RNAi-Mediated Silencing of Putative Halloween Gene Phantom Affects the Performance of Rice Striped Stem Borer, Chilo suppressalis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Total RNA Isolation and cDNA Synthesis
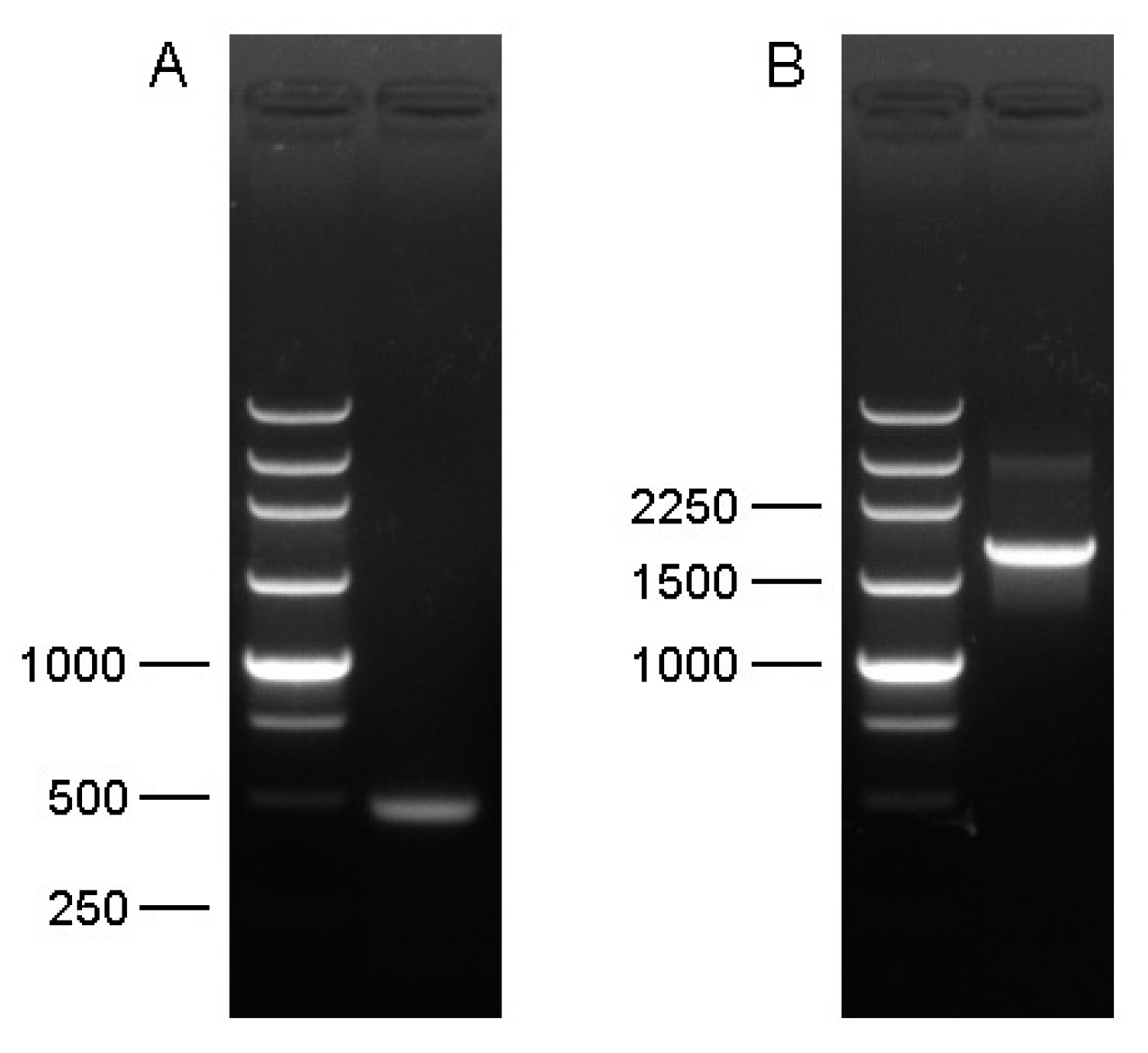
2.3. Amplification of cDNA Fragment
2.4. Rapid Amplification of cDNA Ends (3′ RACE and 5′ RACE)
2.5. Phylogenetic Analysis
| Primer | Sequence (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|
| Primers used in RT-PCR | ||
| Phm-RT-F | AGCAACCTCATTTGACCCTA | 483 |
| Phm-RT-R | GGCACCCTTCTTCTTGGA | |
| Primers used in 5′-RACE | ||
| Phm-5GSP | CACCCTTCTTCTTGGATCTGACGAAACC | |
| Phm-5NGSP | CAGCACATATGATACCATTGCCACGC | |
| Primers used in 3′-RACE | ||
| Phm-3GSP | ACTGGAAAAACGCATCGCTGCTGGC | |
| Phm-3NGSP | GGTTTCGTCAGATCCAAGAAGAAGGGTG | |
| Primers used in PCR for End to End | ||
| Phm-F | ATGGGGAGTCGCTGGTCA | 1717 |
| Phm-R | CCCGCACCGACGTGGTAT | |
| Primers used for synthesizing the dsRNAs Phm-dsRNA-F Phm-dsRNA-R GFP-dsRNA-F GFP-dsRNA-R Primers used in qRT-PCR | taatacgactcactataggTGACACCAAAAGGAGCGGAA taatacgactcactataggGCCCACTGGAGAGGTATCAC taatacgactcactataggTCACGGATACAACCTCTTT taatacgactcactataggAGTTCAGCGTGTCCG | 544 414 |
| Phm-qF | GCCAGGTGATAGGTTGTGTT | 204 |
| Phm-qR | ATGAGGTTGCTTGGGATCTATG |
2.6. Quantitative Reverse Transcription Analysis
2.7. dsRNA Preparation and Microinjection
2.8. Cs-Phm Relative Expression Level in dsRNA-Treated Insects
2.9. 20-Hydroxyecdysone Titer Measurement
2.10. Rescue Analysis
2.11. Statistical Analysis
3. Results
3.1. Molecular Cloning and Sequence Analysis of Cs-Phm
3.2. Spatial Transcript Profiles
3.3. Temporal Transcript Profiles
3.4. Effects of dsRNA Injection
3.5. Ecdysteroid Titer Measurement
3.6. Rescue Experiment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Danielsen, E.T.; Moeller, M.E.; Rewitz, K.F. Chapter Two—Nutrient Signaling and Developmental Timing of Maturation. In Current Topics in Developmental Biology; Ann, E.R., Michael, B.O.C., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 105, pp. 37–67. [Google Scholar]
- Miller, W.L.; Auchus, R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2010, 32, 81–151. [Google Scholar] [CrossRef]
- Thummel, C.S. Molecular Mechanisms of Developmental Timing in C. elegans and Drosophila. Dev. Cell 2001, 1, 453–465. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Rybczynski, R.; Warren, J.T. Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 2002, 47, 883–916. [Google Scholar] [CrossRef] [PubMed]
- Thummel, C.S.; Chory, J. Steroid signaling in plants and insects—Common themes, different pathways. Genes Dev. 2002, 16, 3113–3129. [Google Scholar] [CrossRef] [PubMed]
- Schumann, I.; Kenny, N.; Hui, J.; Hering, L.; Mayer, G. Halloween genes in panarthropods and the evolution of the early moulting pathway in Ecdysozoa. R. Soc. Open Sci. 2018, 5, 180888. [Google Scholar] [CrossRef]
