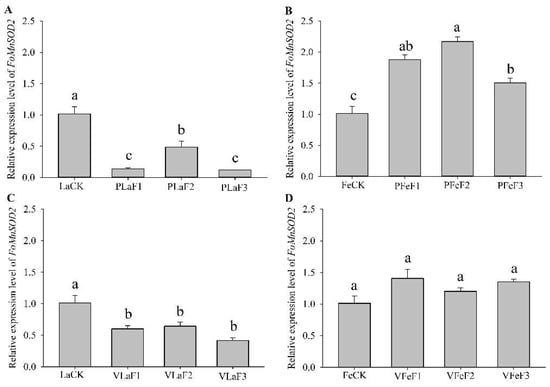
Figure 5.
Expression of FoMnSOD2 in Frankliniella occidentalis feeding on kidney bean and broad bean plants. (A), second-instar larvae feeding on kidney bean plants; (B), adult females feeding on kidney bean plants; (C), second-instar larvae feeding on broad bean plants; (D), adult females feeding on broad bean plants. LaCK, second-instar larvae reared on kidney bean pods; FeCk, adult females reared on kidney bean pods; PLaF1–PLaF3, second larvae of F1–F3 generation reared on kidney bean plants; PFeF1–PfeF3, adult females of F1–F3 generation reared on kidney bean plants; VLaF1–VlaF3, second larvae of F1–F3 generation reared on broad bean plants; VFeF1–VFeF3, adult females of F1–F3 generation reared on broad bean plants. Data are expressed as mean ± standard error of the mean (n = 3). Different lowercase letters indicate significant difference among all treatments (Tukey’s test, p < 0.05).
3.6. FoCCS1 Expression Level and SOD Activity Downregulated by RNAi
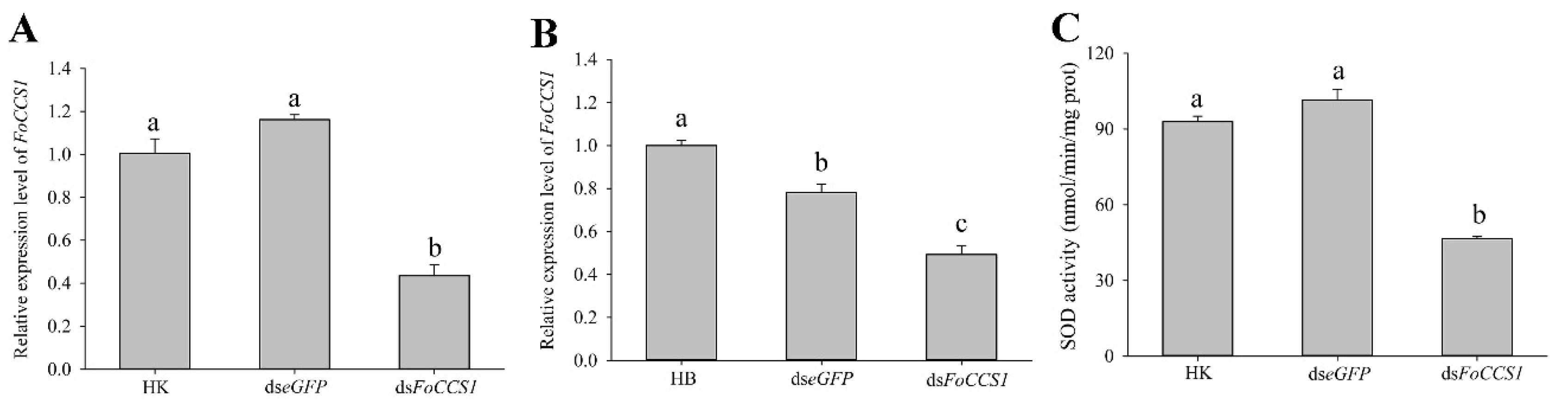
The results of gene expression and enzyme activity assay indicated that feeding a dsFoCCS1 solution significantly downregulated the expression level of FoCCS1 and SOD activity in adult females of F. occidentalis (Figure 6). When F. occidentalis shifted to kidney bean plants were treated with the dsRNA solution, the mRNA expression levels of FoCCS1 was significantly downregulated by 56.64% and 62.54%, respectively, compared with those in those fed with dseGFP and HK. After RNAi, the mRNA expression of FoCCS1 in F. occidentalis reared on broad bean plants was significantly downregulated by 50.73% and 36.93%, respectively. The SOD activity in adult F. occidentalis females in the dsFoCCS1 treatment group was 49.93% and 54.15% lower, respectively, than that in the HK and dseGFP treatment groups.
