Reference Genes for Expression Analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Samples through Developing Stages
2.3. Specimens among Various Tissues
2.4. Collections during Varied Temperature Incubation
2.5. Samples for the Expression Analysis of PoChSA
2.6. Selection and Authentication of Candidate HKGs
2.7. Quantitative Real-Time PCR (qRT-PCR)
2.8. Evaluation of Reference Gene Selection
2.9. Data Processing
3. Results
3.1. Selection of Candidate HKGs
3.2. Expression Variations of the Ten HKGs
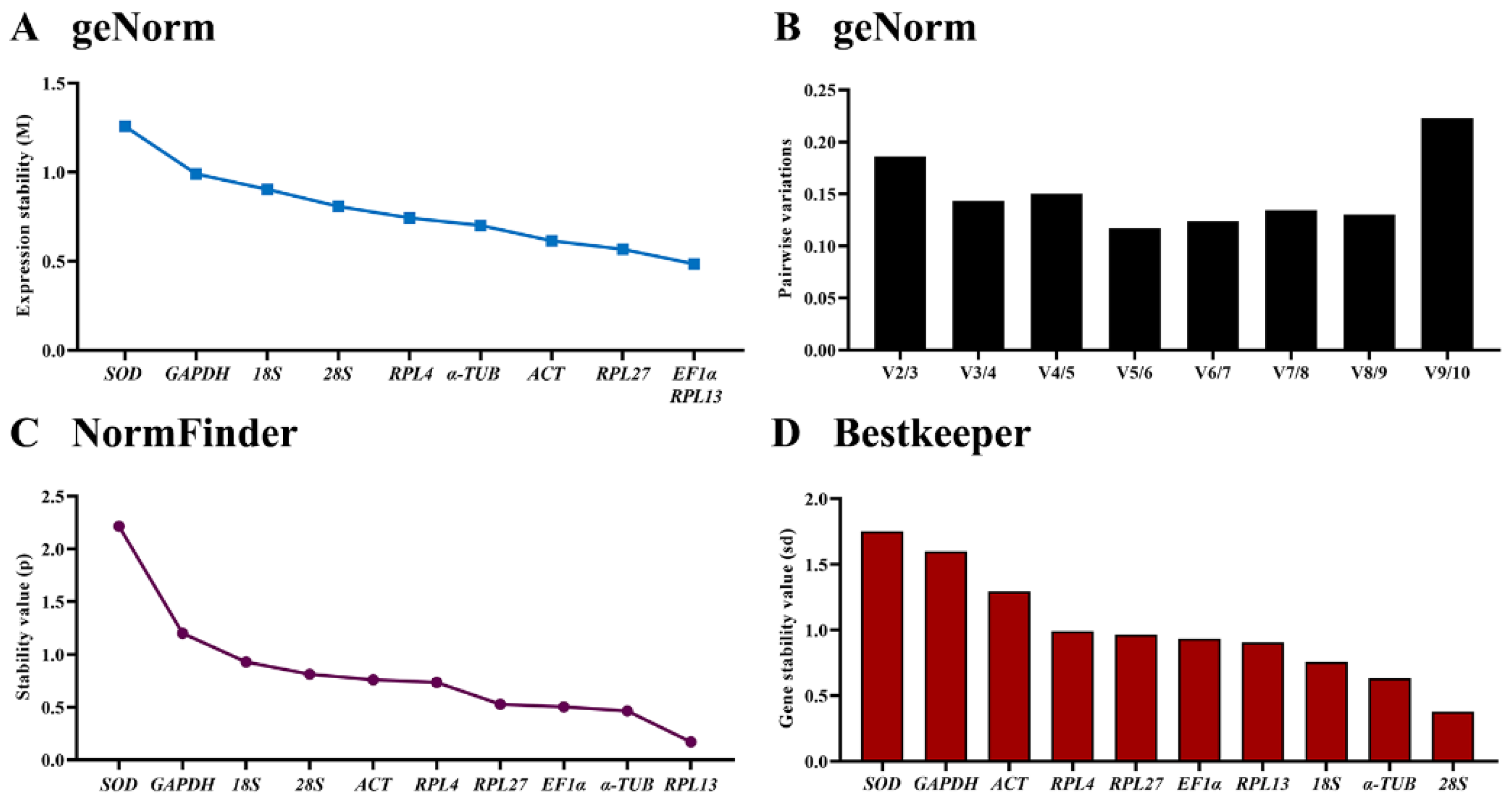
3.3. Expression Stability of the Ten HKGs during Developmental Stages
3.4. Expression Stability of the Ten HKGs among Different Tissues
3.5. Stability of the Ten HKGs under Different Temperatures
3.6. Validation of the Selected Reference Genes after Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.M.A.; Diab, M.R.; Abdelsattar, M.; Khalil, S.M.S. Characterization and RNAi-mediated knockdown of Chitin Synthase A in the potato tuber moth, Phthorimaea operculella. Sci. Rep. 2017, 7, 9502. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Cai, Y.; Zhu, J.; Zhang, M.; Zhang, Y.; Ge, Y.; Zhu, Z.; Zhou, W.; Wang, G.; Gao, Y. Identification and functional char-acterization of two putative pheromone receptors in the potato tuber moth, Phthorimaea operculella. Front. Physiol. 2021, 11, 618983. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Cao, J.; Zhang, S.; Zhang, H.; Wu, X.; Zhang, Q.; Liu, X. Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS ONE 2014, 9, e98164. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, Y.; Xu, K.; Wang, Y.; Yang, W. Selection and validation of reference genes for gene expression analysis in Tuta absoluta Meyrick (Lepidoptera: Gelechiidae). Insects 2021, 12, 589. [Google Scholar] [CrossRef]
- Xu, J.; Welker, D.L.; James, R.R. Variation in expression of reference genes across life stages of a bee, Megachile rotundata. Insects 2021, 12, 36. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.; Bustin, S. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Rondon, S.I. Decoding Phthorimaea operculella (Lepidoptera: Gelechiidae) in the new age of change. J. Integr. Agric. 2020, 19, 316–324. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Nam, H.; Hopkins, R.G.; Morrison, R.F. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE 2010, 5, e15208. [Google Scholar] [CrossRef] [PubMed]
- Chapuis, M.P.; Tohidiesfahani, D.; Dodgson, T.; Blondin, L.; Ponton, F.; Cullen, D.; Simpson, S.J.; Sword, G.A. Assessment and validation of a suite of reverse transcription-quantitative PCR reference genes for analyses of density dependent behavioural plasticity in the Australian plague locust. BMC Mol. Biol. 2011, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Chen, J.Y.; Lu, M.X.; Gao, Y.; Tian, Z.H.; Gong, W.R.; Zhu, W.; Du, Y.Z. Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS ONE 2017, 12, e0181862. [Google Scholar] [CrossRef] [PubMed]
