Genetic Variability of Polypedilum (Diptera: Chironomidae) from Southwest Ecuador
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Specimen Collection
2.2. Field and Laboratory Procedures
2.3. DNA Extraction and PCR Amplification
2.4. Species Delimitation and Phylogenetic Reconstruction
2.5. Environmental Data Analysis of El Oro Rivers as Drivers of Polypedilum Assemblage Structure
3. Results
3.1. Environmental Characteristics of El Oro Watersheds
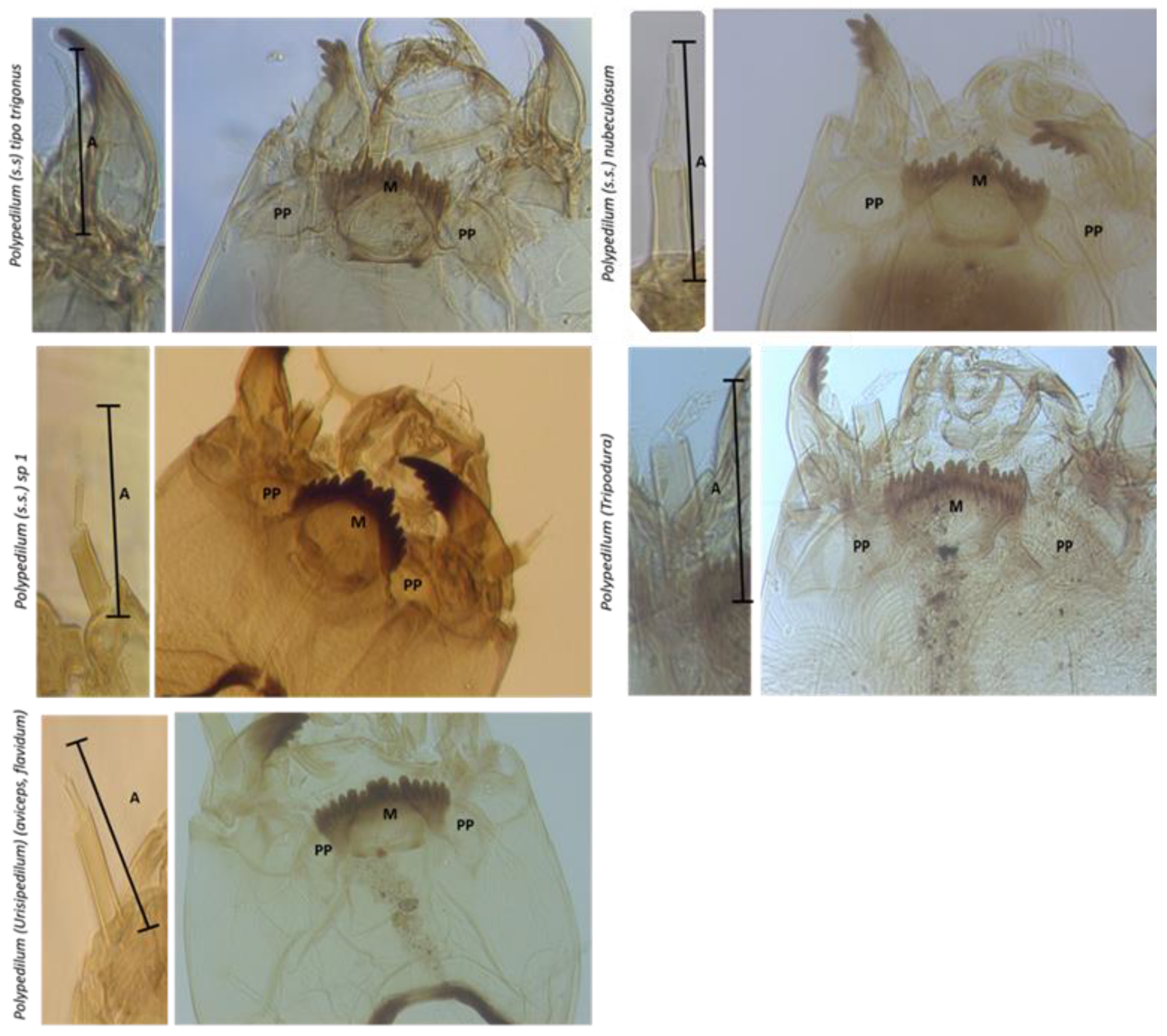
3.2. El Oro Polypedilum Morphotypes
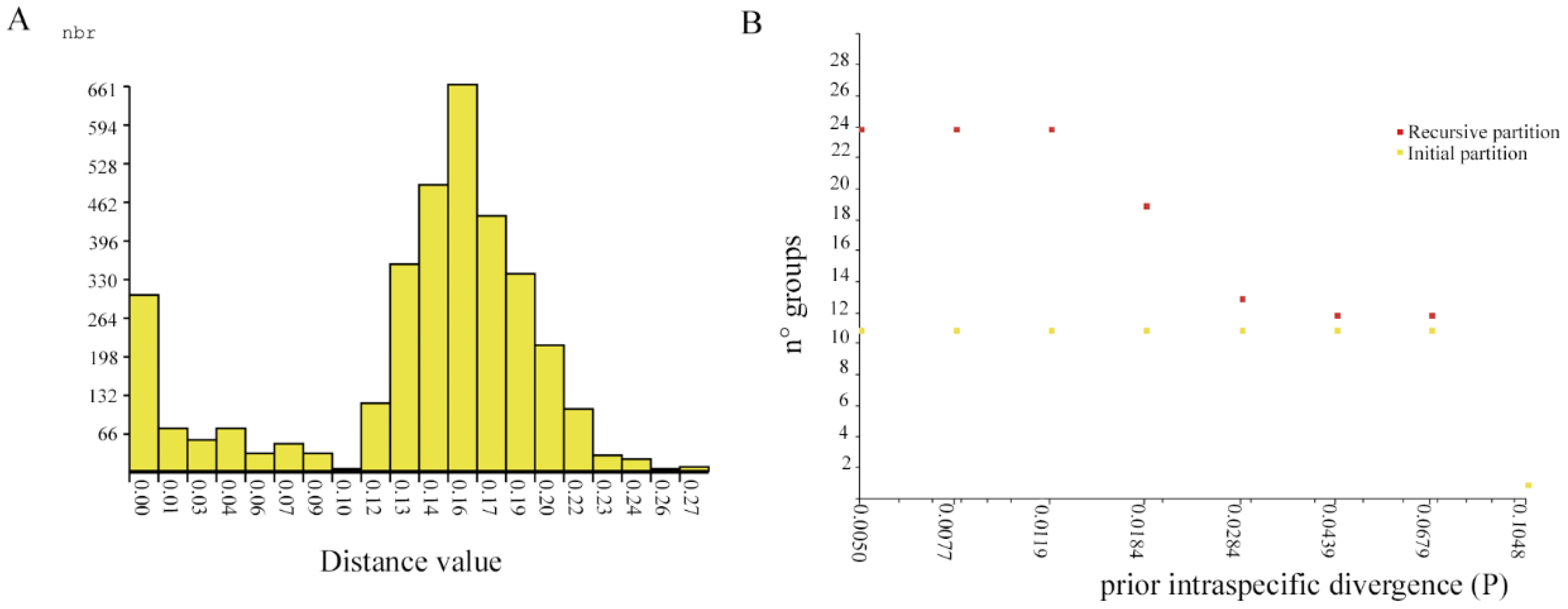
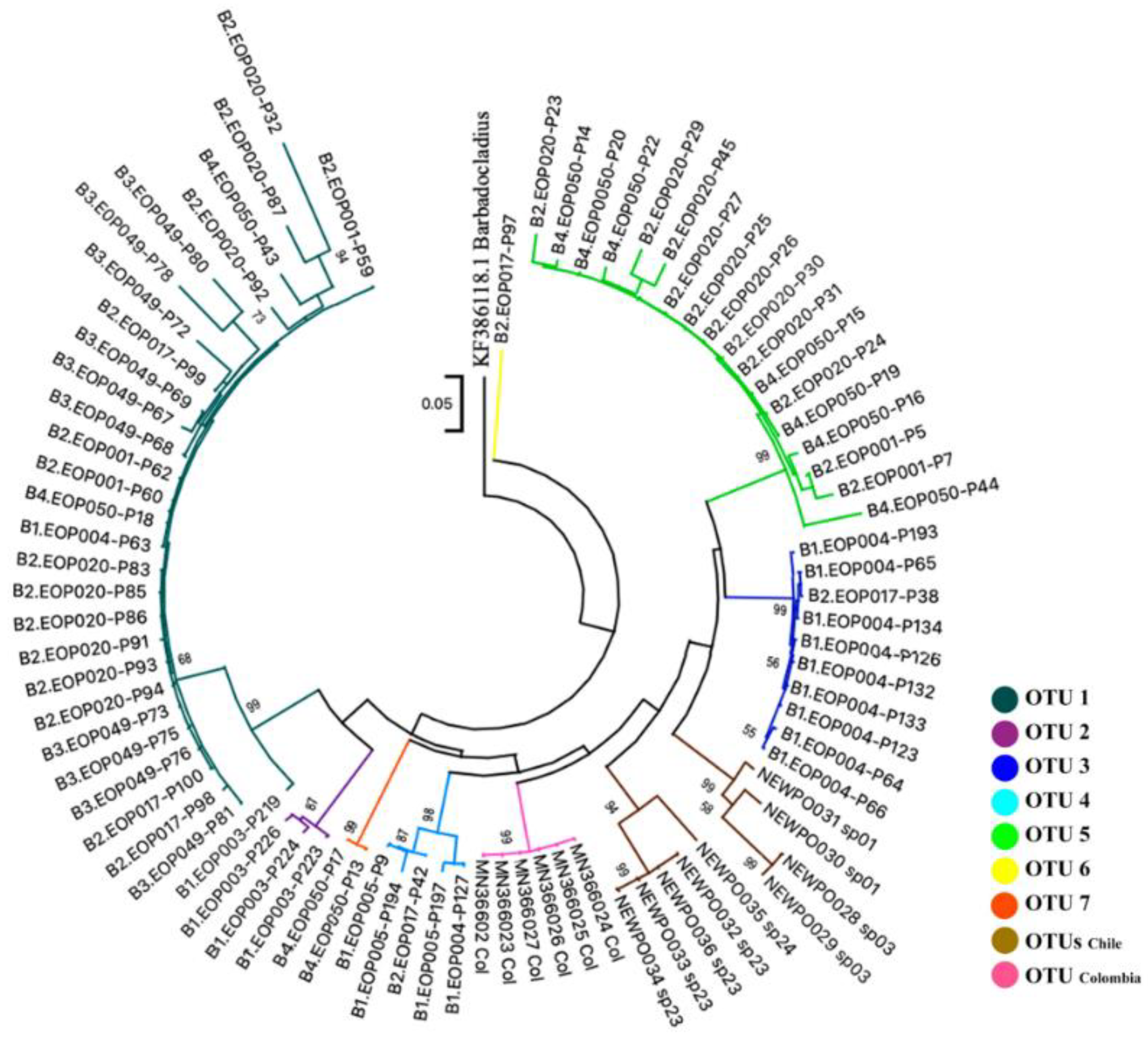
3.3. Species Delimitation and Phylogenetic Analysis
3.4. Environmental Characteristics and Polypedilum OTUs Population Composition
3.5. Polypedilum Genus Diversity Indices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashe, P.; Murray, D.A.; Reiss, F. The zoogeographical distribution of Chironomidae (Insecta: Diptera). Annls Limnol. 1987, 23, 27–60. [Google Scholar] [CrossRef]
- Lencioni, V.; Cranston, P.; Makarchenko, E.A. Recent advances in the study of Chironomidae: An overview. J. Limnol. 2018, 77, 1–6. [Google Scholar] [CrossRef]
- Acosta, R.; Prat, N. Chironomid assemblages in high altitude streams of the Andean region of Peru. Fundam. Appl. Limnol. 2010, 177, 57–79. [Google Scholar] [CrossRef]
- Koperski, P. Phylogenetic diversity of larval Chironomidae (Diptera) in lowland rivers as a potential tool in assessment of environmental quality. Hydrobiologia 2019, 836, 83–96. [Google Scholar] [CrossRef]
