Transcriptome Analyses Provide Insights into the Aggressive Behavior toward Conspecific and Heterospecific in Thitarodes xiaojinensis (Lepidoptera: Hepialidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Insects and Assays
2.2. Laboratory Trials
2.2.1. Experimental Design and Procedures
Assay for Testing Aggressive Behavior
- Hit (by head or tail);
- Bite (by jaw);
- Chase (the attacker pursues the victim).
Assay for Testing Survival Rate
2.2.2. Statistical Analysis
2.3. RNA-Seq Experiment
2.3.1. Library Construction
2.3.2. Preparation of RNA, Library Construction, and Sequencing
2.3.3. Mapping and Transcriptome Annotation
2.3.4. Differentially Expressed Gene Analysis
2.3.5. Functional Enrichment Analysis
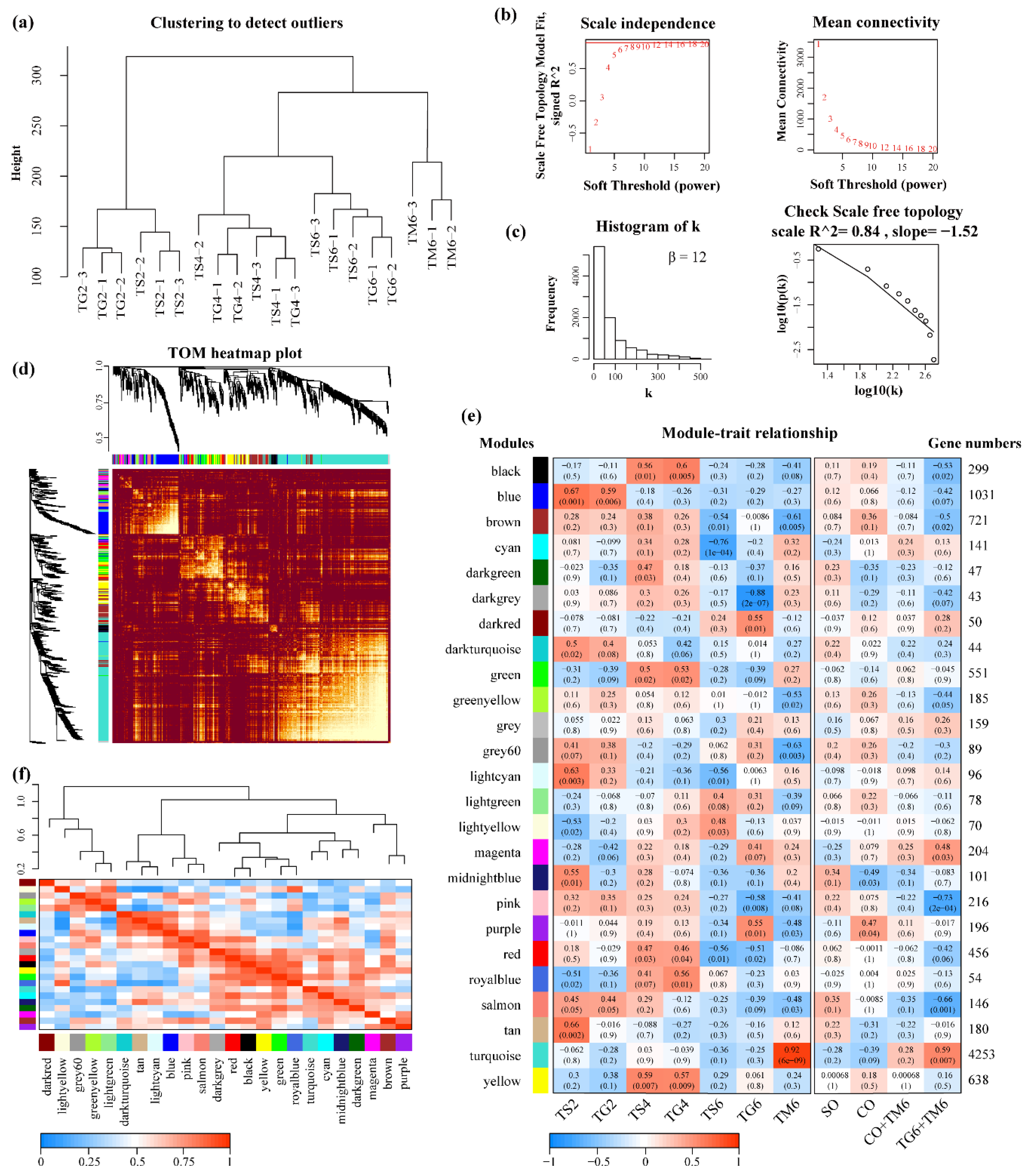
2.4. WGCNA Analysis
2.4.1. Construction of Weighted Gene Co-Expression Network
2.4.2. Interactions Analysis of Co-Expression Modules
2.4.3. Identification of Hub Genes