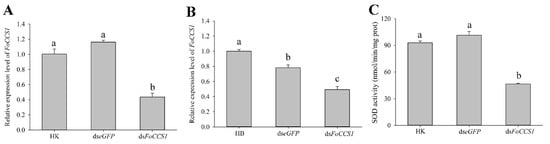
Figure 6.
Effects of feeding dsRNA on the FoCCS1 mRNA expression and SOD activity in Frankliniella occidentalis. (A,B), Efficiency of FoCCS1 gene silencing in F2 adult females of thrips transferred to kidney bean and broad bean plants for feeding, respectively. (C), SOD activity of F. occidentalis reared on kidney bean plants after downregulation of FoCCS1 expression levels by RNAi. HK and HB: F. occidentalis reared on kidney bean and broad bean plants and fed with dsRNA-free solution, respectively. Data are expressed as mean ± SE (n = 3). Different lowercase letters indicate significant differences among all treatments (Tukey’s test, p < 0.05).
3.7. RNAi Reduced the Survival Rate and Fecundity of Adult F. occidentalis Females
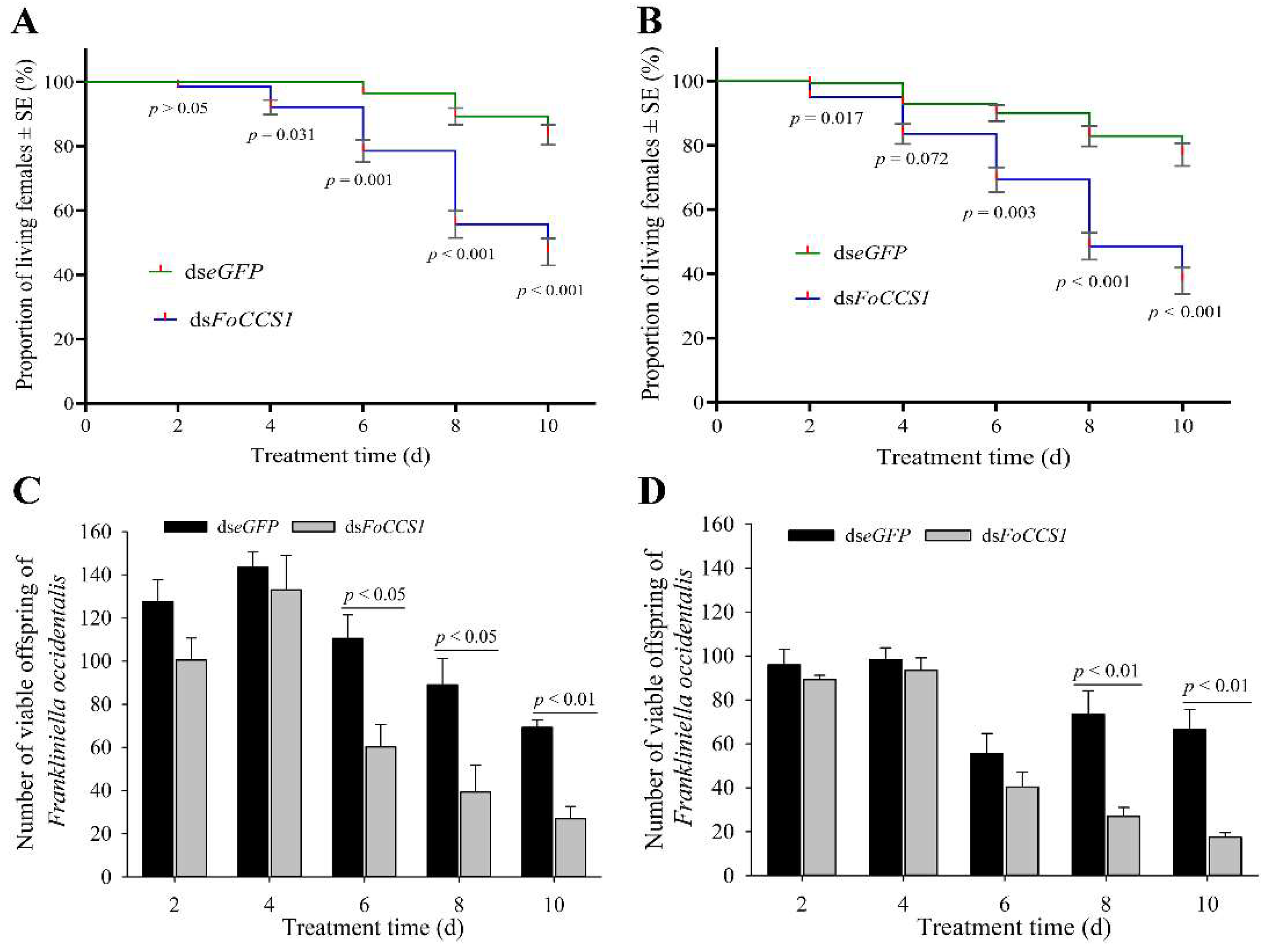
After the mRNA expression of FoCCS1 was downregulated by RNAi, the survival rate of adult females provided with kidney bean and broad bean leaves for rearing significantly decreased with rearing time (Figure 7A,B). After feeding with dsFoCCS1 solution, the survival rates of adult females fed on the leaves of kidney bean significantly decreased by 7.86%, 18.52%, 37.60%, and 43.59%, respectively, on days 4, 6, 8, and 10 compared with the dseGFP-treated group. After RNAi treatment, the survival rates of adult females fed on broad bean leaves on days 2, 6, 8, and 10 were 4.32%, 23.02%, 40.52%, and 50.00% lower than those in the dseGFP-treated group, respectively.