- Ponton, F.; Chapuis, M.P.; Pernice, M.; Sword, G.A.; Simpson, S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011, 57, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Scharlaken, B.; Graaf, D.C.D.; Goossens, K.; Brunain, M.; Peelman, L.J.; Jacobs, F.J. Reference gene selection for insect expression studies using quantitative real-time PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 2008, 8, 1–10. [Google Scholar] [CrossRef]
- Sinha, D.K.; Smith, C.M. Selection of reference genes for expression analysis in Diuraphis noxia (Hemiptera: Aphididae) fed on resistant and susceptible wheat plants. Sci. Rep. 2014, 4, 5059. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Zhang, Z.; He, G.; Yang, L.; Li, F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J. Insect Sci. 2012, 12, 1–17. [Google Scholar] [CrossRef]
- Van Hiel, M.B.; Van Wielendaele, P.; Temmerman, L.; Van Soest, S.; Vuerinckx, K.; Huybrechts, R.; Broeck, J.V.; Simonet, G. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 2009, 10, 56. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P.; Palli, S.R. RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae). BMC Mol. Biol. 2019, 20, 6. [Google Scholar] [CrossRef]
- Liu, G.; Qiu, X.; Li, C.; Zhang, Y.; Zhan, Z.; Han, R. Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth, Thitarodes armoricanus (Lepidoptera, Hepialidae). PLoS ONE 2016, 11, e0159060. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Meng, Q.Q.; Zhu, X.; Sun, S.W.; Liu, A.Q.; Gao, S.F.; Gou, Y.F. Identification and evaluation of reference genes for normalization of gene expression in developmental stages, sexes, and tissues of Diaphania caesalis (Lepidoptera, Pyralidae). J. Insect Sci. 2020, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lu, M.X.; Tang, X.T.; Du, Y.Z. Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS ONE 2015, 10, e0115979. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.M.; Chahad-Ehlers, S.; Lima, A.L.A.; Taniguti, C.H.; Sobrinho, I.; Torres, F.R.; Torres, F.R. Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. 2015, 6, 17480. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Szabo, A.; Perou, C.M.; Karaca, M.; Perreard, L.; Bernard, P.S. Statistical modeling for selecting housekeeper genes. Genome Biol. 2008, 9, 405. [Google Scholar] [CrossRef]
- Yang, C.; Pan, H.; Liu, Y.; Zhou, X. Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite, Tetranychus urticae. PLoS ONE 2015, 10, e0120833. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Dhandapani, R.K.; Duan, J.J.; Palli, S.R. RNA interference in the Asian longhorned beetle: Identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 2017, 7, 8913. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.Q.; Guo, W.C.; Wan, P.J.; Zhou, L.T.; Ren, X.L.; Ahmat, T.; Fu, K.Y.; Li, G.Q. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 2013, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Yang, X.; Siegfried, B.D.; Zhou, X. A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS ONE 2015, 10, e0125868. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Pan, H.; Noland, J.E.; Hang, D.; Zhang, Z.; Liu, Y.; Zhou, X. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 2015, 5, 18201. [Google Scholar] [CrossRef]
