Lipidomic Profiling Reveals Distinct Differences in Sphingolipids Metabolic Pathway between Healthy Apis cerana cerana larvae and Chinese Sacbrood Disease
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
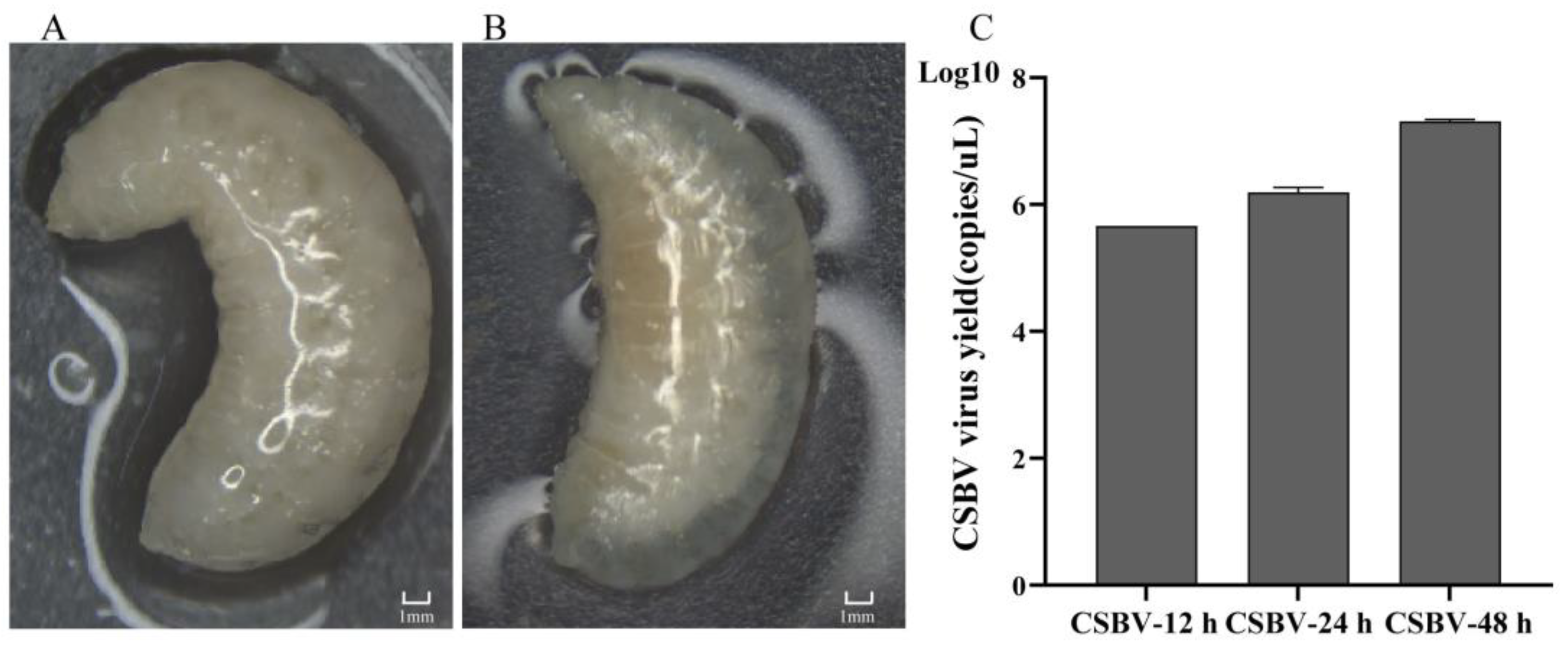
2.1. Sample Collection
2.2. Lipidomics
2.3. Data Processing, Lipid Identification and Quantification
2.4. Statistical Analysis
2.5. Quantitative Real-Time PCR
3. Results and Discussion
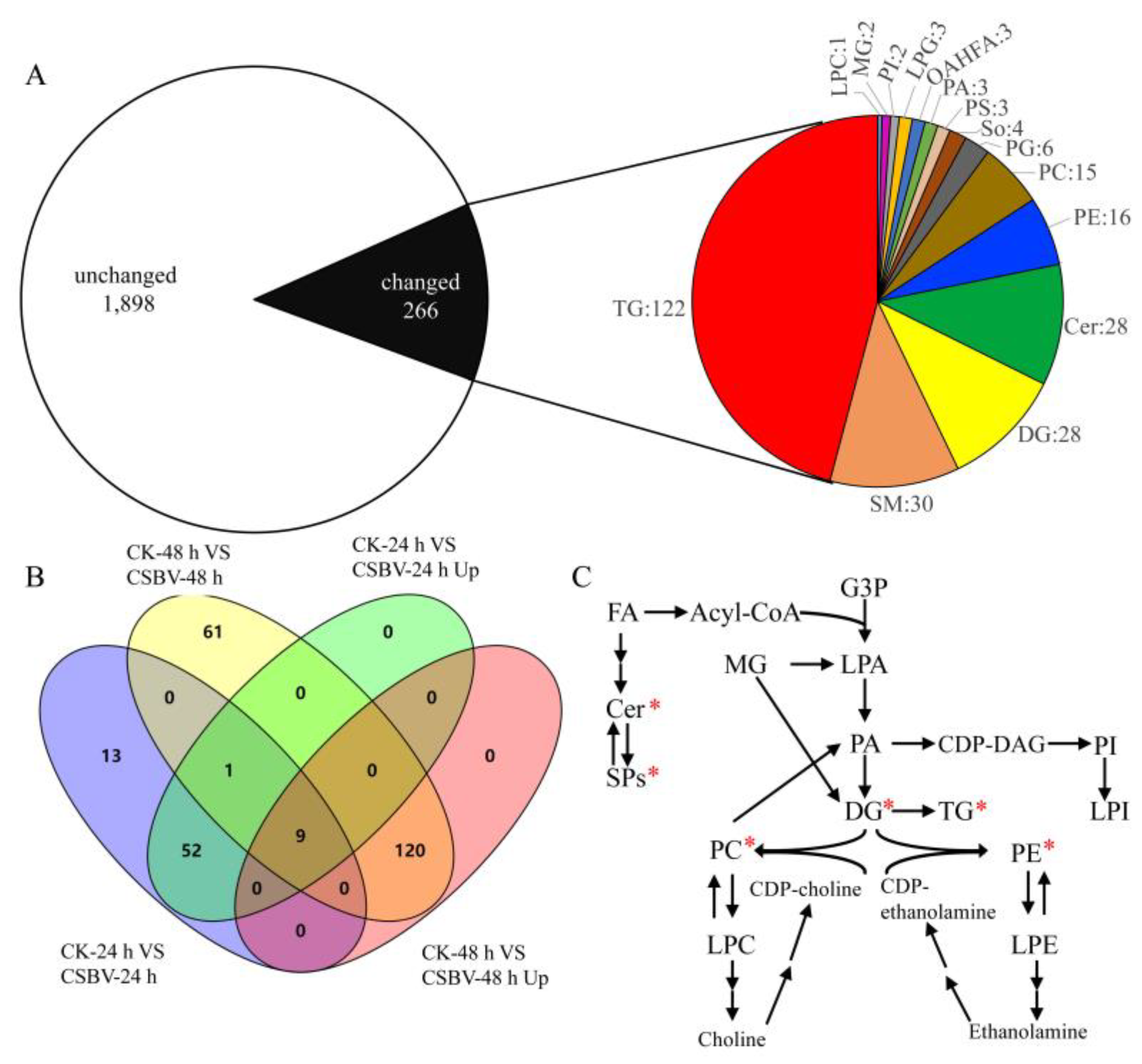
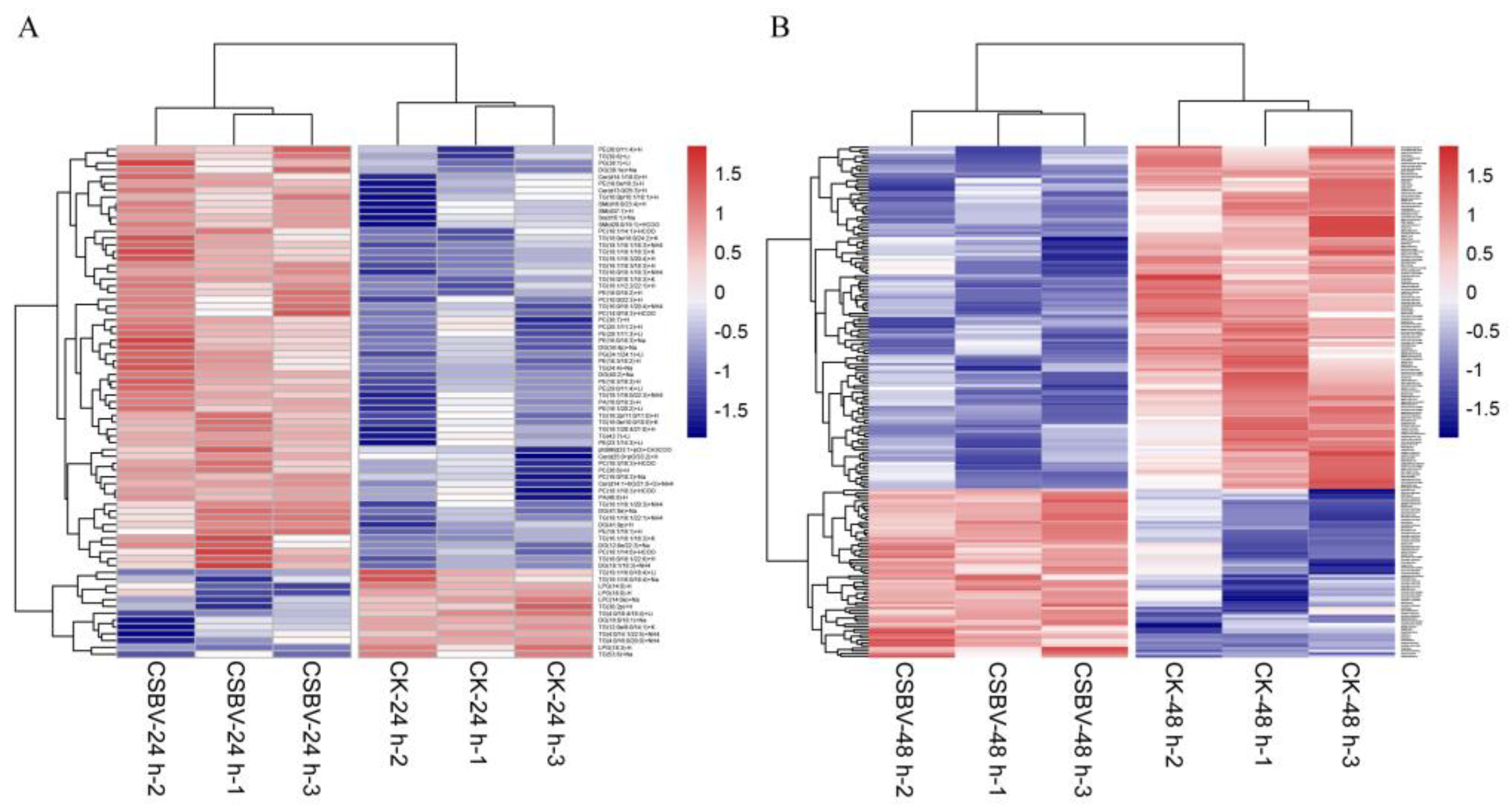
3.1. General Description of Lipidome Data
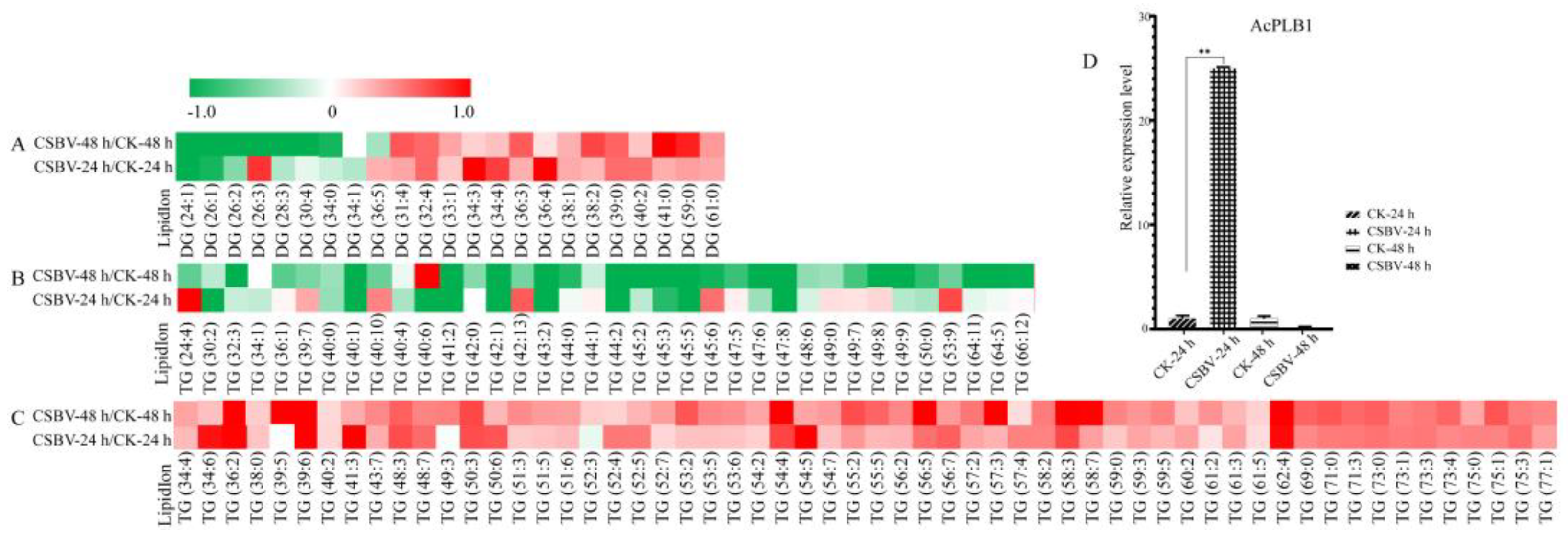
3.2. Glycerolipids Showed Maximal Remodeling after CSBV Infection
3.3. Alterations in Phospholipids Homeostasis during CSBV Infection
3.4. CSBV Infection Causes an Accumulation of Sphingolipids
3.5. Biomarkers Screening of the CSBV-Infected Larvae
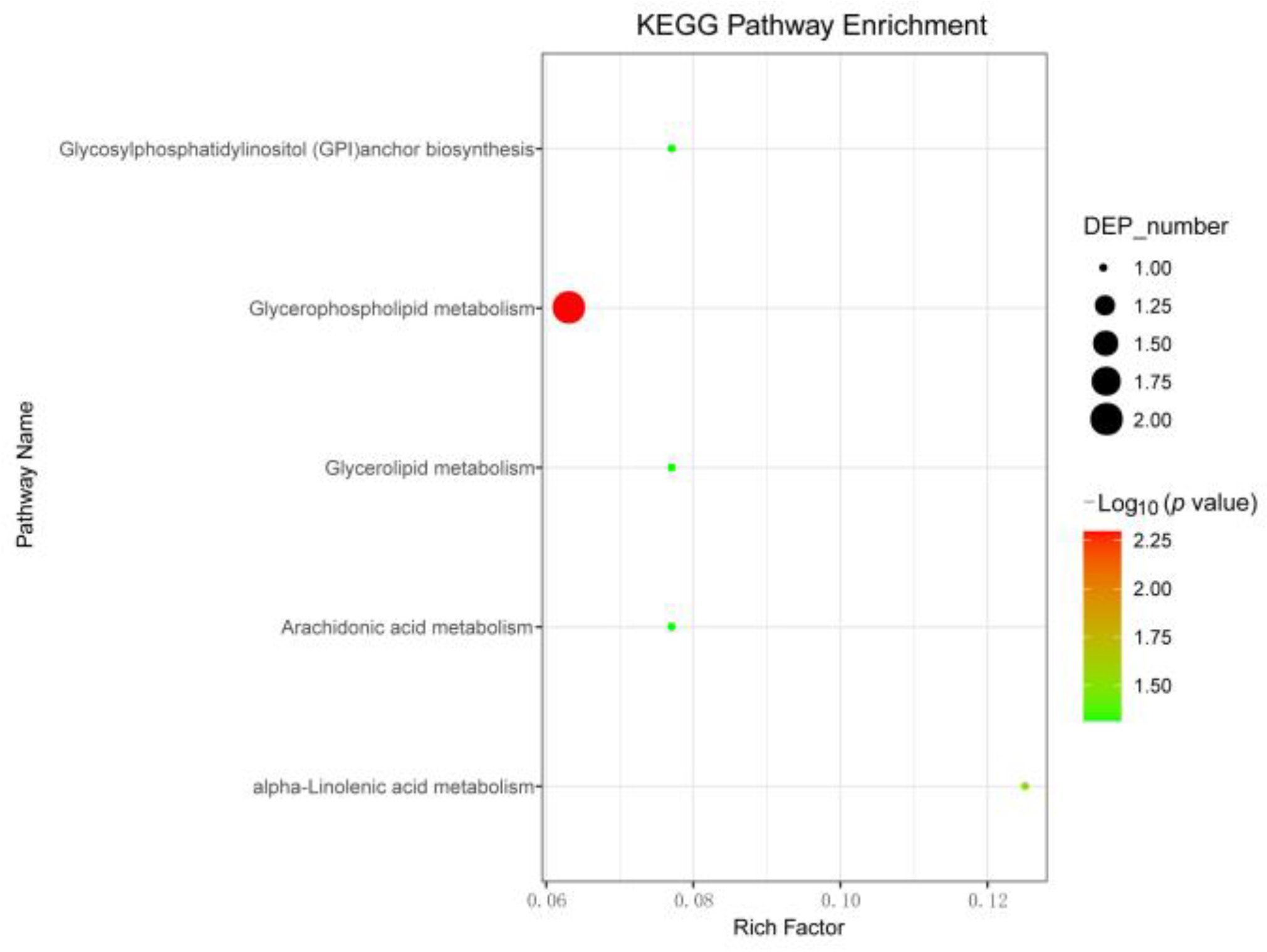
3.6. Pathway Analysis of CSBV-Infected A. c. cerana larvae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, P.; Wu, J.; Li, H. Economic value assessment of agricultural bee pollination in China. Sci. Agric. Sin. 2012, 44, 5117–5123. [Google Scholar]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef] [PubMed]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Max, B. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [Green Version]
- Bailey, L. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 1969, 63, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Gibbs, A.J.; Woods, R.D. Sacbrood Virus of the Larval Honey Bee (Apis Mellifera Linnaeus). Virology 1964, 23, 425–429. [Google Scholar] [CrossRef]
- Zhang, G.Z.; Han, R.C. Advances on Sacbrood of Honeybees. Chin. J. Biol. Control. 2008, 24, 130–137. (In Chinese) [Google Scholar]