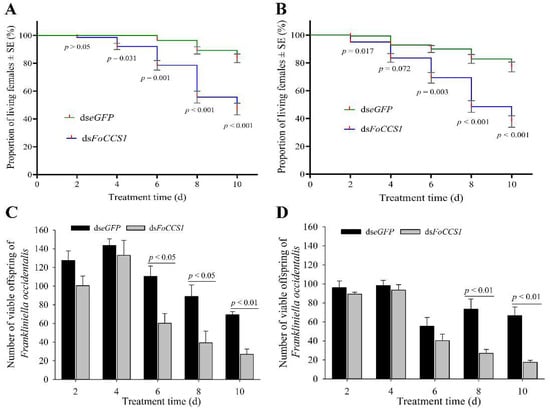
Figure 7.
Effects of RNAi downregulation of FoCCS1 expression on survival and offspring number of the Frankliniella occidentalis adult females shifted to feed on kidney bean and broad bean plants. (A,C), survival curve and number of offspring larvae of F. occidentalis reared on kidney bean plants after being fed with dsRNA solution. (B,D), survival curve and number of offspring larvae of F. occidentalis reared on broad bean plants after being fed with dsRNA-solution. Data are presented as the mean ± standard error of the mean (n = 4) and compared using the independent sample t-test. p < 0.05 indicates statistical significance.
With the gradual decrease in the survival rate, the number of offspring larvae hatched on the kidney bean and broad bean leaves showed a decreasing trend (Figure 7C,D). On days 6, 8, and 10 after RNAi treatment, the number of offspring larvae that continued to be fed with kidney bean leaves significantly reduced by 45.48%, 55.90%, and 61.01%, respectively, compared with the control. However, in the treatment group fed with broad bean leaves, the number of offspring larvae decreased significantly by 46.50% and 73.68% on days 8 and 10, respectively. The results indicate that the downregulation of FoCCS1 mRNA expression by RNAi was not conducive to the feeding adaptation of F. occidentalis after host shifting.