- Moulos, P.; Samiotaki, M.; Panayotou, G.; Dedos, S.G. Combinatory annotation of cell membrane receptors and signalling pathways of Bombyx mori prothoracic glands. Sci. Data 2016, 3, 1–12. [Google Scholar] [CrossRef]
- Petryk, A.; Warren, J.T.; Marqués, G.; Jarcho, M.P.; Gilbert, L.I.; Kahler, J.; Parvy, J.-P.; Li, Y.; Dauphin-Villemant, C.; O’Connor, M.B. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. USA 2003, 100, 13773–13778. [Google Scholar] [CrossRef]
- Rewitz, K.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. The Halloween genes code for cytochrome P450 enzymes mediating synthesis of the insect moulting hormone. Biochem. Soc. Trans. 2006, 34, 1256–1260. [Google Scholar] [CrossRef]
- Lehmann, M. Endocrine and physiological regulation of neutral fat storage in Drosophila. Mol. Cell. Endocrinol. 2018, 461, 165–177. [Google Scholar] [CrossRef]
- Ruang-Rit, K.; Park, Y. Endocrine system in supernumerary molting of the flour beetle, Tribolium freemani, under crowded conditions. Insect Biochem. Mol. Biol. 2018, 101, 76–84. [Google Scholar] [CrossRef]
- Clark, A.; Bloch, K. The absence of sterol synthesis in insects. J. Biol. Chem. 1959, 234, 2578–2582. [Google Scholar] [CrossRef]
- Sumiya, E.; Ogino, Y.; Toyota, K.; Miyakawa, H.; Miyagawa, S.; Iguchi, T. Neverland regulates embryonic moltings through the regulation of ecdysteroid synthesis in the water flea Daphnia magna, and may thus act as a target for chemical disruption of molting. J. Appl. Toxicol. 2016, 36, 1476–1485. [Google Scholar] [CrossRef]
- Warren, J.T.; O’Connor, M.B.; Gilbert, L.I. Studies on the black box: Incorporation of 3-oxo-7-dehydrocholesterol into ecdysteroids by Drosophila melanogaster and Manduca sexta. Insect Biochem. Mol. Biol. 2009, 39, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.I.; Warren, J.T. A Molecular Genetic Approach to the Biosynthesis of the Insect Steroid Molting Hormone. In Vitamins & Hormones; Gerald, L., Ed.; Academic Press: Cambridge, MA, USA, 2005; Volume 73, pp. 31–57. [Google Scholar]
- Chávez, V.M.; Marqués, G.; Delbecque, J.P.; Kobayashi, K.; Hollingsworth, M.; Burr, J.; Natzle, J.E.; O’Connor, M.B. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 2000, 127, 4115–4126. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.T.; Petryk, A.; Marqués, G.; Jarcho, M.; Parvy, J.-P.; Dauphin-Villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Matsuda, T.; Yoshiyama, T.; Namiki, T.; Mita, K.; Fujimoto, Y.; Kataoka, H. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 2004, 279, 35942–35949. [Google Scholar] [CrossRef]