- Ma, M.X.; Ma, C.; Li, M.; Wang, S.Q.; Yang, S.; Wang, S.D. Loop-mediated isothermal amplification for rapid detection of Chinese sacbrood virus. J. Virol. Methods 2011, 176, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Qin, H.R.; Wu, J.; Sadd, B.M.; Wang, X.H.; Evans, J.D.; Peng, W.J.; Chen, Y.P. The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS ONE 2012, 7, e47955. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Han, R.C. Diagnostic technologies of common pathogens of honeybees in China. Chin. Bull. Entomol. 2008, 45, 483–488. [Google Scholar]
- Ghosh, R.C.; Ball, B.V.; Willcocks, M.M. The nucleotide sequence of sacbrood virus of the honey bee: An insect picorna-like virus. J. Gen. Virol. 1999, 80, 1541–1549. [Google Scholar] [CrossRef]
- Skubnik, K.; Novacek, J.; Fuzik, T.; Pridal, A.; Paxton, R.J.; Plevka, P. Structure of deformed wing virus, a major honey bee pathogen. Proc. Natl. Acad. Sci. USA 2017, 114, 3210–3215. [Google Scholar] [CrossRef] [Green Version]
- De Miranda, J.R.; Dainat, B.; Locke, B.; Cordoni, G.; Berthoud, H.; Gauthier, L.; Neumann, P.; Budge, G.E.; Ball, B.V.; Stoltz, D.B. Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L.). J. Gen. Virol. 2010, 91, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Feng, J.; Liang, Y.; Chen, D.; Zhou, Z.H.; Zhang, Q.; Lu, X. Three-dimensional structure of the Chinese Sacbrood bee virus. Sci. China C Life Sci. 2001, 44, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, M.; Fuzik, T.; Skubnik, K.; Moravcova, J.; Ubiparip, Z.; Pridal, A.; Plevka, P. Virion structure and genome delivery mechanism of sacbrood honeybee virus. Proc. Natl. Acad. Sci. USA 2018, 115, 7759–7764. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; He, G.; Filipowicz, N.A.; Randall, G.; Belov, G.A.; Kopek, B.G.; Wang, X. Host Lipids in Positive-Strand RNA Virus Genome Replication. Front. Microbiol. 2019, 10, 286. [Google Scholar] [CrossRef] [PubMed]