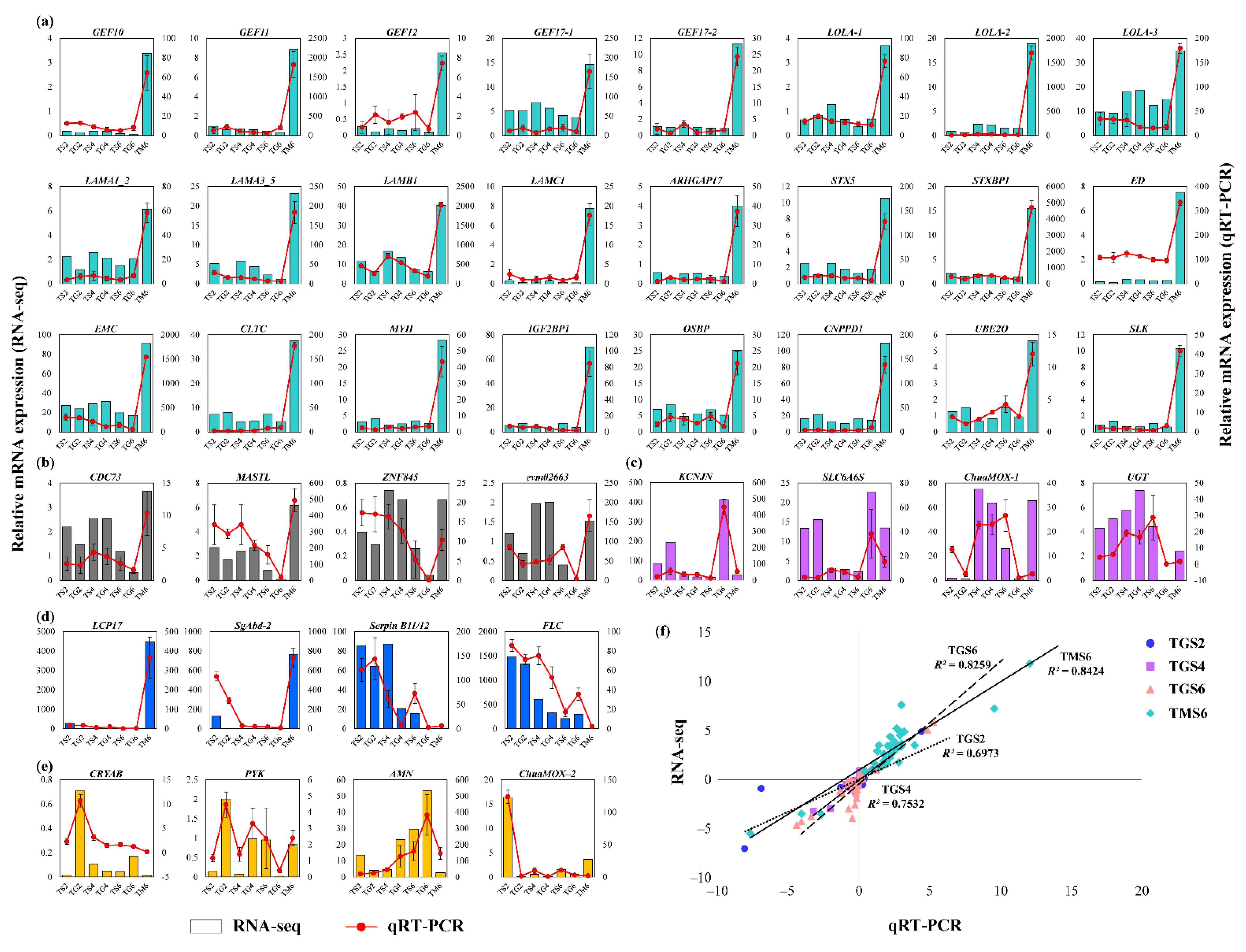
2.5. Quantitative Real-Time PCR
3. Results
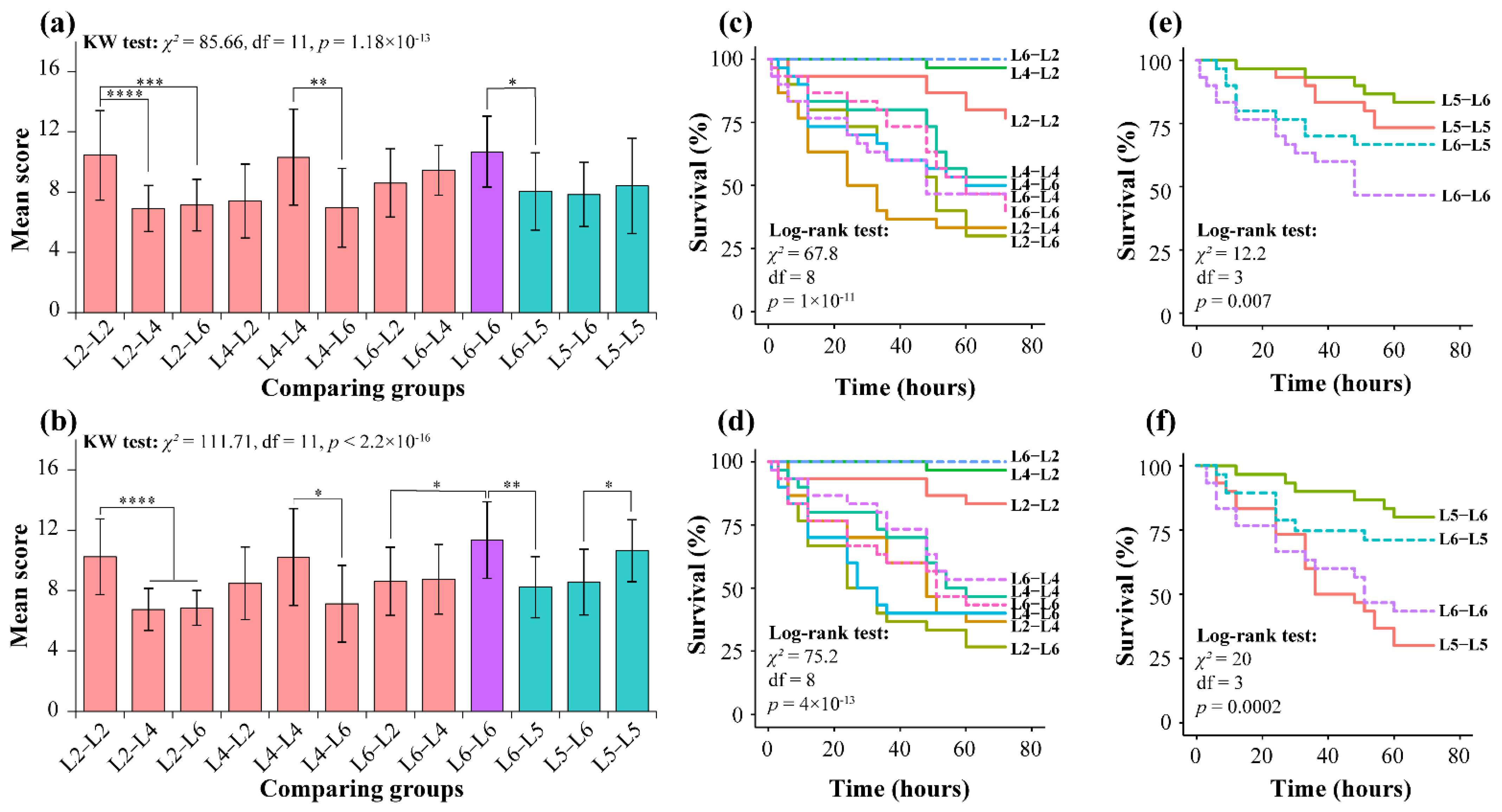
3.1. Behavior Observation
3.1.1. Aggressive Behavior of T. xiaojinensis toward Conspecific
3.1.2. T. xiaojinensis and H. armigera Larvae Caged with Same Instar Conspecifics or Similar-Sized Heterospecifics