- Villamarín, C.; Villamarín-Cortez, S.; Salcido, D.M.; Herrera-Madrid, M.; Ríos-Touma, B. Drivers of diversity and altitudinal distribution of chironomids (Diptera: Chironomidae) in the Ecuadorian Andes. Rev. Biol. Trop. 2021, 69, 113–126. [Google Scholar] [CrossRef]
- Miller, M.P.; Blinn, D.W.; Keim, P. Correlations between observed dispersal capabilities and patterns of genetic differentiation in populations of four aquatic insect species from the Arizona White Mountains, U.S.A. Freshw. Biol. 2002, 47, 1660–1673. [Google Scholar] [CrossRef]
- Finn, D.S.; Encalada, A.C.; Hampel, H. Genetic isolation among mountains but not between stream types in a tropical high-altitude mayfly. Freshw. Biol. 2016, 61, 702–714. [Google Scholar] [CrossRef]
- Hamerlik, L.; Da Silva, F.L.; Jacobsen, D. Chironomidae (Insecta: Diptera) of Ecuadorian highaltitude streams: A survey and illustrated key. Fla. Èntomol. 2018, 101, 663–675. [Google Scholar] [CrossRef]
- Prat, N.; Ribera, C.; Rieradevall, M.; Villamarín, C.; Acosta, R. Distribution, abundance and molecular analysis of genus Barbadocladius Cranston & Krosch (Diptera, Chironomidae) in tropical, high altitude Andean streams and rivers. Neotrop. Èntomol. 2013, 42, 607–617. [Google Scholar] [CrossRef][Green Version]
- Cranston, P.; Martin, J.; Spies, M. Cryptic species in the nuisance midge Polypedilum nubifer (Skuse) (Diptera: Chironomidae) and the status of Tripedilum Kieffer. Zootaxa 2016, 4079, 429–447. [Google Scholar] [CrossRef] [PubMed]
- Cranston, P. Introduction. In The Chironomidae: Biology and Ecology of Non-Biting Midges; Armitage, P.D., Cranston, P.S., Pinder, L.C., Eds.; Chapman & Hall: London, UK, 1995; pp. 1–7. [Google Scholar]
