Identification of the Short Neuropeptide F and Short Neuropeptide F Receptor Genes and Their Roles of Food Intake in Dendroctonus armandi
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Sample Preparation
2.2. Total RNA Isolation and cDNA Synthesis
2.3. Identification of D. armandi NPF and NPFR Genes
2.3.1. cDNA Amplification and Cloning
2.3.2. 3′ RACE and Cloning of Full-Length cDNA
2.3.3. Analysis of cDNA Sequences
2.4. Expression Patterns of sNPF and sNPFR Genes in Different Life Stages, Body Parts, and Treatments
2.4.1. Insects Sampling and Treatments for RT-qPCR
2.4.2. RT-qPCR
2.5. RNA Interference
2.5.1. The dsRNA Synthesis and Injection
2.5.2. Survival Test and Body Weight Measurement
2.5.3. Determination of Glycogen, Free Fatty Acid, Trehalose
2.6. Statistical Analysis
3. Results
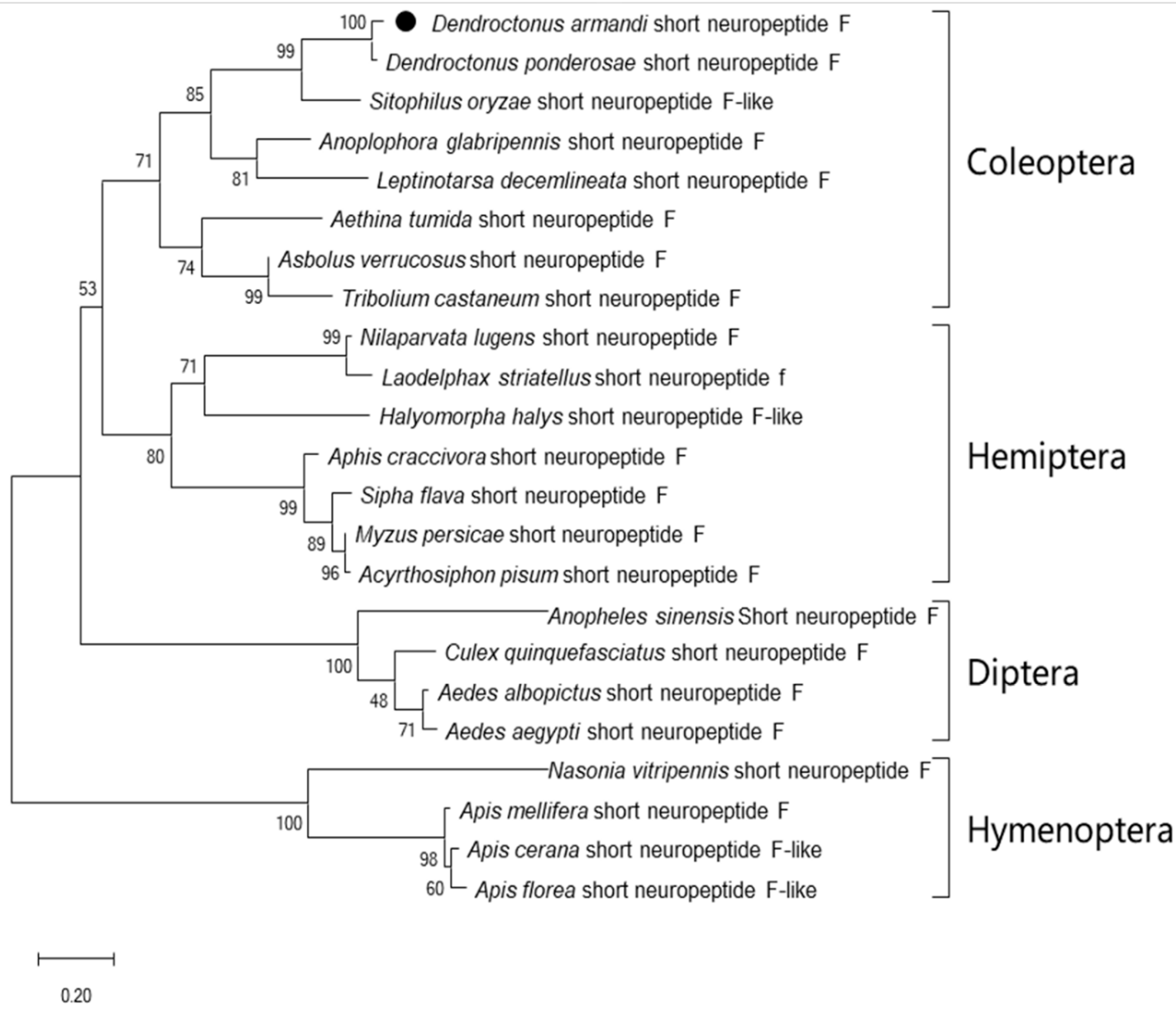
3.1. Sequence Characteristics and Bioinformatics Analysis
3.2. RT-qPCR
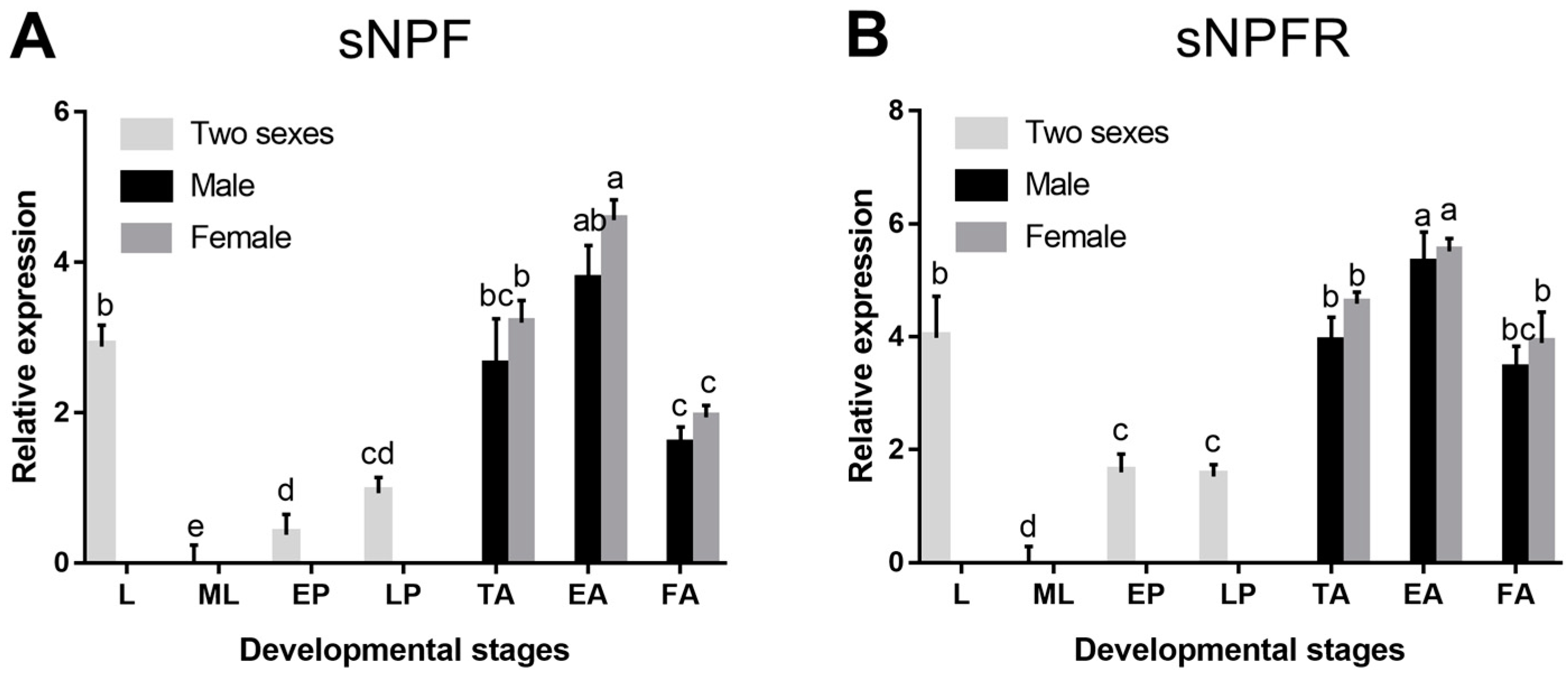
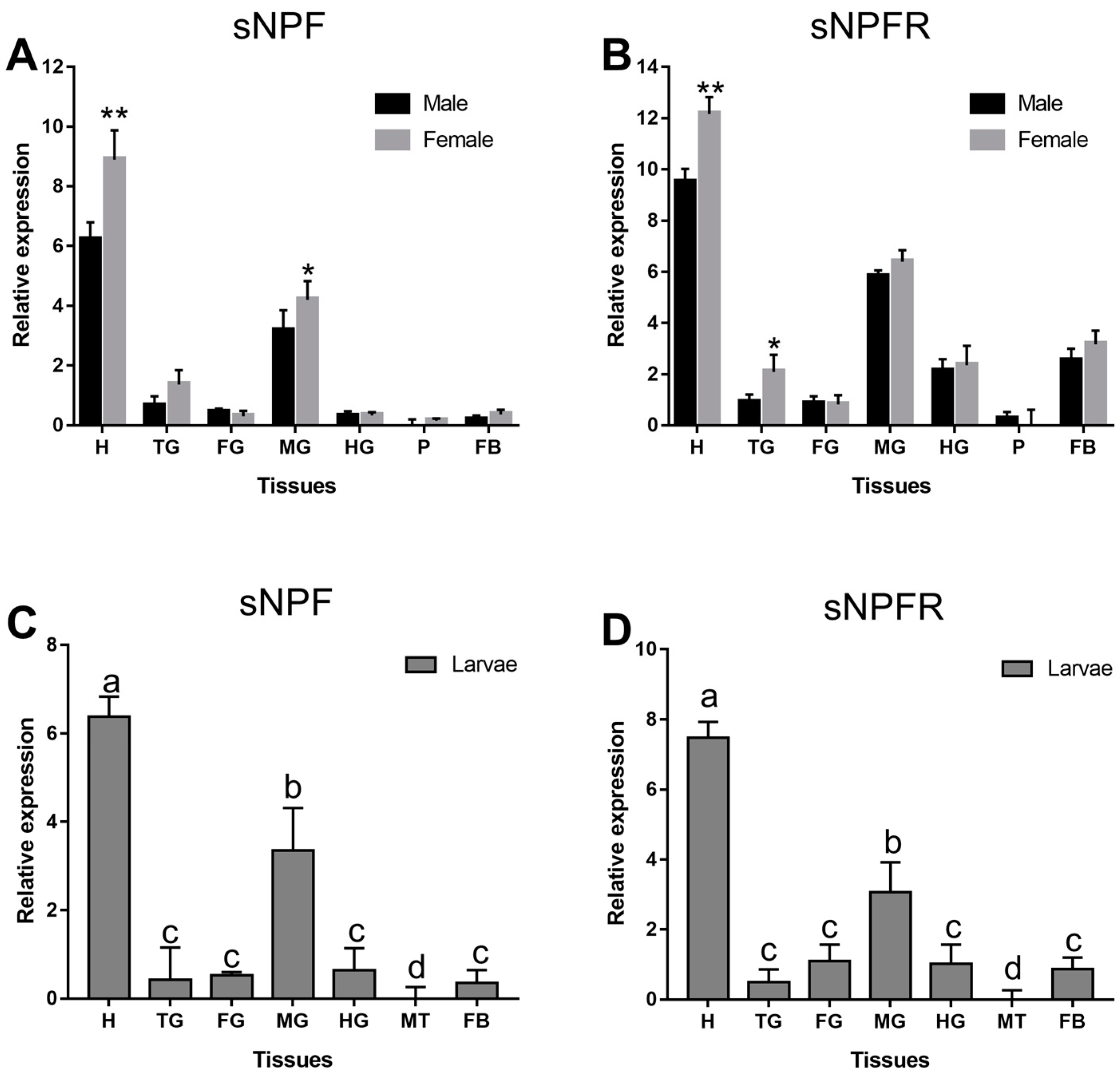
3.2.1. Different Developmental Stages and Body Parts
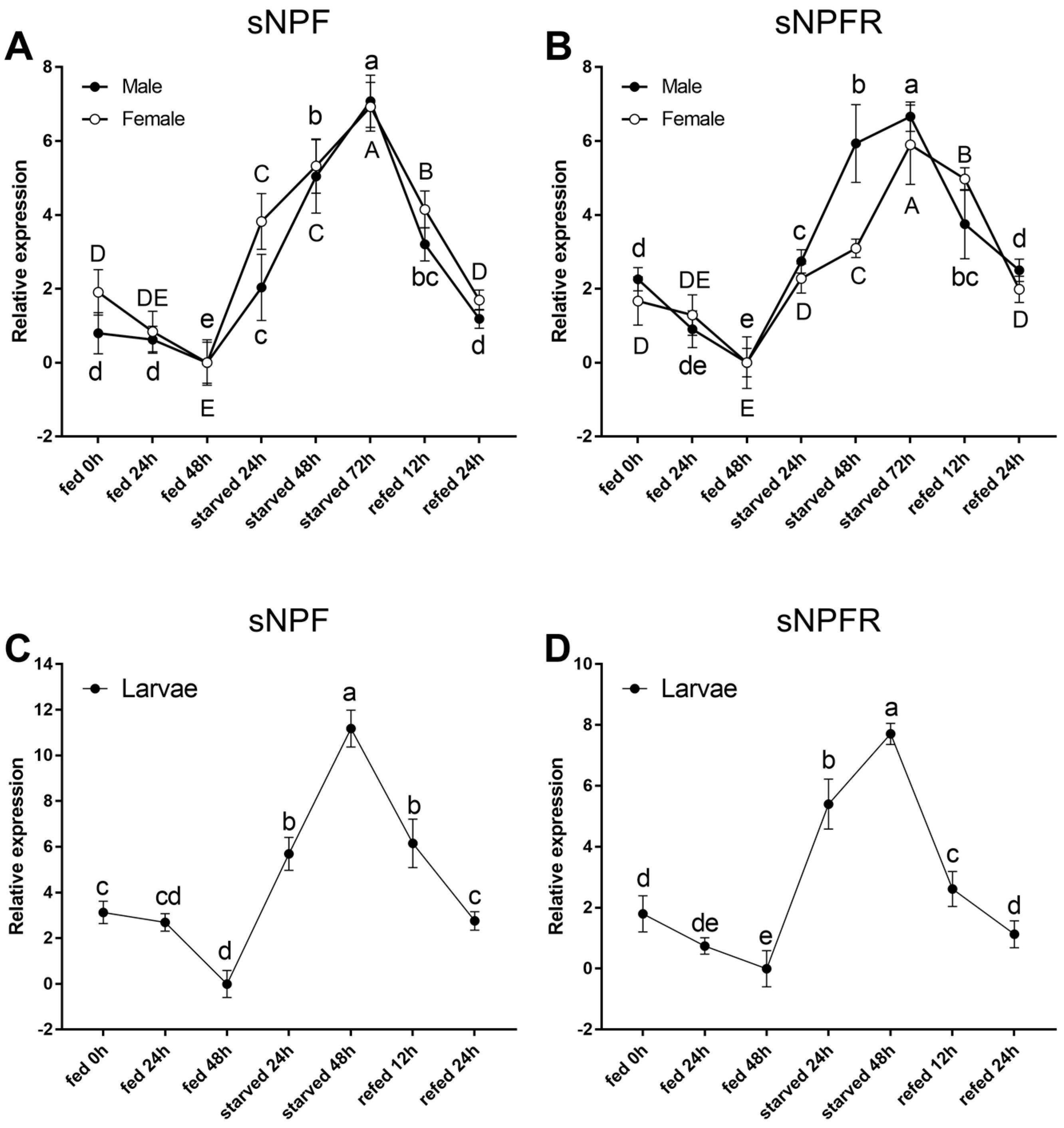
3.2.2. Starvation and Re-Feeding Assays
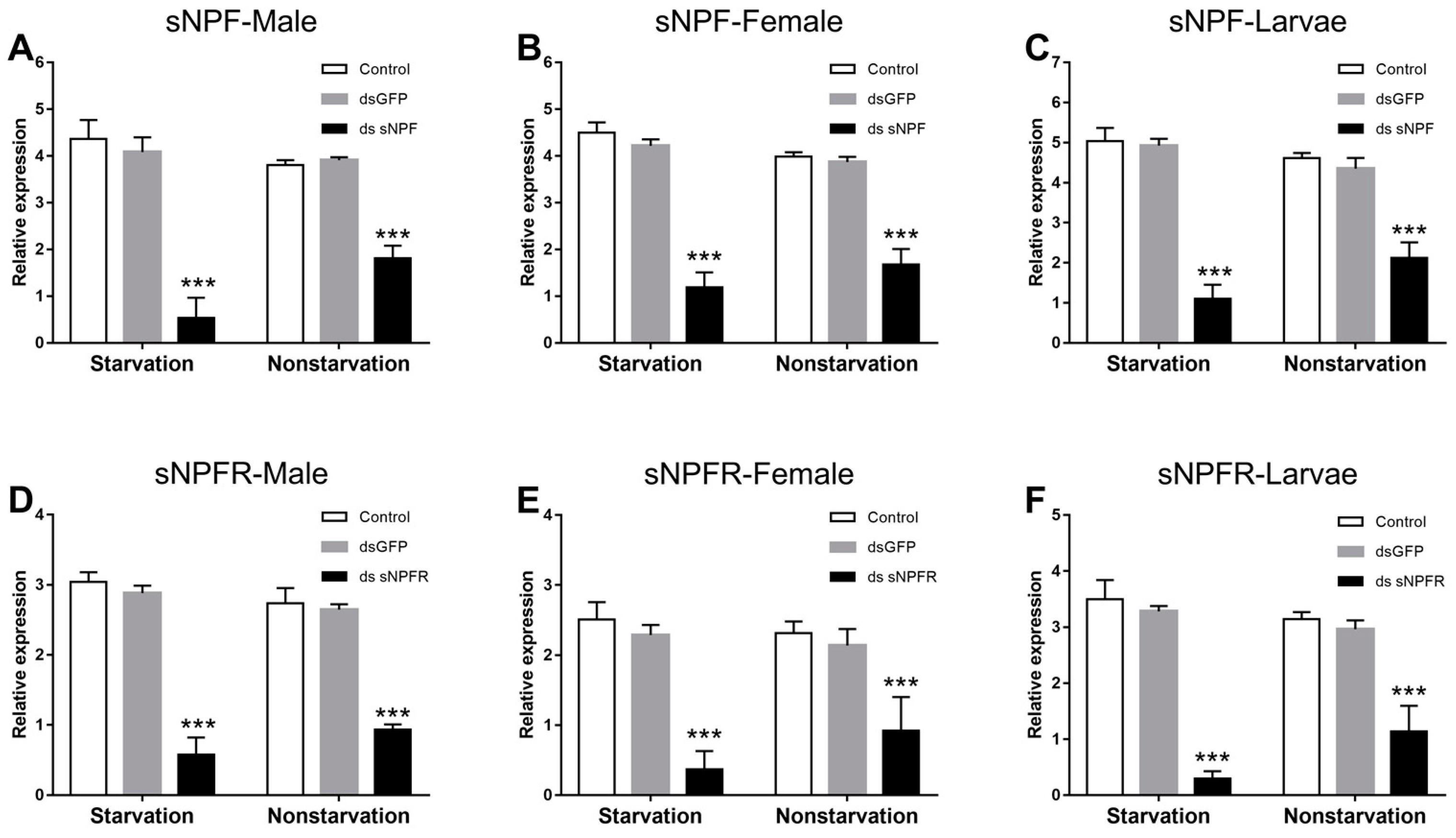
3.3. Efficiency Analysis of RNAi on sNPF and sNPFR
3.3.1. Effect of dsRNA Treatment on sNPF and sNPFR Transcript Level
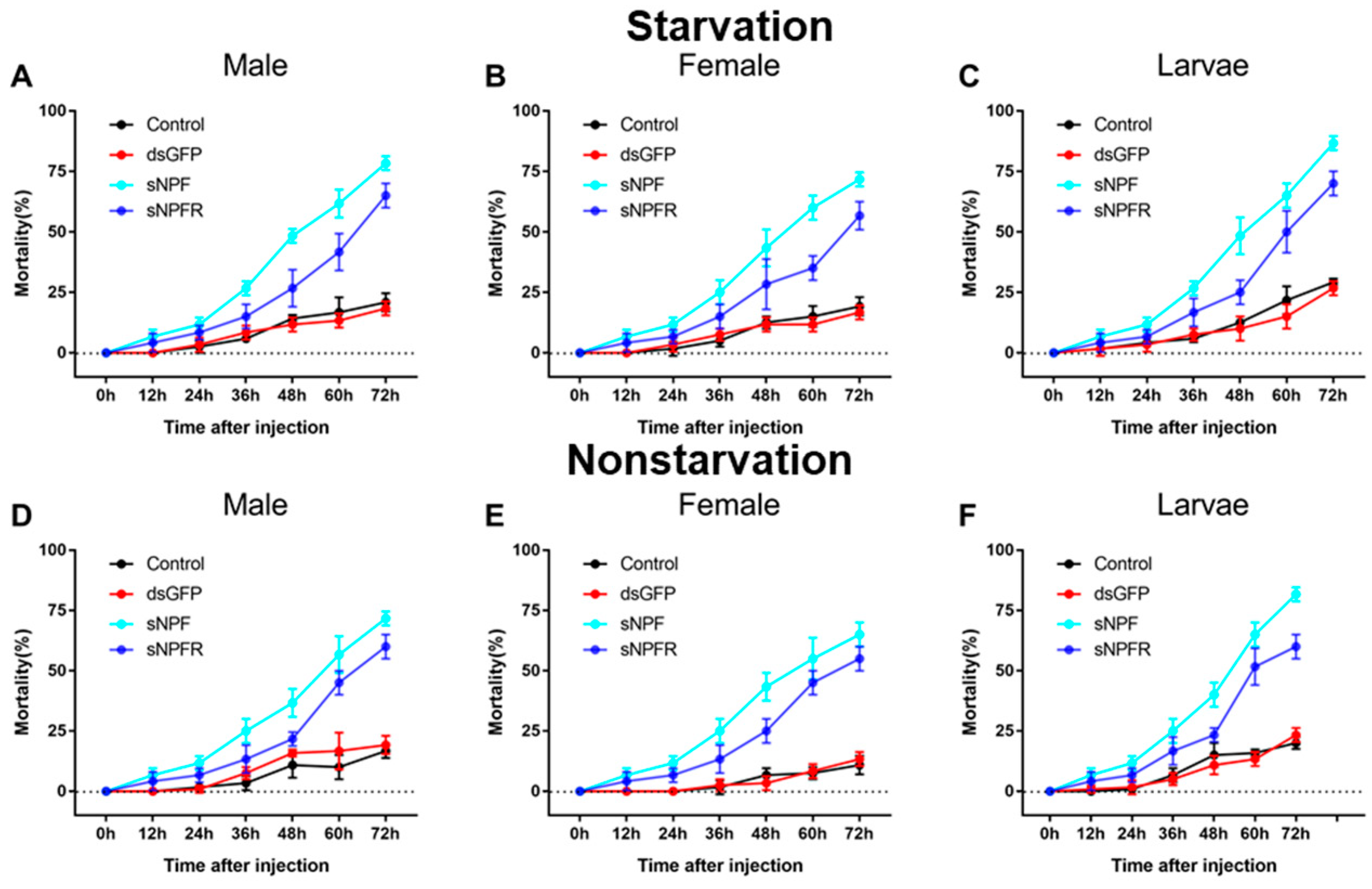
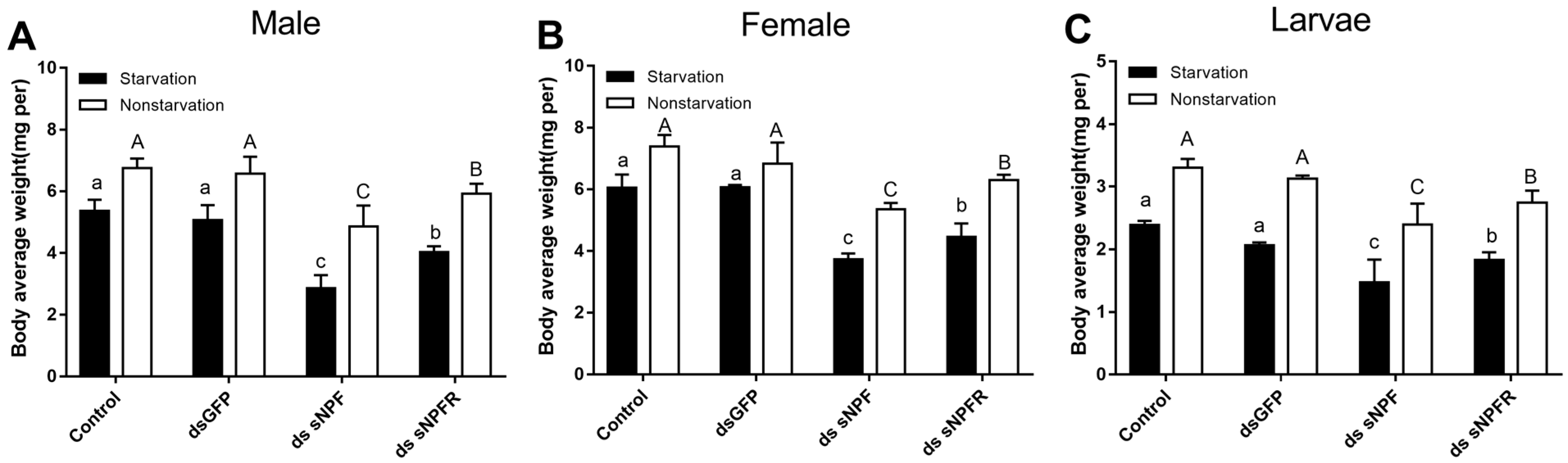
3.3.2. Effect of dsRNA Treatment on Mortality and Body Weight
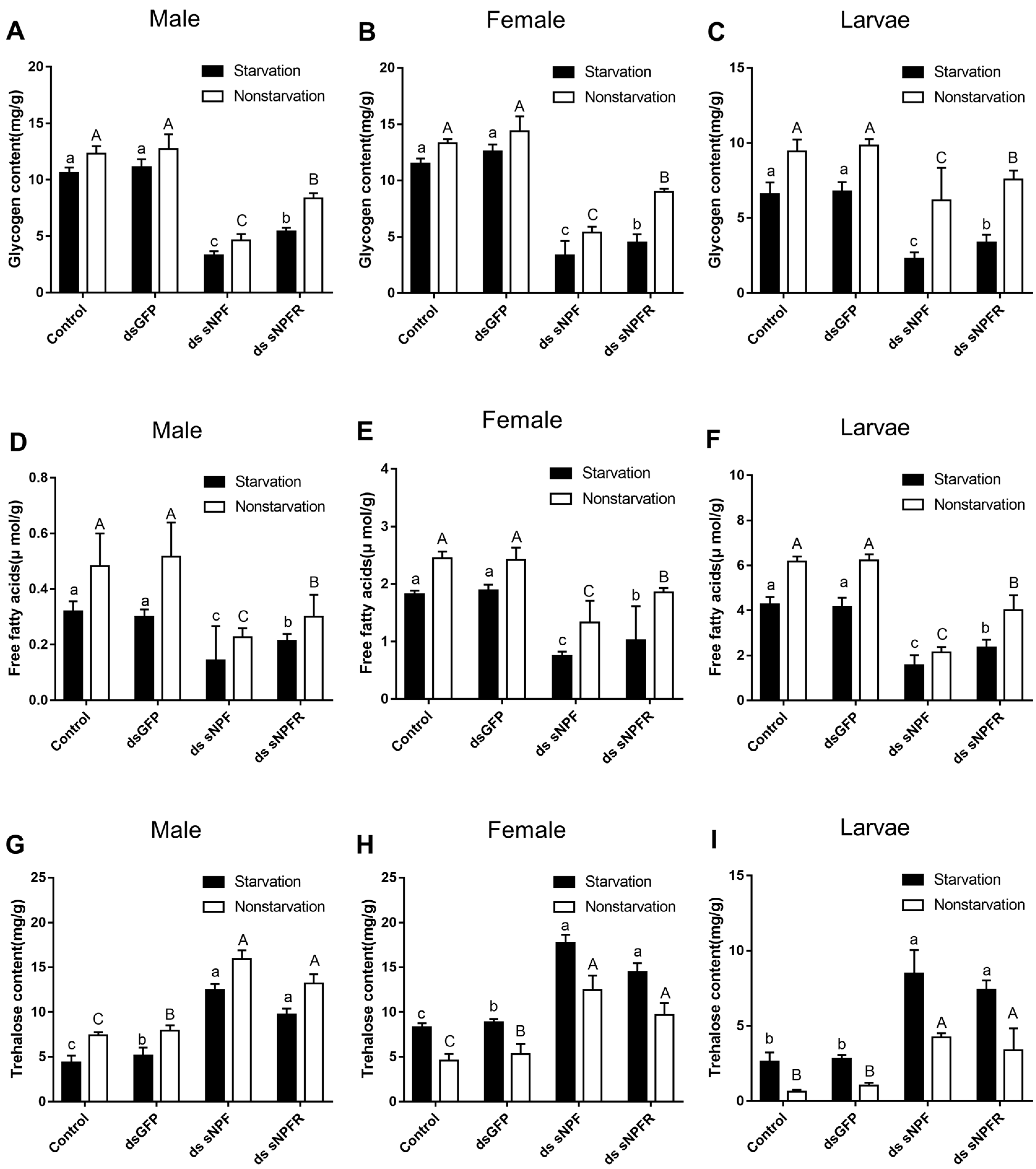
3.3.3. Effects of dsNPF on Regulating Energy Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nässel, D.R.; Wegener, C.A. Comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides 2011, 32, 1335–1355. [Google Scholar] [CrossRef]