- Yang, C.; Preisser, E.L.; Zhang, H.; Liu, Y.; Dai, L.; Pan, H.; Zhou, X. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 2016, 7, e53006. [Google Scholar]
- Lü, J.; Chen, S.; Guo, M.; Ye, C.; Qiu, B.; Wu, J.; Yang, C.; Pan, H. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 2018, 9, 1614. [Google Scholar] [CrossRef]
- Ma, L.; Jiang, T.; Liu, X.; Xiao, H.; Peng, Y.; Zhang, W. Evaluation of candidate reference genes for gene expression analysis in the brassica leaf beetle, Phaedon brassicae (Coleoptera: Chrysomelidae). PLoS ONE 2021, 16, e0251920. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Zhang, S.D.; An, S.H.; Li, Z.; Wu, F.M.; Yang, Q.P.; Liu, Y.C.; Cao, J.J.; Zhang, H.J.; Zhang, Q.W.; Liu, X.X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Rahman, S.; Zhao, Z.; Umair Sial, M.; Zhang, Y.; Jiang, H. Case study using recommended reference genes actin and 18S for reverse-transcription quantitative real-time PCR analysis in Myzus persicae. PLoS ONE 2021, 16, e0258201. [Google Scholar] [CrossRef]
- Jia, H.; Sun, R.; Shi, W.; Yan, Y.; Li, H.; Guo, X.; Xu, B. Characterization of a mitochondrial manganese superoxide dismutase gene from Apis cerana cerana and its role in oxidative stress. J. Insect Physiol. 2014, 60, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y.; Shi, X.; Zhang, J.; Ma, E. Overexpression of Mn-superoxide dismutase in Oxya chinensis mediates increased malathion tolerance. Chemosphere 2017, 181, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Nojima, Y.; Sakamoto, T.; Iwabuchi, K.; Nakazato, T.; Bono, H.; Toyoda, A.; Fujiyama, A.; Kanost, M.R.; Tabunoki, H. Comparative analysis of seven types of superoxide dismutases for their ability to respond to oxidative stress in Bombyx mori. Sci. Rep. 2019, 9, 2170. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Zhu, L.; Li, Y.; Liu, W.; Ma, W.; Lei, C.; Wang, X.-P. A de novo transcriptome and valid reference genes for quantitative Real-Time PCR in Colaphellus bowringi. PLoS ONE 2015, 10, e0118693. [Google Scholar] [CrossRef]
- Li, H.; Dai, C.; Zhang, C.; He, Y.; Ran, H.; Chen, S. Screening potential reference genes for quantitative real time PCR analysis in the oriental armyworm, Mythimna separata. PLoS ONE 2018, 13, e019509. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Chen, G.; Ma, C.; Chen, H.; Gao, X.; Tian, Z.; Cui, S.; Tian, Z.; Guo, J.; et al. Identification and validation of reference genes for quantitative gene expression analysis in Ophraella communa. Front. Physiol. 2020, 11, 355. [Google Scholar] [CrossRef]
- Nicholls, C.; Li, H.; Liu, J. GAPDH: A common enzyme with uncommon functions. Clin. Exp. Pharmacol. Physiol. 2012, 39, 674–679. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.; et al. The need for transparency and good practices in the qPCR literature. Nat. Methods 2013, 10, 1063–1067. [Google Scholar] [CrossRef]
- Hornáková, D.; Matousková, P.; Kindl, J.; Valterová, I.; Pichová, I. Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 2010, 397, 118–120. [Google Scholar] [CrossRef]