- Warren, J.T.; Petryk, A.; Marqués, G.; Parvy, J.-P.; Shinoda, T.; Itoyama, K.; Kobayashi, J.; Jarcho, M.; Li, Y.; O’Connor, M.B. Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: A P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 2004, 34, 991–1010. [Google Scholar] [CrossRef]
- Namiki, T.; Niwa, R.; Sakudoh, T.; Shirai, K.-i.; Takeuchi, H.; Kataoka, H. Cytochrome P450 CYP307A1/Spook: A regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 2005, 337, 367–374. [Google Scholar] [CrossRef]
- Shahzad, M.F.; Li, Y.; Ge, C.; Sun, Y.; Yang, Q.; Li, F. Knockdown of Cs-Spook induces delayed larval molting in Rice Striped Stem Borer Chilo suppressalis. Arch. Insect Biochem. Physiol. 2014, 88, 179–191. [Google Scholar] [CrossRef]
- Wan, P.J.; Jia, S.; Li, N.; Fan, J.M.; Li, G.Q. A Halloween gene shadow is a potential target for RNA-interference-based pest management in the small brown planthopper Laodelphax striatellus. Pest Manag. Sci. 2015, 71, 199–206. [Google Scholar] [CrossRef]
- Rewitz, K.F.; O’Connor, M.B.; Gilbert, L.I. Molecular evolution of the insect Halloween family of cytochrome P450s: Phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 2007, 37, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; He, Q.H.; Zhang, T.T.; Wu, H.H.; Zhang, J.Z.; Ma, E.B. Characteristics of Halloween genes and RNA interference-mediated functional analysis of LmCYP307a2 in Locusta migratoria. Insect Sci. 2022, 29, 51–64. [Google Scholar] [CrossRef]
- Yin, C.; Liu, Y.; LIu, J.; Xiao, H.; Huang, S.; Lin, Y.; Han, Z.; Li, F. ChiloDB: A genomic and transcriptome database for an important rice insect pest Chilo suppressalis. Database 2014, 2014, bau065. [Google Scholar] [CrossRef]
- Ren, L.; Zhu, B.; Zhang, Y.; Wang, H.; Li, C.; Su, Y.; Ba, C. The research of applying primer premier 5.0 to design PCR primer. J. Jinzhou Med. Coll. 2004, 25, 43–46. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Zhang, Z.; He, G.; Yang, L.; Li, F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J. Insect Sci. 2012, 12, 60. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dinan, L.N.; Donnahey, P.L.; Rees, H.H.; Goodwin, T.W. High-performance liquid chromatography of ecdysteroids and their 3-epi, 3-dehydro and 26-hydroxy derivatives. J. Chromatogr. A 1981, 205, 139–145. [Google Scholar] [CrossRef]
- Zhou, J.; Qi, Y.; Hou, Y.; Zhao, J.; Li, Y.; Xue, X.; Wu, L.; Zhang, J.; Chen, F. Quantitative determination of juvenile hormone III and 20-hydroxyecdysone in queen larvae and drone pupae of Apis mellifera by ultrasonic-assisted extraction and liquid chromatography with electrospray ionization tandem mass spectrometry. J. Chromatogr. B 2011, 879, 2533–2541. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999, 369, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, 3003.3001–3003.3009. [Google Scholar] [CrossRef] [PubMed]