- Nässel, D.R.; Enell, L.E.; Santos, J.G.; Wegener, C.; Johard, H.A. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008, 9, 1. [Google Scholar] [CrossRef]
- Johard, H.A.; Yoishii, T.; Dircksen, H.; Cusumano, P.; Rouyer, F.; Helfrich-Forster, C.; Nassel, D.R. Peptidergic clock neurons in Drosophila: Ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 2009, 516, 59–73. [Google Scholar] [CrossRef]
- Chen, W.; Shi, W.; Li, L.; Zheng, Z.; Li, T.; Bai, W.; Zhao, Z. Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2013, 43, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Donelson, N.C.; Vecsey, C.G.; Guo, F.; Rosbash, M.; Griffith, L.C. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 2013, 80, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahsai, L.; Kapan, N.; Dircksen, H.; Winther, A.M.; Nassel, D.R. Metabolic stress responses in Drosophila are modulated by brain neurosecretory cells that produce multiple neuropeptides. PLoS ONE 2010, 5, e11480. [Google Scholar] [CrossRef] [PubMed]
- Kapan, N.; Lushchak, O.V.; Luo, J.; Nassel, D.R. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol. Life Sci. 2012, 69, 4051–4066. [Google Scholar] [CrossRef]
- Johard, H.A.; Enell, L.E.; Gustafsson, E.; Trifilieff, P.; Veenstra, J.A.; Nassel, D.R. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: Relations to extrinsic neurons expressing different neurotransmitters. J. Comp. Neurol. 2008, 507, 1479–1496. [Google Scholar] [CrossRef] [PubMed]