- Zheng, Y.T.; Li, H.B.; Lu, M.X.; Du, Y.Z. Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLoS ONE 2014, 9, e111369. [Google Scholar]
- Zhou, X.; Liao, W.; Liao, J.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Kyre, B.R.; Rodrigues, T.B.; Rieske, L.K. RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 2019, 9, 5640. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.A.; Németh, Z.; Juhász, E.; Póliska, S.; Rácz, R.; Kosztolányi, A.; Barta, Z. Evaluation of potential reference genes for real-time qPCR analysis in a biparental beetle, Lethrus apterus (Coleoptera: Geotrupidae). PeerJ 2017, 5, e4047. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Huang, Y.; Liao, Y.; Yin, Y. Identification of suitable reference genes for gene expression studies by qRT-PCR in the blister beetle Mylabris cichorii. J. Insect Sci. 2014, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lord, J.C.; Hartzer, K.; Toutges, M.; Oppert, B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 2010, 80, 219–221. [Google Scholar] [CrossRef]
- Sang, W.; He, L.; Wang, X.-P.; Zhu-Salzman, K.; Lei, C.-L. Evaluation of reference genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) under UVB stress. Environ. Entomol. 2015, 44, 418–425. [Google Scholar] [CrossRef]
- Gao, X.K.; Zhang, S.; Luo, J.Y.; Wang, C.Y.; Lü, L.M.; Zhang, L.J.; Zhu, X.Z.; Wang, L.; Cui, J.J. Identification and validation of reference genes for gene expression analysis in Aphidius gifuensis (Hymenoptera: Aphidiidae). PLoS ONE 2017, 12, e0188477. [Google Scholar] [CrossRef]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Rathore, P.; Palli, S.R. Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida). PLoS ONE 2018, 13, e0191116. [Google Scholar] [CrossRef]
- Yang, C.; Pan, H.; Liu, Y.; Zhou, X. Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS ONE 2015, 10, e0130593. [Google Scholar] [CrossRef]
- Mamidala, P.; Rajarapu, S.P.; Jones, S.C.; Mittapalli, O. Identification andvalidation of reference genes for quantitative real-time polymerse chain reaction in Cimex lectularius. J. Med. Entomol. 2011, 48, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Koramutla, M.K.; Aminedi, R.; Bhattacharya, R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 2016, 6, 25883. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Jain, G.; Upadhyay, S.K.; Singh, H.; Dixit, S.; Verma, P.C. Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 2017, 7, 13520. [Google Scholar] [CrossRef] [PubMed]
- Majerowicz, D.; Alves-Bezerra, M.; Logullo, R.; Fonseca-de-Souza, A.L.; Meyer-Fernandes, J.R.; Braz, G.R.; Gondim, K.C. Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae). Insect Mol. Biol. 2011, 20, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jin, Y.; He, L.; Lu, W.C.; Li, M. Suitable reference gene selection for the different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci. 2010, 10, 208. [Google Scholar] [CrossRef]
- Morales, M.A.; Mendoza, B.M.; Lavine, L.C.; Lavine, M.D.; Walsh, D.B.; Zhu, F. Selection of reference genes for expression studies of xenobiotic adaptation in Tetranychus urticae. Int. J. Biol. Sci. 2016, 12, 1129–1139. [Google Scholar] [CrossRef]