- Luoto, T.P. The relationship between water quality and chironomid distribution in Finland—A new assemblage-based tool for assessments of long-term nutrient dynamics. Ecol. Indic. 2011, 11, 255–262. [Google Scholar] [CrossRef]
- Song, C.; Wang, Q.; Zhang, R.; Sun, B.; Wang, X. Exploring the utility of DNA barcoding in species delimitation of Polypedilum (Tripodura) non-biting midges (Diptera: Chironomidae). Zootaxa 2016, 4079, 534. [Google Scholar] [CrossRef] [PubMed]
- Previšić, A.; Gelemanović, A.; Urbanič, G.; Ternjej, I. Cryptic diversity in the Western Balkan endemic copepod: Four species in one? Mol. Phylogenetics Evol. 2016, 100, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Lin, X.-L.; Wang, Q.; Wang, X.-H. DNA barcodes successfully delimit morphospecies in a superdiverse insect genus. Zool. Scr. 2018, 47, 311–324. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Boil. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef]
- Epler, J. Identification manual for the larval Chironomidae (Diptera) of North and South Carolina. In A Guide to the Taxonomy of the Midges of the Southeastern United States, including Florida; Special Publication SJ2001-SP13; North Carolina Department of Environment and Natural Resources: Palatka, FL, USA, 2001. [Google Scholar]
- Prat, N.; Rieradevall, M.; Acosta, R.; Villamarín, C. Guía para el Reconocimiento de las Larvas de Chironomidae (Diptera) de los Ríos Altoandinos de Ecuador y Perú. Clave para la Determinación de los Géneros; 2011; p. 78. Available online: http://www.ub.edu/riosandes/docs/CLAVE_LARVAS_PERU_ECUADORvfoto3_v7.pdf (accessed on 22 March 2022).