4. Discussion
Insect SODs include two classes, namely Cn/ZnSOD and MnSOD (SOD2). The molecular conformation of Cn/ZnSOD involves a β-sheet structure, whereas that of MnSOD includes many α-helical structures [31]. CCS is a member of the Cu/ZnSOD family [18]. In the amino acid sequence of FoCCS1 cloned in this study, α-helical structures accounted for only 0.66%, whereas tertiary structure prediction indicated that this protein mostly comprises β-sheet structures (Figures S1 and S2). In the FoMnSOD2 amino acid sequence, α-helical structures accounted for 56.44%. These findings are consistent with the basic structural features of Cn/ZnSOD and MnSOD. The amino acid sequence of CCS1 of F. occidentalis is the same as that of CCS of Coptotermes formosanus (GenBank: GFG34083.1) [32]. L. striatellus (GenBank: RZF38044.1) [33], and Apolygus lucorum (GenBank: KAF6204931.1) [34], with a specific Cu/ZnSOD signature 2 (GNSGhRlACgiI) and a specific conserved domain PLN02957 (copper, zinc superoxide dismutase) (Table 1 and Figure 1). Similar to the MnSOD2 of insects, such as F. vespiformis [35], FoMnSOD2 contains a specific manganese and iron SOD signature sequence (DvWEHAYY) and a specific conserved domain SodA (Table 1 and Figure 1). Homology alignment and phylogenetic analysis showed that FoCCS1 exhibits high sequence homology with CCS in other insects, such as T. palmi (GenBank: XP_034249013.1) and L. striatellus. FoMnSOD2 exhibits high sequence homology to MnSODs in other insects, such as T. palmi (GenBank: XP_034256995.1), F. vespiformis (GenBank: QBH73415.1), and H. vitessoides (GenBank: AWX63642.1). Therefore, the cloned FoCCS1 and FoMnSOD2 in this study belong to the CCS (Cu/ZnSOD family) and MnSOD classes, respectively.
Analysis of expression profiles at different developmental stages of F. occidentalis showed that the mRNA expression levels of FoCCS1 and FoMnSOD2 in adult females were higher than those in adult males, pupae, and larvae. Previous studies have shown that the mRNA levels of Cu/ZnSOD and MnSOD in Chironomus riparius are also higher in adult females than that in eggs, larvae, pupae, and male adults [36]. The expression levels of Drosophila melanogaster SOD3 were also higher in females than males [37]. In F. occidentalis, the higher mRNA levels of FoCCS1 and FoMnSOD2 in adult females than in males may be associated with the significantly longer lifespan of females [38]; the antioxidant systems of long-lived animals exhibit higher SOD activity than shorter-lived animals [39]. In addition, the expression level of Cu/ZnSOD in Bemisia tabaci was also higher in the adult stage than in the nymphal and pupal stages [40]. However, the expression level of Bt-mMnZnSOD was significantly higher in the fourth instar than in eggs, nymphs, and adults [41]. This may be because SOD is a key enzyme of the antioxidant defense system in insects.