- Han, S.; Qin, Q.; Wang, D.; Zhou, Y.; He, Y. Selection and evaluation of reference genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2021, 12, 902. [Google Scholar] [CrossRef]
- Shi, J.-F.; Mu, L.-L.; Chen, X.; Guo, W.-C.; Li, G.-Q. RNA interference of chitin synthase genes inhibits chitin biosynthesis and affects larval performance in Leptinotarsa decemlineata (Say). Int. J. Biol. Sci. 2016, 12, 1319–1331. [Google Scholar] [CrossRef]
- Yang, W.-J.; Xu, K.-K.; Cong, L.; Wang, J.-J. Identification, mRNA expression, and functional analysis of Chitin Synthase 1 gene and its two alternative splicing variants in Oriental fruit fly, Bactrocera dorsalis. Int. J. Biol. Sci. 2013, 9, 331–342. [Google Scholar] [CrossRef]
- Tetreau, G.; Cao, X.; Chen, Y.-R.; Muthukrishnan, S.; Jiang, H.; Blissard, G.W.; Kanost, M.R.; Wang, P. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 2015, 62, 114–126. [Google Scholar] [CrossRef]
- Shirk, P.D.; Perera, O.P.; Shelby, K.S.; Furlong, R.B.; LoVullo, E.D.; Popham, H.J.R. Unique synteny and alternate splicing of the chitin synthases in closely related heliothine moths. Gene 2015, 574, 121–139. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Primer Sequences (5′ to 3′) | Amplicon Size (bp) | Accession Number |
|---|---|---|---|
| ACT | Forward GTGTTCCCCTCCATCGTC Reverse ACATCGCCTGGAAAGTAG | 979 | OL675412 |
| α-TUB | Forward GCCGTGTTTGTGGACTTG Reverse TGATGGAGGATACGATTTGA | 523 | OL690519 |
| 18S | Forward ATGCCCTTAGATGTCCTGG Reverse GGATTTCTAACCCGTCTGC | 557 | OL655414 |
| 28S | Forward ACGTCGTTGTCGATGTCC Reverse CAAGCCTTCACTTTCGTT | 212 | OL672488 |
| GAPDH | Forward GACCACTGTCCACGCTAC Reverse GATGACACGGCTGGAGTA | 451 | OL675413 |
| EF1α | Forward CTTCTCGCCTTCACCCTT Reverse GGCGAATCTACCCAGAGG | 864 | OL690518 |
| RPL4 | Forward TGAGAAGAGCGAGCAAGT Reverse TTTTCCCTCAGTTTCTCG | 1098 | OL652885 |
| RPL13 | Forward ACAAGGATTGGCAAAGATT Reverse ACCCTTGAGGACCTTCTT | 365 | OL690517 |
| RPL27 | Forward GAAGAACTACGACGAGGGG Reverse TGTTCTTTCCGCTCTTGTAT | 299 | OL675414 |
| SOD | Forward ATGGTTGCTTTGCTGAAT Reverse AGATAGCTTTGACATAGTCGG | 370 | OL675415 |
| Gene | Primer Sequences (5′ to 3′) | Length (bp) | Slope | R2 | Efficiency (%) |
|---|---|---|---|---|---|
| ACT | F-AATTGTGCGAGACGTCAAGG | 239 | −3.480 | 0.998 | 93.80 |
| R-CGTCGCACTTCATGATGGAG | |||||
| α-TUB | F-CACTGGTAAAGAAGACGCGG | 194 | −3.241 | 0.999 | 103.49 |
| R-AGAGACGTTCCATCAGCAGG | |||||
| 18S | F-CGTTTGCTGGGAAGTTGACC | 199 | −3.289 | 0.997 | 101.39 |
| R-GACACGACCGTAAACCCATC | |||||
| 28S | F-GATTCAGTTTCGGGCACTCG | 154 | −3.232 | 0.999 | 103.89 |
| R-CTAGACCGACGCTCCATCC | |||||
| GAPDH | F-TGCCACCCAAAAGACTGTTG | 240 | −3.338 | 0.998 | 99.33 |
| R-ACCTTGGCTTTGATCGCATC | |||||
| EF1α | F-TGTCAAGCAGCTGATCGTTG | 164 | −3.286 | 0.999 | 101.52 |
| R-CTCCGTGCCATCCAGAAATG | |||||
| RPL4 | F-GGTCTGACGTGCTCAAGGTA | 183 | −3.452 | 0.991 | 94.84 |
| R-GCAGGTTCAGCTTGTCAACA | |||||
| RPL13 | F-AACCAACCCGCTAGAAGACA | 97 | −3.294 | 0.999 | 101.18 |
| R-CCACAGGTCTCAATGGTCCA | |||||
| RPL27 | F-TGAAGAACTACGACGAGGG | 199 | −3.384 | 0.992 | 97.47 |