- Iga, M.; Smagghe, G. Identification and expression profile of Halloween genes involved in ecdysteroid biosynthesis in Spodoptera littoralis. Peptides 2010, 31, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Marchal, E.; Badisco, L.; Verlinden, H.; Vandersmissen, T.; Van Soest, S.; Van Wielendaele, P.; Vanden Broeck, J. Role of the Halloween genes, Spook and Phantom in ecdysteroidogenesis in the desert locust, Schistocerca gregaria. J. Insect Physiol. 2011, 57, 1240–1248. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Iga, M.; Velarde, R.; Rougé, P.; Smagghe, G. Halloween genes and nuclear receptors in ecdysteroid biosynthesis and signalling in the pea aphid. Insect Mol. Biol. 2010, 19, 187–200. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. Developmental expression of Manduca shade, the P450 mediating the final step in molting hormone synthesis. Mol. Cell. Endocrinol. 2006, 247, 166–174. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Kiuchi, M.; Takeuchi, H.; Kubo, T. Ecdysteroid biosynthesis in workers of the European honeybee Apis mellifera L. Insect Biochem. Mol. Biol. 2011, 41, 283–293. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Gilbert, L.I. Daphnia Halloween genes that encode cytochrome P450s mediating the synthesis of the arthropod molting hormone: Evolutionary implications. BMC Evol. Biol. 2008, 8, 60. [Google Scholar] [CrossRef]
- Iga, M.; Kataoka, H. Recent studies on insect hormone metabolic pathways mediated by cytochrome P450 enzymes. Biol. Pharm. Bull. 2012, 35, 838–843. [Google Scholar] [CrossRef]
- Ono, H.; Rewitz, K.F.; Shinoda, T.; Itoyama, K.; Petryk, A.; Rybczynski, R.; Jarcho, M.; Warren, J.T.; Marqués, G.; Shimell, M.J. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 2006, 298, 555–570. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect cytochrome P450. Compr. Mol. Insect Sci. 2005, 4, 1–77. [Google Scholar]
- Luan, J.-B.; Ghanim, M.; Liu, S.-S.; Czosnek, H. Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 2013, 43, 740–746. [Google Scholar] [CrossRef]
- Hentze, J.L.; Moeller, M.E.; Jørgensen, A.F.; Bengtsson, M.S.; Bordoy, A.M.; Warren, J.T.; Gilbert, L.I.; Andersen, O.; Rewitz, K.F. Accessory gland as a site for prothoracicotropic hormone controlled ecdysone synthesis in adult male insects. PLoS ONE 2013, 8, e55131. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Sakudoh, T.; Namiki, T.; Saida, K.; Fujimoto, Y.; Kataoka, H. The ecdysteroidogenic P450 Cyp302a1/disembodied from the silkworm, Bombyx mori, is transcriptionally regulated by prothoracicotropic hormone. Insect Mol. Biol. 2005, 14, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Zhou, X.; Ye, Y.-Z.; Ogihara, M.H.; Takeshima, M.; Fujinaga, D.; Liu, C.-W.; Zhu, Z.; Kataoka, H.; Bao, Y.-Y. Functional analysis of ecdysteroid biosynthetic enzymes of the rice planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 2020, 123, 103428. [Google Scholar] [CrossRef]