Transcript level analysis of insects after seasonal host shifting showed that antioxidant enzyme genes represented by SOD are actively involved in regulating the anti-adaptation ability of insects to new hosts [8,40,41]. The upregulation of SOD activity and expression of SOD genes effectively helps insects avoid the toxic effects of ROS, such as H2O2, from plants, thereby protecting the insects against oxidative damage [13,16,17]. As a member of the Cu/ZnSOD1 family, CCS is a key factor that regulates copper homeostasis and promotes all levels of SOD1 activity in organisms [19,20,21,22]. The results of this study showed that the FoCCS1 expression levels were significantly upregulated in the F2 generation adult females that were shifted to the kidney bean and broad bean plants for rearing. The expression levels of FoCCS1 were also significantly upregulated in the F1 and F2 generation larvae that were shifted to the kidney bean plants and the F1 generation larvae that were shifted to the broad bean plants. The level of FoMnSOD2 expression was significantly upregulated only in F1, F2, and F3 generation for adult females that were shifted to kidney bean plants for rearing. Previous studies have shown that SOD plays an active role in regulating the feeding adaptations of B. tabaci and aphids after host shifting [42]. For example, the mRNA levels of Bt-mMnSOD and Bt-ecCuZnSOD were significantly upregulated 24 h after the MEAM1 and Asia II 3 whiteflies were transferred from cotton to tobacco, and the corresponding MnSOD activity and Cu/ZnSOD activity were significantly increased [40]. The activities of Cu/ZnSOD and MnSOD also increased significantly after MEAM1 B. tabaci were shifted for feeding from cotton to tobacco for 5 days [41,43]. Transcript level analysis of the aphids Hyalopterus persikonus feeding on winter and summer hosts also found that genes, such as SOD, were significantly expressed in aphids feeding on summer hosts [8]. This indicates that SOD plays an important role in regulating the feeding adaptation after insect host shifting. In addition, the present study found that the expression level of FoMnSOD2 was significantly upregulated only in F1, F2, and F3 generation for adult females that were shifted to kidney bean plants and was significantly downregulated in other treatments, or not different from the control. This may be related to the specific insect state of F. occidentalis and the targeted role of the host plant’s antioxidant defense. Only under some special conditions can the MnSOD located on the mitochondria fully exert the function of peroxidase [44].
Decreased SOD activity reportedly reduces antioxidant defense and decreases lifespan in Drosophila [45,46]. In addition, inhibiting the mRNA levels and SOD activity of SOD2 (MnSOD class gene) and SOD1 (Cu/ZnSOD class gene) by RNAi technology reduced the survival of Drosophila [39,47]. In this study, the differences in the mRNA expression levels of FoCCS1 and FoMnSOD2 in F. occidentalis reared on kidney bean and broad bean plants suggest that FoCCS1 plays an active role in regulating the feeding adaptation in F. occidentalis after host shifting. Therefore, FoCCS1 mRNA level was further downregulated by RNAi, which showed that SOD activity was also inhibited to a certain extent. This may be because the downregulation of FoCCS1 mRNA expression reduced the activation of Cu/Zn superoxide dismutase, resulting in the inhibition of SOD1 maturation [19,22]. Moreover, F. occidentalis with downregulated FoCCS1 mRNA levels released in kidney bean and broad bean leaves significantly decreased the survival rate and fecundity of adult females. This finding suggests that the decreased mRNA expression of FoCCS1 and SOD activity resulted in the inability of F. occidentalis to resist a series of plant defense responses related to ROS, which reduced the feeding adaptation ability after host shifting. By interfering with the function of FoCCS1, the adaptability of F. occidentalis to host shift can decrease. In practice, how to reduce the function of FoCCS1 will be the focus and direction of future research. The results of this study also provide a reference for the selection of control targets for F. occidentalis. Secondly, in the early process of thrips transferring to new hosts is the key time to control, and with the higher control effect, measures should be taken to destroy the adaptability of thrips to the new host as soon as thrips are found in the new host.
5. Conclusions
Previous studies on insects, such as aphids and B. tabaci, reported that SODs respond positively to a new host, leading to feeding adaptations after host shifting [8,40,41,43]. In the present study, the full-length CDS region of CCS1 and MnSOD2 of F. occidentalis was cloned and the protein structures were predicted. The expression profiles of FoCCS1 and FoMnSOD2 in different developmental stages of F. occidentalis were analyzed. Moreover, the differences in the FoCCS1 and FoMnSOD2 mRNA expression levels in F. occidentalis that had shifted to the kidney bean and broad bean plants for rearing were examined. RNAi downregulation of FoCCS1 mRNA expression levels and SOD activity decreased the survival rate and fecundity of F. occidentalis adult females. These results indicate that FoCCS1 plays an important role in regulating the feeding adaptation of F. occidentalis after host shifting. These findings provide a reference for future research on indirectly improving insect resistance of crops by reducing the antioxidant capacity of insects.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13090782/s1. Table S1: Primer sequences used in CDS cloning; Table S2: Primer sequences used in quantitative reverse-transcription PCR (RT-qPCR) and double-stranded (ds) RNA synthesis; Figure S1: Secondary structure of the protein sequence of FoCCS1 and FoMnSOD2; Figure S2: Predicted tertiary structure of FoCCS1 and FoMnSOD2 protein sequences.