| R-TCGAAGCTGAAGTCTACGGA | |||||
| SOD | F-CAACCTGTCTCCCTGCAAAA | 159 | −3.328 | 0.998 | 99.75 |
| R-TTCGCCAACTTGTTGTAGCC |
| Conditions | CRGs * | geNorm | Normfinder | BestKeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Developmental stages | ACT | 1.542 | 9 | 3.751 | 10 | 2.987 | 10 | 3.819 | 10 |
| EF | 0.208 | 1 | 0.374 | 2 | 0.289 | 3 | 1.065 | 1 | |
| 18S | 0.973 | 8 | 1.214 | 9 | 1.097 | 9 | 1.728 | 9 | |
| 28S | 0.429 | 2 | 0.759 | 7 | 0.284 | 2 | 1.298 | 6 | |
| SOD | 0.769 | 6 | 0.740 | 5 | 0.617 | 6 | 1.351 | 7 | |
| GAPDH | 0.721 | 5 | 0.951 | 8 | 0.878 | 8 | 1.362 | 8 | |
| α-TUB | 0.551 | 3 | 0.753 | 6 | 0.545 | 4 | 1.240 | 4 | |
| RPL4 | 0.818 | 7 | 0.573 | 4 | 0.699 | 7 | 1.278 | 5 | |
| RPL13 | 0.208 | 1 | 0.294 | 1 | 0.141 | 1 | 1.069 | 2 | |
| RPL27 | 0.650 | 4 | 0.560 | 3 | 0.609 | 5 | 1.211 | 3 | |
| Larvae tissues | ACT | 0.799 | 7 | 1.150 | 9 | 0.696 | 8 | 1.344 | 9 |
| EF | 0.338 | 1 | 0.007 | 1 | 0.200 | 1 | 0.833 | 1 | |
| 18S | 0.706 | 6 | 1.004 | 8 | 0.656 | 7 | 1.252 | 7 | |
| 28S | 0.445 | 3 | 0.383 | 5 | 0.328 | 3 | 0.934 | 4 | |
| SOD | 1.161 | 9 | 2.131 | 10 | 2.088 | 10 | 2.217 | 10 | |
| GAPDH | 0.897 | 8 | 0.972 | 7 | 0.997 | 9 | 1.302 | 8 | |
| α-TUB | 0.607 | 5 | 0.608 | 6 | 0.493 | 5 | 1.043 | 6 | |
| RPL4 | 0.389 | 2 | 0.272 | 2 | 0.373 | 4 | 0.869 | 3 | |
| RPL13 | 0.338 | 1 | 0.293 | 3 | 0.279 | 2 | 0.868 | 2 | |
| RPL27 | 0.517 | 4 | 0.370 | 4 | 0.542 | 6 | 0.946 | 5 | |
| Temparature treatment | ACT | 0.614 | 3 | 0.759 | 6 | 1.293 | 8 | 1.137 | 5 |
| EF | 0.485 | 1 | 0.502 | 3 | 0.934 | 5 | 0.991 | 2 | |
| 18S | 0.903 | 7 | 0.928 | 8 | 0.755 | 3 | 1.322 | 8 | |
| 28S | 0.808 | 6 | 0.812 | 7 | 0.377 | 1 | 1.194 | 7 | |
| SOD | 1.257 | 9 | 2.215 | 10 | 1.751 | 10 | 2.324 | 10 | |
| GAPDH | 0.990 | 8 | 1.200 | 9 | 1.599 | 9 | 1.451 | 9 | |
| α-TUB | 0.701 | 4 | 0.464 | 2 | 0.630 | 2 | 1.026 | 3 | |
| RPL4 | 0.743 | 5 | 0.734 | 5 | 0.989 | 7 | 1.144 | 6 | |
| RPL13 | 0.485 | 1 | 0.170 | 1 | 0.906 | 4 | 0.945 | 1 | |
| RPL27 | 0.567 | 2 | 0.527 | 4 | 0.963 | 6 | 1.0.36 | 4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, C.-H.; Peng, L.-J.; Zhang, Y.-X.; Zeng, H.-R.; Yu, H.-F.; Jin, L.; Li, G.-Q. Reference Genes for Expression Analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects 2022, 13, 140. https://doi.org/10.3390/insects13020140
Shen C-H, Peng L-J, Zhang Y-X, Zeng H-R, Yu H-F, Jin L, Li G-Q. Reference Genes for Expression Analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects. 2022; 13(2):140. https://doi.org/10.3390/insects13020140
Chicago/Turabian StyleShen, Chen-Hui, Li-Juan Peng, Yu-Xing Zhang, Hua-Rui Zeng, Hong-Fei Yu, Lin Jin, and Guo-Qing Li. 2022. "Reference Genes for Expression Analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae)" Insects 13, no. 2: 140. https://doi.org/10.3390/insects13020140
APA StyleShen, C.-H., Peng, L.-J., Zhang, Y.-X., Zeng, H.-R., Yu, H.-F., Jin, L., & Li, G.-Q. (2022). Reference Genes for Expression Analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects, 13(2), 140. https://doi.org/10.3390/insects13020140