- Kahsai, L.; Martin, J.R.; Winther, A.M. Neuropeptides in the Drosophila central complex in modulation of locomotor behavior. J. Exp. Biol. 2010, 213, 2256–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.B.; Gui, S.H.; Xu, L.; Pei, Y.X.; Smagghe, G.; Wang, J.J. The short neuropeptide F modulates olfactory sensitivity of Bactrocera dorsalis upon starvation. J. Insect Physiol. 2017, 99, 78–85. [Google Scholar] [CrossRef]
- Root, C.M.; Ko, K.I.; Jafari, A.; Wang, J.W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 2011, 145, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; You, K.H.; Choo, J.K.; Han, Y.M.; Yu, K. Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 2004, 279, 50781–50789. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Chen, C.; Huang, Y.; Wang, S.; Su, S.; Chen, M. Expression patterns and functional analysis of the short neuropeptide F and NPF receptor genes in Rhopalosiphum padi. Insect Sci. 2021, 28, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Ament, S.A.; Velarde, R.A.; Kolodkin, M.H.; Moyse, D.; Robinson, G.E. Neuropeptide Y-like signalling and nutritionally mediated gene expression and behaviour in the honey bee. Insect Mol. Biol. 2011, 20, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Nagata, S.; Morooka, N.; Matsumoto, S.; Kawai, T.; Nagasawa, H. Effects of neuropeptides on feeding initiation in larvae of the silkworm, Bombyx mori. Gen. Comp. Endocrinol. 2011, 172, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.; Klowden, M.J.; Crim, J.W.; Young, L.; Shrouder, L.A.; Lea, A.O. Endogenous regulation of mosquito host-seeking behavior by a neuropeptide. J. Insect Physiol. 1994, 40, 399–406. [Google Scholar] [CrossRef]