Author Contributions
Conceptualization, J.-R.Z., T.Z. and L.L.; data curation: T.Z. and J.-R.Z.; formal analysis: T.Z., L.L. and W.-B.Y.; funding acquisition: J.-R.Z.; investigation: T.Z., L.L., G.Z. and Y.-L.J.; methodology: J.-R.Z., T.Z. and L.L.; software: W.-B.Y., D.-Y.L. and G.Z.; supervision: J.-R.Z.; validation: T.Z. and L.L.; writing—original draft: T.Z. and W.-B.Y.; writing—review and editing: J.-R.Z., L.L., W.-B.Y., G.Z. and D.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (NSFC) (31660516), and the Talent Platform of Guizhou (No. [2017] 5788).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Reitz, S.R.; Gao, Y.L.; Kirk, W.D.J.; Hoddle, M.S.; Leiss, K.A.; Funderburk, J.E. Invasion biology, ecology, and management of western flower thrips. Annu. Rev. Entomol. 2020, 65, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Reitz, S.R. Biology and ecology of the western flower thrips (Thysanoptera: Thripidae): The making of a pest. Flarida Entomol. 2009, 92, 7–13. [Google Scholar] [CrossRef]
- Zhang, B.; Qian, W.Q.; Qiao, X.; Xi, Y.; Wan, F.H. Invasion biology, ecology, and management of Frankliniella occidentalis in China. Arch. Insect Biochem. Physiol. 2019, 102, e21613. [Google Scholar] [CrossRef] [PubMed]
- Mouden, S.; Sarmiento, K.F.; Klinkhamer, P.G.; Leiss, K.A. Integrated pest management in western flower thrips: Past, present and future. Pest Manag. Sci. 2017, 73, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Zhi, J.R.; Gai, H.T. Effects of host plants and temperature on Frankliniella occidentalis growth and development. Chin. J. Ecol. 2011, 30, 558–563. [Google Scholar] [CrossRef]
- Mason, P.A. On the role of host phenotypic plasticity in host shifting by parasites. Ecol. Lett. 2016, 19, 121–132. [Google Scholar] [CrossRef]
- Forbes, A.A.; Devine, S.N.; Hippee, A.C.; Tvedte, E.S.; Ward, A.; Widmayer, H.A.; Wilson, C.J. Revisiting the particular role of host shifts in initiating insect speciation. Evolution 2017, 71, 1126–1137. [Google Scholar] [CrossRef]
- Cui, N.; Yang, P.C.; Guo, K.; Kang, L.; Cui, F. Large-scale gene expression reveals different adaptations of Hyalopterus persikonus to winter and summer host plants. Insect Sci. 2017, 24, 431–442. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant. Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Maffei, M.E.; Mithöfer, A.; Boland, W. Insects feeding on plants: Rapid signals and responses preceding the induction of phytochemical release. Phytochemistry 2007, 68, 2946–2959. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Kant, M.R.; Jonckheere, W.; Knegt, B.; Lemos, F.; Liu, J.; Schimmel, B.C.J.; Villarroel, C.A.; Ataide, L.M.S.; Dermauw, W.; Glas, J.J.; et al. Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Ann. Bot. 2015, 115, 1015–1051. [Google Scholar] [CrossRef]
- Argandoña, V.H.; Chaman, M.; Cardemil, L.; Muñoz, O.; Zúñiga, G.E.; Corcuera, L.J. Ethylene production and peroxidase activity in aphid-infested barley. J. Chem. Ecol. 2001, 27, 53–68. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; War, M.Y.; Ignacimuthu, S. Jasmonic acid-mediated-induced resistance in groundnut (Arachis hypogaea L.) against Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). J. Plant Growth Regul. 2011, 30, 512–523. [Google Scholar] [CrossRef]