- Epler, J.; Ekrem, T.; Cranston, P. The larvae of Chironominae (Diptera: Chironomidae) of the Holarctic region—Keys and diagnoses. In The Larvae of Chironomidae of the Holarctic Region—Keys and Diagnoses; Insect Systematics & Evolution: Lund, Sweden, 2013; pp. 387–556. [Google Scholar]
- Saether, O.A.; Andersen, T.; Pinho, L.C.; Mendes, H.F. The problems with Polypedilum Kieffer (Diptera: Chironomidae), with the description of Probolum subgen. n. Zootaxa 2010, 2497, 1–36. [Google Scholar] [CrossRef]
- Bolton, M.J. Ohio EPA Supplemental Keys to the Larval Chironomidae (Diptera) of Ohio and Ohio Chironomidae Checklist; Ohio Environmental Protection Agency, Groveport: Groveport, OH, USA, 2012. [Google Scholar]
- Cerda-Granados, D.; Díaz, V. Optimización de un protocolo de extracción de ADN genómico para Pinus tecunumanii. Encuentro 2013, 94, 82–92. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals Èntomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcode of life data system: Barcoding. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Silvestro, D.; Michalak, I. RaxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2011, 12, 335–337. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Clarke, K.; Gorley, R. PRIMER v6: User Manual/Tutorial (Plymouth Routines in Multivariate Ecological Research). PRIMER-E.; Plymouth Marine Laboratory: Plymouth, UK, 2006. [Google Scholar]
- Braak Ter, C.; Barendregt, P. CANOCO. Reference and Manual User’s Guide to Canoco for Windows: Software for Canonical Community Ordination (Version 4); Microcomputer Power: Ithaca, NY, USA, 1998. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2011; ISBN 0123266505. [Google Scholar]