- Chen, M.E.; Pietrantonio, P.V. The short neuropeptide F-like receptor from the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Arch. Insect Biochem. Physiol. 2006, 61, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Matsumoto, S.; Nakane, T.; Ohara, A.; Morooka, N.; Konuma, T. Effects of starvation on brain short neuropeptide F-1, -2, and -3 levels and short neuropeptide F receptor expression levels of the silkworm, Bombyx mori. Front. Endocrinol. 2012, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Mikani, A.; Wang, Q.S.; Takeda, M. Brain-midgut short neuropeptide F mechanism that inhibits digestive activity of the American cockroach, Periplaneta americana upon starvation. Peptides 2012, 34, 135–144. [Google Scholar] [CrossRef]
- Dillen, S.; Verdonck, R.; Zels, S.; Van Wielendaele, P.; Vanden Broeck, J. Identification of the short neuropeptide F precursor in the desert locust: Evidence for an inhibitory role of sNPF in the control of feeding. Peptides 2014, 53, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tang, M. Spatial and temporal dynamics of bark beetles in Chinese white pine in Qinling Mountains of Shaanxi Province, China. Environ. Entomol. 2007, 36, 1124–1130. [Google Scholar] [CrossRef]
- Spittaels, K.; Verhaert, P.; Shaw, C.; Johnston, R.N.; Devreese, B.; Van Beeumen, J.; De Loof, A. Insect neuropeptide F (NPF)-related peptides: Isolation from Colorado potato beetle (Leptinotarsa decemlineata) brain. Insect Biochem. Mol. Biol. 1996, 26, 375–382. [Google Scholar] [CrossRef]
- Huybrechts, J.; De Loof, A.; Schoofs, L. Diapausing Colorado potato beetles are devoid of short neuropeptide F I and II. Biochem. Biophys. Res. Commun. 2004, 317, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, P.; Szymczak, M.; Rogalska, L.; Rosinski, G. Developmental and myotropic efects of the Led-NPF-I peptide in tenebrionid beetles. Invertebr. Reprod. Dev. 2013, 57, 309–315. [Google Scholar] [CrossRef]