- Villareal, V.A.; Rodgers, M.A.; Costello, D.A.; Yang, P.L. Targeting host lipid synthesis and metabolism to inhibit dengue and hepatitis C viruses. Antiviral. Res. 2015, 124, 110–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, S.; Krajewski, M.; Scherer, C.; Scholz, V.; Mordhorst, V.; Truschow, P.; Schobel, A.; Reimer, R.; Schwudke, D.; Herker, E. Complex lipid metabolic remodeling is required for efficient hepatitis C virus replication. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Guedan, A.; Mousnier, A.; Swieboda, D.; Zhang, Q.; Horkai, D.; Le Novere, N.; Solari, R.; Wakelam, M.J.O. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J. Lipid Res. 2018, 59, 1671–1684. [Google Scholar] [CrossRef] [Green Version]
- Queiroz, A.; Pinto, I.F.D.; Lima, M.; Giovanetti, M.; de Jesus, J.G.; Xavier, J.; Barreto, F.K.; Canuto, G.A.B.; do Amaral, H.R.; de Filippis, A.M.B.; et al. Lipidomic analysis reveals serum alteration of plasmalogens in patients infected with ZIKA Virus. Front. Microbiol. 2019, 10, 753. [Google Scholar] [CrossRef] [Green Version]
- Chotiwan, N.; Andre, B.G.; Sanchez-Vargas, I.; Islam, M.N.; Grabowski, J.M.; Hopf-Jannasch, A.; Gough, E.; Nakayasu, E.; Blair, C.D.; Belisle, J.T.; et al. Dynamic remodeling of lipids coincides with dengue virus replication in the midgut of Aedes aegypti mosquitoes. PLoS Pathog. 2018, 14, e1006853. [Google Scholar] [CrossRef] [Green Version]
- Perera, R.; Riley, C.; Isaac, G.; Hopf-Jannasch, A.S.; Moore, R.J.; Weitz, K.W.; Pasa-Tolic, L.; Metz, T.O.; Adamec, J.; Kuhn, R.J. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012, 8, e1002584. [Google Scholar] [CrossRef]
- Feng, J.X.; Zhang, Q.Z.; Ma, Z.W.; Zhang, J.Q.; Huang, W.Z.; Zhang, X.F. Purification, crystallization and structure of the Chinese Sacbrood virus. J. Chin. Electron. Microsc. Soc. 1998, 17, 387–388. [Google Scholar]