- Van Raamsdonk, J.M.; Hekimi, S. Superoxide dismutase is dispensable for normal animal lifespan. Proc. Natl. Acad. Sci. USA 2012, 109, 5785–5790. [Google Scholar] [CrossRef]
- Colinet, D.; Cazes, D.; Belghazi, M.; Gatti, J.L.; Poirié, M. Extracellular superoxide dismutase in insects: Characterization, function, and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 2011, 286, 40110–40121. [Google Scholar] [CrossRef]
- Corona, M.; Robinson, G.E. Genes of the antioxidant system of the honey bee: Annotation and phylogeny. Insect Mol. Biol. 2006, 15, 687–701. [Google Scholar] [CrossRef]
- Price, D.L.; Rothstein, J.; Gitlin, J.D. Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA 2000, 97, 2886–2891. [Google Scholar] [CrossRef]
- Schmidt, P.J.; Ramos-Gomez, M.; Culotta, V.C. A gain of superoxide dismutase (SOD) activity obtained with CCS, the copper metallochaperone for SOD1. Biol. Chem. 1999, 274, 36952–36956. [Google Scholar] [CrossRef]
- Miyayama, T.; Ishizuka, Y.; Iijima, T.; Hiraoka, D.; Ogra, Y. Roles of copper chaperone for superoxide dismutase 1 and metallothionein in copper homeostasis. Metallomics 2011, 3, 693–701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boyd, S.D.; Calvo, J.S.; Liu, L.; Ullrich, M.S.; Skopp, A.; Meloni, G.; Winkler, D.D. The yeast copper chaperone for copper-zinc superoxide dismutase (CCS1) is a multifunctional chaperone promoting all levels of SOD1 maturation. J. Biol. Chem. 2019, 294, 1956–1966. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, A.; Reveillaud, I.; Fleming, J.E. Changes in superoxide dismutase and catalase in aging heat-shocked Drosophila. Free Radic. Res. Commun. 1992, 17, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Gao, Y.F.; Liu, Y.J.; Ge, X.D.; Li, X.; Li, F.S. Identification of the gene MdSOD3 and its role in resisting heavy metal stress in housefly (Musca domestica). Acta Entomol. Sin. 2012, 55, 267–275. [Google Scholar] [CrossRef]
- Liu, L.; Hou, X.L.; Yue, W.B.; Xie, W.; Zhang, T.; Zhi, J.R. Response of protective enzymes in western flower thrips (Thysanoptera: Thripidae) to two leguminous plants. Environ. Entomol. 2020, 49, 1191–1197. [Google Scholar] [CrossRef]
- Dietz, K.J.; Mittler, R.; Noctor, G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 2016, 171, 1535–1539. [Google Scholar] [CrossRef]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef]
- Zheng, Y.T.; Li, H.B.; Lu, M.X.; Du, Y.Z. Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLoS ONE 2014, 9, e111369. [Google Scholar] [CrossRef]
- Zhang, T.; Zhi, J.R.; Li, D.Y.; Liu, L.; Zeng, G. Effect of different double-stranded RNA feeding solutions on the RNA interference of V-ATPase-B in Frankliniella occidentalis. Entomol. Exp. Appl. 2022, 170, 427–436. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bannister, J.V.; Bannister, W.H.; Rotilio, G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit. Rev. Biochem. 1987, 22, 111–180. [Google Scholar] [CrossRef]
- Zhu, J.J.; Jiang, F.; Wang, X.H.; Yang, P.C.; Bao, Y.Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.S.; Cui, N.; et al. Genome sequence of the small brown planthopper, Laodelphax striatellus. GigaScience 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Itakura, S.; Yoshikawa, Y.; Togami, Y.; Umezawa, K. Draft genome sequence of the termite, Coptotermes formosanus: Genetic insights into the pyruvate dehydrogenase complex of the termite. J. Asia-Pac. Entomol. 2020, 23, 666–674. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.W.; Wang, H.; Huang, T.Y.; Liu, B.; Yang, B.; Yin, L.J.; Li, B.; Zhang, Y.; Zhang, S.; et al. Apolygus lucorum genome provides insights into omnivorousness and mesophyll feeding. Mol. Ecol. Resour. 2021, 21, 287–300. [Google Scholar] [CrossRef]