- Marciniak, P.; Urbanski, A.; Kudlewska, M.; Szymczak, M.; Rosinski, G. Peptide hormones regulate the physiological functions of reproductive organs in Tenebrio molitor males. Peptides 2017, 98, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, P.; Urbanski, A.; Lubawy, J.; Szymczak, M.; Pacholska-Bogalska, J.; Chowanski, S.; Kuczer, M.; Rosinski, G. Short neuropeptide F signaling regulates functioning of male reproductive system in Tenebrio Molitor beetle. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2020, 190, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fu, D.; Gao, H.; Ning, H.; Sun, Y.; Chen, H.; Tang, M. Cloning and expression of the neuropeptide F and neuropeptide F receptor genes and their regulation of food intake in the Chinese white pine beetle Dendroctonus armandi. Front. Physiol. 2021, 12, 662651. [Google Scholar] [CrossRef]
- Dai, L.; Wang, C.; Zhang, X.; Yu, J.; Zhang, R.; Chen, H. Two CYP4 genes of the Chinese white pine beetle, Dendroctonus armandi (Curculionidae: Scolytinae), and their transcript levels under different development stages and treatments. Insect Mol. Biol. 2014, 23, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Dai, L.; Fu, D.; Gao, J.; Chen, H. Electrophysiological and behavioral responses of Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae) to two candidate pheromone components: Frontalin and exo-brevicomin. Chemoecology 2017, 27, 91–99. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.S.G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Ma, M.; Wang, C.; Shi, Q.; Zhang, R.; Chen, H. Cytochrome P450s from the Chinese white pine beetle, Dendroctonus armandi (Curculionidae: Scolytinae): Expression profiles of different stages and responses to host allelochemicals. Insect Biochem. Mol. Biol. 2015, 65, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]
- Fu, D.; Dai, L.; Gao, H.; Sun, Y.; Liu, B.; Chen, H. Identification, expression patterns and RNA interference of aquaporins in Dendroctonus armandi (Coleoptera: Scolytinae) larvae during overwintering. Front. Physiol. 2019, 10, 967. [Google Scholar] [CrossRef] [Green Version]
- Costanzi, S. Homology modeling of class A G protein-coupled receptors. Methods Mol. Biol. 2012, 857, 259–279. [Google Scholar]
- Urbanski, A.; Lubawy, J.; Marciniak, P.; Rosinski, G. Myotropic activity and immunolocalization of selected neuropeptides of the burying beetle Nicrophorus vespilloides (Coleoptera: Silphidae). Insect Sci. 2019, 26, 656–670. [Google Scholar] [CrossRef]
- Pandit, A.A.; Ragionieri, L.; Marley, R.; Yeoh, J.; Inward, D.; Davies, S.A.; Predel, R.; Dow, J.A.T. Coordinated RNA-Seq and peptidomics identify neuropeptides and G-protein coupled receptors (GPCRs) in the large pine weevil Hylobius abietis, a major forestry pest. Insect Biochem. Mol. Biol. 2018, 101, 94–107. [Google Scholar] [CrossRef]
- Reiher, W.; Shirras, C.; Kahnt, J.; Baumeister, S.; Isaac, R.E.; Wegener, C. Peptidomics and peptide hormone processing in the Drosophila midgut. J. Proteo. Res. 2011, 10, 1881–1892. [Google Scholar] [CrossRef]