- Grueso-Gilaberth, R.N.; Jaramillo-Timarán, K.S.; Ospina-Pérez, E.M.; Richardi, V.S.; Ossa-López, P.A.; Rivera-Páez, F.A. Histological description and histopathology in Polypedilum sp. (Diptera: Chironomidae): A potential biomarker for the impact of mining on tributaries. Annals Èntomol. Soc. Am. 2020, 113, 359–372. [Google Scholar] [CrossRef]
- Geraci, C.J.; Al-Saffar, M.A.; Zhou, X. DNA barcoding facilitates description of unknown faunas: A case study on Trichoptera in the headwaters of the Tigris River, Iraq. J. N. Am. Benthol. Soc. 2011, 30, 163–173. [Google Scholar] [CrossRef]
- Carew, M.; Pettigrove, V.; Hoffmann, A.A. Identifying chironomids (Diptera: Chironomidae) for biological monitoring with PCR–RFLP. Bull. Èntomol. Res. 2003, 93, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Wang, X.; Bu, W.; Qi, X. Morphology lies: A case-in-point with a new non-biting midge species from Oriental China (Diptera, Chironomidae). ZooKeys 2020, 909, 67–77. [Google Scholar] [CrossRef]
- Rossaro, B.; Marziali, L.; Montagna, M.; Magoga, G.; Zaupa, S.; Boggero, A. Factors controlling morphotaxa distributions of diptera Chironomidae in freshwaters. Water 2022, 14, 1014. [Google Scholar] [CrossRef]
- Villamarín, C.; Rieradevall, M.; Prat, N. Macroinvertebrate diversity patterns in tropical highland Andean rivers. Limnetica 2020, 39, 677–691. [Google Scholar] [CrossRef]
- González-Trujillo, J.D.; Petsch, D.K.; Córdoba-Ariza, G.; Rincón-Palau, K.; Donato-Rondon, J.C.; Castro-Rebolledo, M.I.; Sabater, S. Upstream refugia and dispersal ability may override benthic-community responses to high-Andean streams deforestation. Biodivers. Conserv. 2019, 28, 1513–1531. [Google Scholar] [CrossRef]