- Schmeh, D.R.; Tomé, H.V.V.; Mortensen, A.N.; Martins, G.F.; Ellis, J.D. Protocol for the in vitro rearing of honey bee (Apis mellifera L.) workers. J. Apicult. Res. 2016, 55, 113–129. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.H.; Chen, Y.C.; Kao, T.Y.; Lin, Y.C.; Hsu, T.E.; Wu, Y.C.; Ja, W.W.; Brummel, T.J.; Kapahi, P.; Yuh, C.H.; et al. Diacylglycerol lipase regulates lifespan and oxidative stress response by inversely modulating TOR signaling in Drosophila and C. elegans. Aging Cell 2014, 13, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Merida, I.; Avila-Flores, A.; Merino, E. Diacylglycerol kinases: At the hub of cell signalling. Biochem. J. 2008, 409, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Pennington, J.E.; Nussenzveig, R.H.; Van Heusden, M.C. Lipid transfer from insect fat body to lipophorin: Comparison between a mosquito triacylglycerol-rich lipophorin and a sphinx moth diacylglycerol-rich lipophorin. J. Lipid Res. 1996, 37, 1144–1152. [Google Scholar] [CrossRef]
- Cheon, H.M.; Shin, S.W.; Bian, G.; Park, J.H.; Raikhel, A.S. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J. Biol. Chem. 2006, 281, 8426–8435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, W.; Gill, M.; Esposito, A.; Kaminski, C.F.; Courousse, N.; Chwetzoff, S.; Trugnan, G.; Keshavan, N.; Lever, A.; Desselberger, U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010, 84, 6782–6798. [Google Scholar] [CrossRef] [Green Version]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Gondim, K.C.; Atella, G.C.; Pontes, E.G.; Majerowicz, D. Lipid metabolism in insect disease vectors. Insect. Biochem. Mol. Biol. 2018, 101, 108–123. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Chukkapalli, V.; Nchoutmboube, J.A.; Li, J.; Randall, G.; Belov, G.; Wang, X. Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc. Natl. Acad. Sci. USA 2016, 113, E1064–E1073. [Google Scholar]
- Castorena, K.M.; Stapleford, K.A.; Miller, D.J. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genom. 2010, 11, 183. [Google Scholar]