- Lu, H.L.; Pietrantonio, P.V. Immunolocalization of the short neuropeptide F receptor in queen brains and ovaries of the red imported fire ant (Solenopsis invicta Buren). BMC Neurosci. 2012, 12, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mertens, I.; Meeusen, T.; Huybrechts, R.; De Loof, A.; Schoofs, L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2002, 297, 1140–1148. [Google Scholar] [CrossRef]
- Dillen, S.; Zels, S.; Verlinden, H.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Functional characterization of the short neuropeptide F receptor in the desert locust, Schistocerca gregaria. PLoS ONE 2013, 8, e53604. [Google Scholar] [CrossRef] [Green Version]
- Bigot, L.; Beets, I.; Dubos, M.P.; Boudry, P.; Schoofs, L.; Favrel, P. Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J. Exp. Biol. 2014, 217, 2974–2982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillen, S.; Chen, Z.; Vanden Broeck, J. Nutrient-dependent control of short neuropeptide F transcript levels via components of the insulin/IGF signaling pathway in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2016, 68, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.H.; Jin, L.; Fu, K.Y.; Guo, W.C.; Li, G.Q. Crustacean cardioactive peptide as a stimulator of feeding and a regulator of ecdysis in Leptinotarsa decemlineata. Pest. Biochem. Physiol. 2021, 175, 104838. [Google Scholar] [CrossRef]

| Gene Name | Accession No. | ORF Size (aa/bp) a | MW (KDa) a | IP a | Signal Peptide Prediction b |
|---|---|---|---|---|---|
| sNPF | MW174224 | 102/309 | 11.48 | 8.06 | SP 0.957 mTP 0.027 other 0.066 |
| sNPFR | MW174225 | 428/1287 | 49.27 | 8.20 | SP 0.039 mTP 0.680 other 0.703 |
| Gene | BLAST Matches in GenBank | |||
|---|---|---|---|---|