- Lin, X.; Stur, E.; Ekrem, T. Exploring genetic divergence in a species-rich insect genus using 2790 DNA barcodes. PLoS ONE 2015, 10, e0138993. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Cáceres, A.G.; Arrunátegui-Jiménez, M.J.; Lañas-Rosas, M.F.; Yañez-Trujillano, H.H.; Luna-Caipo, D.V.; Holguín-Mauricci, C.E.; Katakura, K.; Hashiguchi, Y.; Kato, H. DNA barcoding for identification of sand fly species (Diptera: Psychodidae) from leishmaniasis-endemic areas of Peru. Acta Trop. 2015, 145, 45–51. [Google Scholar] [CrossRef]
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef]
- Callisto, M.; Linares, M.S.; Kiffer, W.P., Jr.; Hughes, R.M.; Moretti, M.S.; Macedo, D.R.; Solar, R. Beta diversity of aquatic macroinvertebrate assemblages associated with leaf patches in neotropical montane streams. Ecol. Evol. 2021, 11, 2551–2560. [Google Scholar] [CrossRef]
- Dos Santos, D.A.; Molineri, C.; Nieto, C.; Zuñiga, M.C.; Emmerich, D.; Fierro, P.; Pessacq, P.; Rios-Touma, B.; Márquez, J.; Gomez, D.; et al. Cold/Warm stenothermic freshwater macroinvertebrates along altitudinal and latitudinal gradients in Western South America: A modern approach to an old hypothesis with updated data. J. Biogeogr. 2018, 45, 1571–1581. [Google Scholar] [CrossRef]

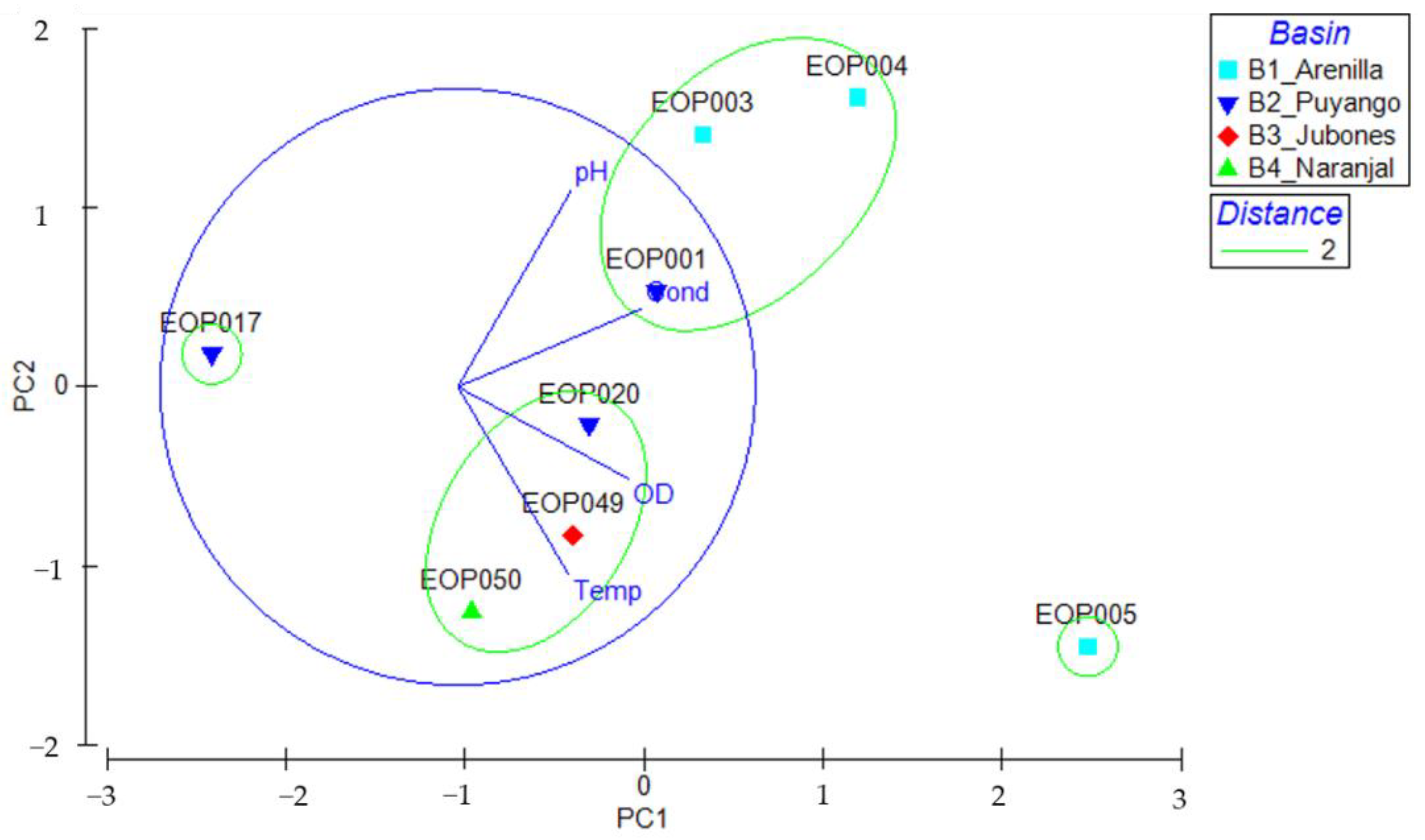
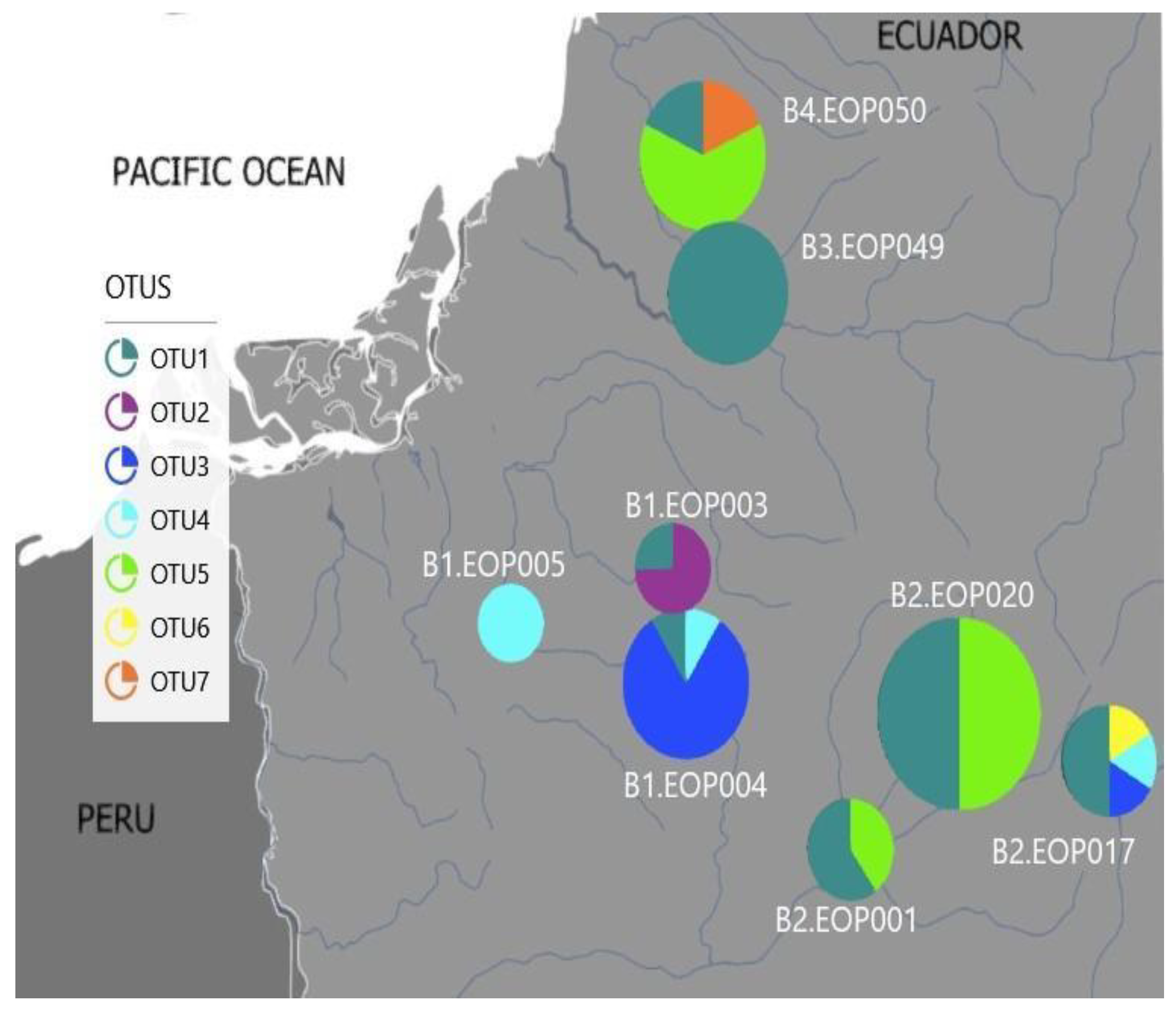
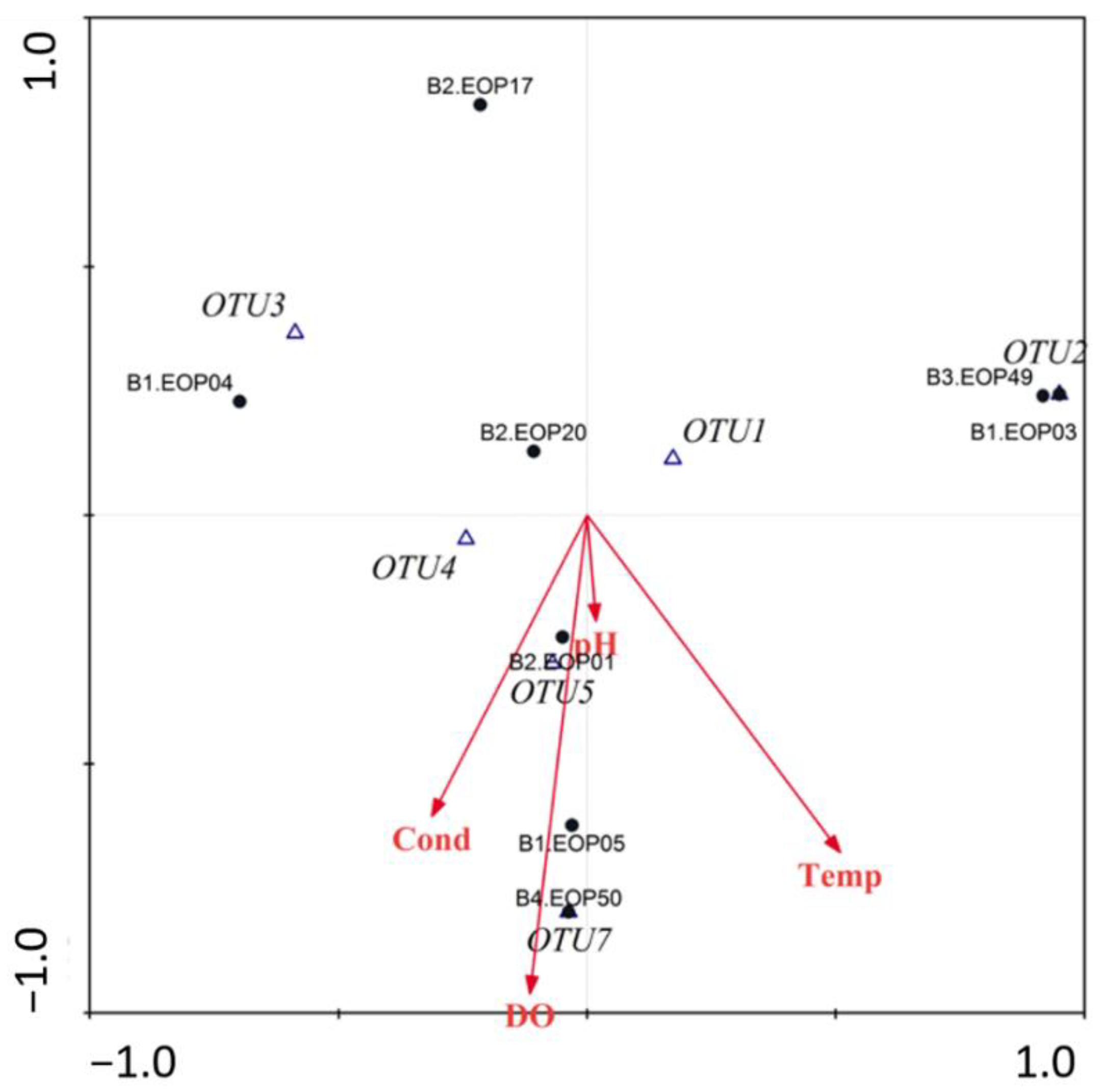
| Code | Basin | pH | Dissolved Oxygen (mg/L) | Conductivity (μS/cm) | Temperature (°C) | Altitude (m.a.s.l.) |
|---|---|---|---|---|---|---|
| EOP003 | Arenilla | 7.8 | 11.69 | 92.6 | 20.8 | 1260 |
| EOP004 | Arenilla | 8.2 | 14.69 | 92.6 | 20.8 | 1015 |
| EOP005 | Arenilla | 7.16 | 18.76 | 113.2 | 24.3 | 252 |
| EOP001 | Puyango | 7.2 | 12.69 | 92.6 | 20.8 | 529 |
| EOP017 | Puyango | 6.75 | 8.76 | 31.8 | 20 | 1293 |
| EOP020 | Puyango | 7.37 | 10.98 | 58.1 | 22.8 | 991 |