- Nchoutmboube, J.A.; Viktorova, E.G.; Scott, A.J.; Ford, L.A.; Pei, Z.; Watkins, P.A.; Ernst, R.K.; Belov, G.A. Increased long chain acyl-Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 2013, 9, e1003401. [Google Scholar]
- Xu, K.; Nagy, P.D. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc. Natl. Acad. Sci. USA 2015, 112, E1782–E1791. [Google Scholar]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [PubMed]
- Schneider-Schaulies, J.; Schneider-Schaulies, S. Viral infections and sphingolipids. Handb. Exp. Pharmacol. 2013, 216, 321–340. [Google Scholar]
- Heung, L.J.; Luberto, C.; Del Poeta, M. Role of sphingolipids in microbial pathogenesis. Infect. Immun. 2006, 74, 28–39. [Google Scholar]
- Hirata, Y.; Ikeda, K.; Sudoh, M.; Tokunaga, Y.; Suzuki, A.; Weng, L.; Ohta, M.; Tobita, Y.; Okano, K.; Ozeki, K.; et al. Self-Enhancement of Hepatitis C Virus Replication by Promotion of Specific Sphingolipid Biosynthesis. PLoS Pathog. 2012, 8, e1002860. [Google Scholar]
- Aktepe, T.E.; Pham, H.; Mackenzie, J.M. Differential utilisation of ceramide during replication of the flaviviruses West Nile and dengue virus. Virology 2015, 484, 241–250. [Google Scholar] [PubMed]
- Umehara, T.; Sudoh, M.; Yasui, F.; Matsuda, C.; Hayashi, Y.; Chayama, K.; Kohara, M. Serine palmitoyltransferase inhibitor suppresses HCV replication in a mouse model. Biochem. Biophys. Res. Commun. 2006, 346, 67–73. [Google Scholar] [PubMed]
- Katsume, A.; Tokunaga, Y.; Hirata, Y.; Munakata, T.; Saito, M.; Hitohisa, H.; Okamoto, K.; Ohmori, Y.; Kusanagi, I.; Fujiwara, S.; et al. A serine palmitoyltransferase inhibitor blocks hepatitis C virus replication in human hepatocytes. Gastroenterology 2013, 145, 865–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider-Schaulies, J.; Schneider-Schaulies, S. Sphingolipids in viral infection. Biol. Chem. 2015, 396, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Nemoto-Sasaki, Y.; Tanikawa, T.; Oka, S.; Tsuchiya, K.; Zama, K.; Mitsutake, S.; Sugiura, T.; Yamashita, A. Sphingomyelin synthase 2, but not sphingomyelin synthase 1, is involved in HIV-1 envelope-mediated membrane fusion. J. Biol. Chem. 2014, 289, 30842–30856. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.H.; Hsu, F.F.; Crumrine, D.; Meyer, J.; Elias, P.M.; Miner, J.H. Fatty acid transport protein 4 is required for incorporation of saturated ultralong-chain fatty acids into epidermal ceramides and monoacylglycerols. Sci. Rep. 2019, 9, 13254. [Google Scholar] [CrossRef] [PubMed]


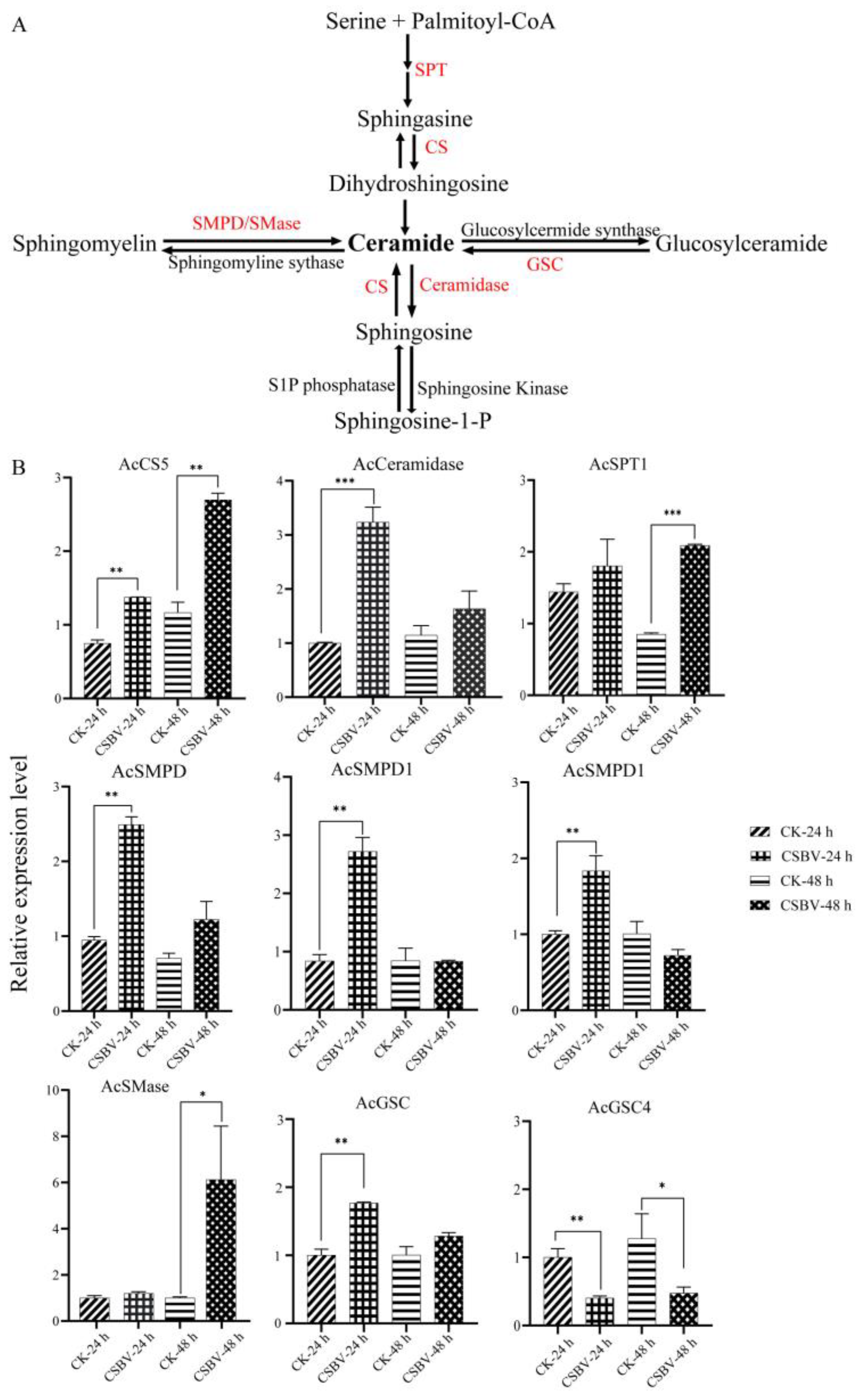
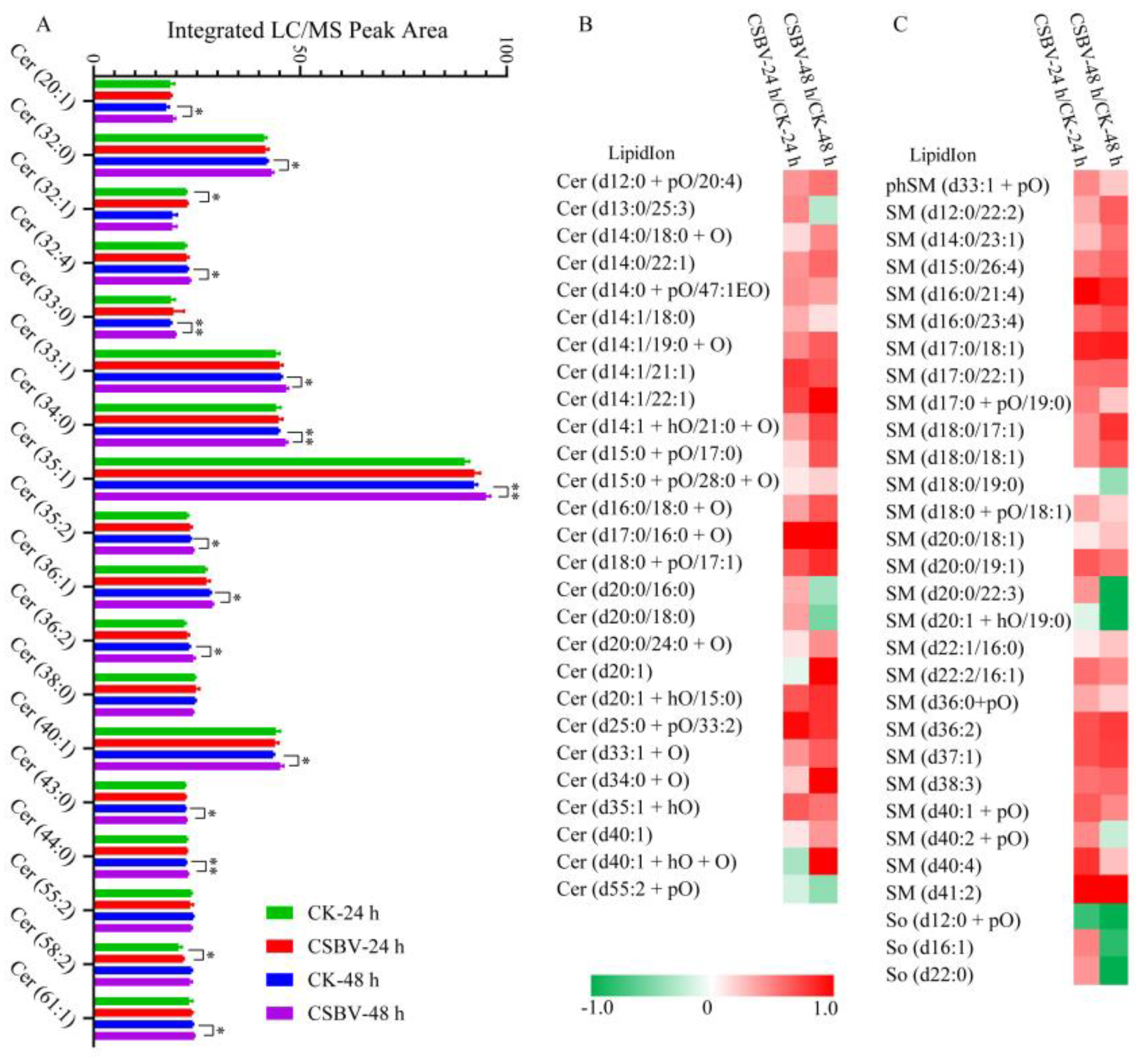
| Name | Aberration | Accession No. | Primer | Sequence (5′–3′) |
|---|---|---|---|---|
| phospholipase B1 | AcPLB1 | XM_017059237.2 | F | TTGACTGACAATGGATATTATG |
| R | GTCTTTCAGAAGTAGGACACAA | |||
| serine palmitoyltransferase 1 | AcSPT1 | XM_017049187.1 | F | AAGGATTGGATGCAACTAAAGC |
| R | CTGGTAATGGACAGATATTTCCAG | |||
| Ceramide synthase5 | AcCS5 | XM_017049042.1 | F | AATAGTGCCTATGTTCCCAGCA |
| R | ATCCTCACTGCTACTACTACGA | |||
| Ceramidase | AcCeramidase | XM_017052678.1 | F | GCACAACGGTTACTATCGTTAC |
| R | CATGTAGCTATATATTAAGGAG | |||
| Sphingomyelinase | AcSMase | XM_017050328.1 | F | GTGGAGTTATTGGATCTGGACT |
| R | TTGAAGTTTACAAAGTCCGACA | |||
| sphingomyelin phosphodiesterase | AcSMPD | XM_017055261.1 | F | CCGTTATGCTGCAGATTGACAA |
| R | GAGCATATGTTCAACGGTTCTC | |||
| sphingomyelin phosphodiesterase 1 | AcSMPD1 | XM_017060020.1 | F | GGAATGGTGGTAGCATAACAGC |
| R | TTTGCTGCTATAGAGAGCCAAT | |||
| sphingomyelin phosphodiesterase 4 | AcSMPD4 | XM_017064091.1 | F | ATGAGGACAGATCTTGTAGCCC |
| R | CGTACAATTGAAGCCCATTGAT | |||
| glucosylceramidase-like | AcGSC | XM_017065397.1 | F | ATTTAAGATTCTTTAGCGCCGC |
| R | GAATGGCTCGTTTCCAGTTGAA | |||
| glucosylceramidase 4 | AcGSC4 | XM_017065381.1 | F | TCCACCGATTTTTCTACGAGAA |
| R | GCTGCGAACAATAGTACTTCAG | |||
| Sacbrood virus CSBV-LN | qVp1 | gi|307148859| | F | TAGAGTTACGTTTTGATTTTGTTT |
| R | GCGCTAGCCGTATTTCTC |