| Name | Species | Name | Accession Number | % Identity a |
| sNPF | Dendroctonus ponderosae | short neuropeptide F-like | XP_019767920.1 | 96 |
| Sitophilus oryzae | short neuropeptide F-like | XP_030755875.1 | 67 | |
| Rhynchophorus ferrugineus | short neuropeptide F-like | QGA72576.1 | 60 | |
| sNPFR | Dendroctonus ponderosae | prolactin-releasing peptide receptor-like | XP_019761868.1 | 97 |
| Rhynchophorus ferrugineus | prolactin-releasing peptide receptor-like | QGA72517.1 | 82 | |
| Anopheles gambiae | short neuropeptide F receptor | ABD96049.1 | 66 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Fu, D.; Ning, H.; Tang, M.; Chen, H. Identification of the Short Neuropeptide F and Short Neuropeptide F Receptor Genes and Their Roles of Food Intake in Dendroctonus armandi. Insects 2021, 12, 844. https://doi.org/10.3390/insects12090844
Liu B, Fu D, Ning H, Tang M, Chen H. Identification of the Short Neuropeptide F and Short Neuropeptide F Receptor Genes and Their Roles of Food Intake in Dendroctonus armandi. Insects. 2021; 12(9):844. https://doi.org/10.3390/insects12090844
Chicago/Turabian StyleLiu, Bin, Danyang Fu, Hang Ning, Ming Tang, and Hui Chen. 2021. "Identification of the Short Neuropeptide F and Short Neuropeptide F Receptor Genes and Their Roles of Food Intake in Dendroctonus armandi" Insects 12, no. 9: 844. https://doi.org/10.3390/insects12090844
APA StyleLiu, B., Fu, D., Ning, H., Tang, M., & Chen, H. (2021). Identification of the Short Neuropeptide F and Short Neuropeptide F Receptor Genes and Their Roles of Food Intake in Dendroctonus armandi. Insects, 12(9), 844. https://doi.org/10.3390/insects12090844