| EOP049 | Jubones | 7.1 | 11.52 | 53.4 | 23.2 | 121 |
| EOP050 | Siete | 6.66 | 14.61 | 31 | 21.7 | 205 |
| Taxa | HC Width (min/max) | AS2/AS3-5 | LS3/AS3 | MC/M1 | M4/M3-5 | PP-lóbpost | PP Wid/Length | PP W/Intp | p-Dist |
|---|---|---|---|---|---|---|---|---|---|
| Polypedilum (s.s.) gr trigonus | 320.88 (309.79–349.82) | 0.54 (0.43–0.72) | >> | 2.19 (1.98–2.67) | 3 > 4 < 5 | No | 2.28 (2.04–2.47) | 2.14 (2.00–2.26) | 0.0272952 |
| Polypedilum (s.s.) nubeculosum | 313.93 (240.41–388.73) | 0.50 (0.43–0.67) | >> | 1.43 (0.85–2.06) | 3 > 4 > 5 | No | 2.40 (2.05–3.19) | 2.30 (2.00–2.79) | 0.0588167 |
| Polypedilum (s.s.) sp1 | 287.73 (301.20–550.14) | 0.88 (0.80–0.98) | >> | 0.44 (0.34–0.53) | 3 > 4 > 5 | No | 2.29 (1.77–2.64) | 2.45 (1.89–3.16) | 0.0963929 |
| Polypedilum (Tripodura) | 209.24 (176.79–240.28) | 0.82 (0.67–1.04) | << | 1.79 (1.47–2.08) | 3 > 4 > 5 | No | 2.49 (2.32–2.60) | 4.69 (3.97–5.13) | 0.0067183 |
| Polypedilum (Urisipedilum) | 329.80 (299.09–380.44) | 1.26 (1.13–1.34) | >> | 1.76 (1.52–1.90) | 3 > 4 > 5 | Si | 1.44 (1.28–1.58) | 1.55 (1.48–1.64 | 0.1136894 |
| Polypedilum sp Colombia | 0 | ||||||||
| Polypedilum sp3 Chile | 0 | ||||||||
| Polypedilum sp1 Chile | 0.0382833 | ||||||||
| Polypedilum sp23 Chile | 0.0393383 |
| OTUs | N | Mean Distance | S.E. |
|---|---|---|---|
| OTU1 | 29 | 0.048 | 0.005 |
| OTU2 | 3 | 0.015 | 0.005 |
| OTU3 | 10 | 0.006 | 0.002 |
| OTU4 | 5 | 0.043 | 0.008 |
| OTU5 | 18 | 0.029 | 0.004 |
| OTU6 | 1 | n/c | n/c |
| OTU7 | 2 | 0.0042 | 0.003 |
| Sampling Site | Number of Sequences | Number of Segregating Site (S) | Number of Haplotype (h) | Haplotype Diversity (Hd) | Nucleotide Diversity (Pi) | Theta_S | s.d. Theta_S | Theta_pi | s.d. Theta_pi |
|---|---|---|---|---|---|---|---|---|---|