| Significant Lipids | Theoretical Mass | Trend in CSBV vs. Control | Detection Mode | Lipid Class | Log2 (FC) aCSBV_24 h vs. CK_24 h) | Pvalue (CSBV_24 h vs. CK_25 h) | VIP (CSBV_24 h vs. CK_24 h) | Log2 (FC) a (CSBV_48 h vs. CK_48 h) | Pvalue (CSBV_48 h vs. CK_48 h) | VIP (CSBV_48 h vs. CK_48 h) |
|---|---|---|---|---|---|---|---|---|---|---|
| Cer (d14:1 + hO/21:0 + O) | 601.551 | Up | pos | Cer | 1.282 | 0.012 | 2.067 | 1.674 | 0.020 | 1.770 |
| Cer (d17:0/16:0 + O) | 542.514 | Up | pos | Cer | 3.809 | NS | 0.875 | 2.358 | 0.0006 | 1.966 |
| Cer (d20:1) | 359.326 | Up | pos | Cer | 0.931 | NS | 0.250 | 3.093 | 0.043 | 1.656 |
| Cer (d40:1 + hO + O) | 652.588 | Up | neg | Cer | 0.789 | NS | 0.796 | 2.642 | 0.046 | 1.642 |
| DG (41:0e) | 703.657 | Up | pos | DG | 1.251 | 0.011 | 2.071 | 2.252 | 0.0001 | 1.989 |
| MG (36:1) | 631.563 | Up | pos | MG | 1.607 | NS | 1.698 | 2.281 | 0.004 | 1.904 |
| PA (38:2) | 727.528 | Up | neg | PA | 2.552 | NS | 1.138 | 13.545 | 0.035 | 1.694 |
| PC (18:1/14:1) | 774.529 | Up | pos | PC | 2.735 | 0.036 | 1.915 | 2.100 | NS | 1.138 |
| PE (18:0/18:2) | 744.553 | Up | pos | PE | 4.856 | 0.017 | 2.026 | 17.333 | NS | 0.987 |
| PE (18:0e/18:3) | 726.544 | Up | neg | PE | 1.200 | 0.044 | 1.873 | 1.270 | 0.006 | 1.872 |
| PE (29:0/11:4) | 802.593 | Up | pos | PE | 2.379 | 0.017 | 2.028 | 7.548 | NS | 1.057 |
| SM (d20:0/19:1) | 817.644 | Up | pos | SM | 1.572 | 0.024 | 1.980 | 1.455 | 0.006 | 1.873 |
| SM (d37:1) | 745.621 | Up | pos | SM | 1.613 | 0.032 | 1.933 | 1.685 | 0.019 | 1.776 |
| SM (d41:2) | 799.668 | Up | pos | SM | 2.641 | NS | 1.298 | 4.188 | 0.0317 | 1.708 |
| TG (16:0/18:1/18:3) | 872.770 | Up | pos | TG | 1.447 | 0.009 | 2.0974 | 1.133 | 0.028 | 1.729 |
| TG (18:1/18:1/18:3) | 898.785 | Up | pos | TG | 2.289 | 0.011 | 2.078 | 1.276 | NS | 1.006 |
| TG (18:1/20:4/21:0) | 951.837 | Up | pos | TG | 1.219 | 0.032 | 1.938 | 1.401 | 0.014 | 1.811 |