- Brandt, A.; Bast, J.; Scheu, S.; Meusemann, K.; Donath, A.; Schütte, K.; Machida, R.; Kraaijeveld, K. No signal of deleterious mutation accumulation in conserved gene sequences of extant asexual hexapods. Sci. Rep. 2019, 9, 5338. [Google Scholar] [CrossRef]
- Park, S.Y.; Nair, P.M.; Choi, J. Characterization and expression of superoxide dismutase genes in Chironomus riparius (Diptera, Chironomidae) larvae as a potential biomarker of ecotoxicity. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012, 156, 187–194. [Google Scholar] [CrossRef]
- Blackney, M.J.; Cox, R.; Shepherd, D.; Parker, J.D. Cloning and expression analysis of Drosophila extracellular Cu Zn superoxide dismutase. Biosci. Rep. 2014, 34, e00164. [Google Scholar] [CrossRef]
- Li, X.H.; Wang, D.J.; Lei, Z.R.; Wang, H.H. Comparison of life tables for experimental populations of individual-rearing and group-rearing Frankliniella occidentalis. Sci. Agric. Sin. 2021, 54, 959–968. [Google Scholar] [CrossRef]
- Deepashree, S.; Shivanandappa, T.; Ramesh, S.R. Genetic repression of the antioxidant enzymes reduces the lifespan in Drosophila melanogaster. J. Comp. Physiol. B 2022, 192, 1–13. [Google Scholar] [CrossRef]
- Gao, X.L.; Li, J.M.; Xu, H.X.; Yan, G.H.; Jiu, M.; Liu, S.S.; Wang, X.W. Cloning of a putative extracellular Cu/Zn superoxide dismutase and functional differences of superoxide dismutases in invasive and indigenous whiteflies. Insect Sci. 2015, 22, 52–64. [Google Scholar] [CrossRef]
- Gao, X.L.; Li, J.M.; Wang, Y.L.; Jiu, M.; Yan, G.H.; Liu, S.S.; Wang, X.W. Cloning, expression and characterization of mitochondrial manganese superoxide dismutase from the Whitefly, Bemisia tabaci. Int. J. Mol. Sci. 2013, 14, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, P.; Escudero-Martinez, C.M.; Eves-van den Akker, S.; Bos, J. Transcriptional changes in the aphid species Myzus cerasi under different host and environmental conditions. Insect Mol. Biol. 2020, 29, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Li, J.M.; Su, Y.L.; Gao, X.L.; He, J.; Liu, S.S.; Wang, X.W. Molecular characterization and oxidative stress response of an intracellular Cu/Zn superoxide dismutase (CuZnSOD) of the whitefly, Bemisia tabaci. Arch. Insect Biochem. Physiol. 2011, 77, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Ansenberger-Fricano, K.; Ganini, D.; Mao, M.; Chatterjee, S.; Dallas, S.; Mason, R.P.; Stadler, K.; Santos, J.H.; Bonini, M.G. The peroxidase activity of mitochondrial superoxide dismutase. Free Radic. Biol. Med. 2013, 54, 116–124. [Google Scholar] [CrossRef]
- Phillips, J.P.; Campbell, S.D.; Michaud, D.; Charbonneau, M.; Hilliker, A.J. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA 1989, 86, 2761–2765. [Google Scholar] [CrossRef]
- Duttaroy, A.; Paul, A.; Kundu, M.; Belton, A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics 2003, 165, 2295–2299. [Google Scholar] [CrossRef]
- Kirby, K.; Hu, J.; Hilliker, A.J.; Phillips, J.P. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. USA 2002, 99, 16162–16167. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).