- Yoshiyama-Yanagawa, T.; Enya, S.; Shimada-Niwa, Y.; Yaguchi, S.; Haramoto, Y.; Matsuya, T.; Shiomi, K.; Sasakura, Y.; Takahashi, S.; Asashima, M. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J. Biol. Chem. 2011, 286, 25756–25762. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Zou, M.-M.; Vasseur, L.; Chu, L.-N.; Qin, Y.-D.; Zhai, Y.-L.; You, M.-S. Identification of halloween genes and RNA interference-mediated functional characterization of a Halloween gene Shadow in Plutella xylostella. Front. Physiol. 2019, 10, 1120. [Google Scholar] [CrossRef]
- Niwa, R.; Namiki, T.; Ito, K.; Shimada-Niwa, Y.; Kiuchi, M.; Kawaoka, S.; Kayukawa, T.; Banno, Y.; Fujimoto, Y.; Shigenobu, S. Non-molting glossy/shroud encodes a short-chain dehydro genase/reductase that functions in the ‘Black Box’of the ecdysteroid biosynthesis pathway. Development 2010, 137, 1991–1999. [Google Scholar] [CrossRef]
- Wan, P.-J.; Jia, S.; Li, N.; Fan, J.-M.; Li, G.-Q. RNA interference depletion of the Halloween gene disembodied implies its potential application for management of planthopper Sogatella furcifera and Laodelphax striatellus. PLoS ONE 2014, 9, e86675. [Google Scholar] [CrossRef] [PubMed]
- Salvia, R.; Nardiello, M.; Scieuzo, C.; Scala, A.; Bufo, S.A.; Rao, A.; Vogel, H.; Falabella, P. Novel factors of viral origin inhibit TOR pathway gene expression. Front. Physiol. 2018, 9, 1678. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahzad, M.F.; Idrees, A.; Afzal, A.; Iqbal, J.; Qadir, Z.A.; Khan, A.A.; Ullah, A.; Li, J. RNAi-Mediated Silencing of Putative Halloween Gene Phantom Affects the Performance of Rice Striped Stem Borer, Chilo suppressalis. Insects 2022, 13, 731. https://doi.org/10.3390/insects13080731
Shahzad MF, Idrees A, Afzal A, Iqbal J, Qadir ZA, Khan AA, Ullah A, Li J. RNAi-Mediated Silencing of Putative Halloween Gene Phantom Affects the Performance of Rice Striped Stem Borer, Chilo suppressalis. Insects. 2022; 13(8):731. https://doi.org/10.3390/insects13080731
Chicago/Turabian StyleShahzad, Muhammad Faisal, Atif Idrees, Ayesha Afzal, Jamshaid Iqbal, Ziyad Abdul Qadir, Azhar Abbas Khan, Ayat Ullah, and Jun Li. 2022. "RNAi-Mediated Silencing of Putative Halloween Gene Phantom Affects the Performance of Rice Striped Stem Borer, Chilo suppressalis" Insects 13, no. 8: 731. https://doi.org/10.3390/insects13080731
APA StyleShahzad, M. F., Idrees, A., Afzal, A., Iqbal, J., Qadir, Z. A., Khan, A. A., Ullah, A., & Li, J. (2022). RNAi-Mediated Silencing of Putative Halloween Gene Phantom Affects the Performance of Rice Striped Stem Borer, Chilo suppressalis. Insects, 13(8), 731. https://doi.org/10.3390/insects13080731