| TG (24:4) | 485.287 | Up | pos | TG | 2.307 | 0.029 | 1.953 | 0.651 | NS | 0.897 |
| TG (39:6) | 675.517 | Up | pos | TG | 10.899 | 0.049 | 1.848 | 23.588 | NS | 0.988 |
| TG (43:7) | 729.563 | Up | pos | TG | 1.251 | 0.031 | 1.943 | 1.383 | 0.015 | 1.804 |
| OAHFA (16:0/26:0) | 649.614 | Down | neg | FA | 1.361 | NS | 1.341 | 1.589 | 0.008 | 1.856 |
| OAHFA (16:0/28:0) | 677.645 | Down | neg | FA | 1.257 | NS | 1.354 | 1.617 | 0.007 | 1.863 |
| OAHFA (18:1/26:0) | 675.629 | Down | neg | FA | 1.433 | NS | 1.409 | 1.673 | 0.002 | 1.926 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, X.; Li, Y.; Li, X.; Wang, C.; Ma, Z.; Wang, L.; Fan, X.; Li, Z.; Huang, D.; Xu, J.; et al. Lipidomic Profiling Reveals Distinct Differences in Sphingolipids Metabolic Pathway between Healthy Apis cerana cerana larvae and Chinese Sacbrood Disease. Insects 2021, 12, 703. https://doi.org/10.3390/insects12080703
Dang X, Li Y, Li X, Wang C, Ma Z, Wang L, Fan X, Li Z, Huang D, Xu J, et al. Lipidomic Profiling Reveals Distinct Differences in Sphingolipids Metabolic Pathway between Healthy Apis cerana cerana larvae and Chinese Sacbrood Disease. Insects. 2021; 12(8):703. https://doi.org/10.3390/insects12080703
Chicago/Turabian StyleDang, Xiaoqun, Yan Li, Xiaoqing Li, Chengcheng Wang, Zhengang Ma, Linling Wang, Xiaodong Fan, Zhi Li, Dunyuan Huang, Jinshan Xu, and et al. 2021. "Lipidomic Profiling Reveals Distinct Differences in Sphingolipids Metabolic Pathway between Healthy Apis cerana cerana larvae and Chinese Sacbrood Disease" Insects 12, no. 8: 703. https://doi.org/10.3390/insects12080703
APA StyleDang, X., Li, Y., Li, X., Wang, C., Ma, Z., Wang, L., Fan, X., Li, Z., Huang, D., Xu, J., & Zhou, Z. (2021). Lipidomic Profiling Reveals Distinct Differences in Sphingolipids Metabolic Pathway between Healthy Apis cerana cerana larvae and Chinese Sacbrood Disease. Insects, 12(8), 703. https://doi.org/10.3390/insects12080703