| EOP001 | 5 | 78 | 4 | 0.900 | 0.095 | 37.440 | 18.967 | 45.200 | 27.780 |
| EOP003 | 4 | 64 | 4 | 1.000 | 0.070 | 34.909 | 19.100 | 33.167 | 22.067 |
| EOP004 | 12 | 113 | 9 | 0.945 | 0.050 | 32.093 | 12.918 | 23.818 | 12.815 |
| EOP005 | 3 | 32 | 3 | 1.000 | 0.046 | 21.333 | 13.103 | 21.667 | 16.591 |
| EOP017 | 6 | 115 | 6 | 1.000 | 0.123 | 50.364 | 23.942 | 58.400 | 34.119 |
| EOP020 | 18 | 141 | 14 | 0.961 | 0.113 | 40.994 | 14.469 | 53.673 | 27.235 |
| EOP049 | 10 | 73 | 9 | 0.978 | 0.042 | 25.804 | 10.728 | 19.800 | 10.837 |
| EOP050 | 10 | 138 | 10 | 1.000 | 0.115 | 48.781 | 19.960 | 54.622 | 29.235 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ballesteros, I.; Bravo-Castro, M.; Villamarín-Cortez, S.; Jijón, G.; Prat, N.; Ríos-Touma, B.; Villamarín, C. Genetic Variability of Polypedilum (Diptera: Chironomidae) from Southwest Ecuador. Insects 2022, 13, 382. https://doi.org/10.3390/insects13040382
Ballesteros I, Bravo-Castro M, Villamarín-Cortez S, Jijón G, Prat N, Ríos-Touma B, Villamarín C. Genetic Variability of Polypedilum (Diptera: Chironomidae) from Southwest Ecuador. Insects. 2022; 13(4):382. https://doi.org/10.3390/insects13040382
Chicago/Turabian StyleBallesteros, Isabel, Mishell Bravo-Castro, Santiago Villamarín-Cortez, Gabriela Jijón, Narcís Prat, Blanca Ríos-Touma, and Christian Villamarín. 2022. "Genetic Variability of Polypedilum (Diptera: Chironomidae) from Southwest Ecuador" Insects 13, no. 4: 382. https://doi.org/10.3390/insects13040382
APA StyleBallesteros, I., Bravo-Castro, M., Villamarín-Cortez, S., Jijón, G., Prat, N., Ríos-Touma, B., & Villamarín, C. (2022). Genetic Variability of Polypedilum (Diptera: Chironomidae) from Southwest Ecuador. Insects, 13(4), 382. https://doi.org/10.3390/insects13040382