3.2. RNA-Sequencing Analysis
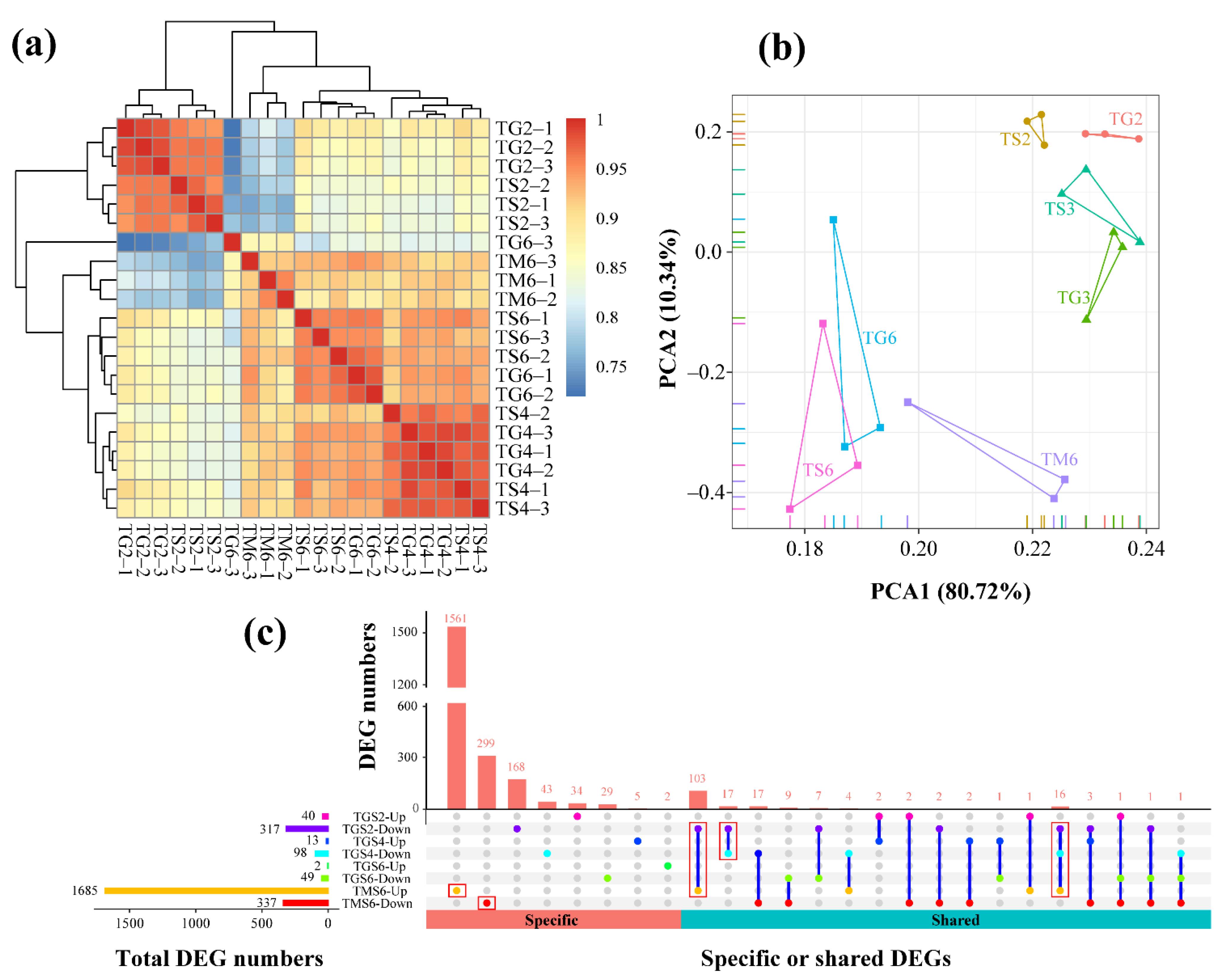
3.2.1. Differential Gene Expression Analysis
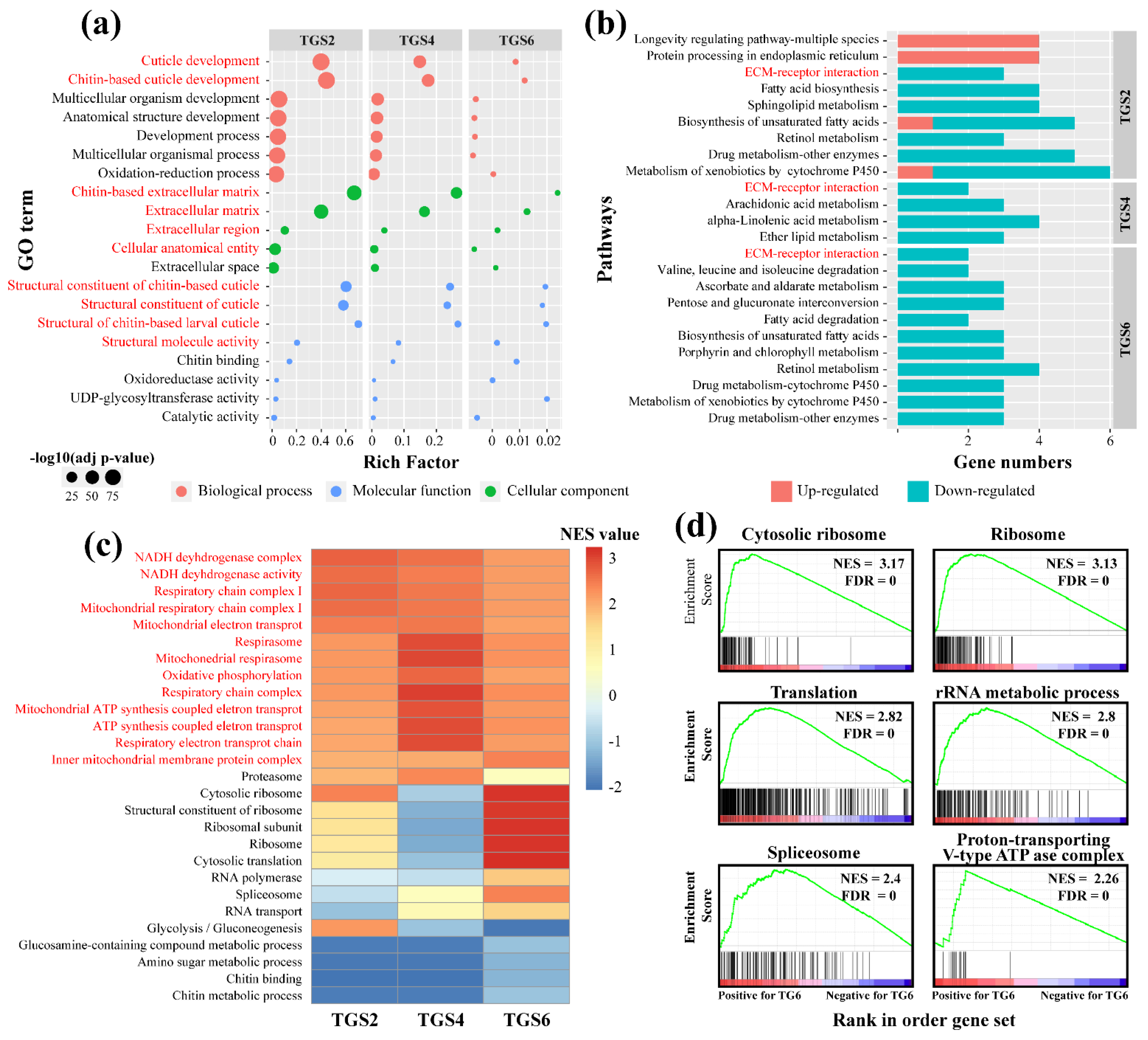
3.2.2. Molecular Responses of T. xiaojinensis Larvae When Caged with Conspecific of the Same Instar Stage
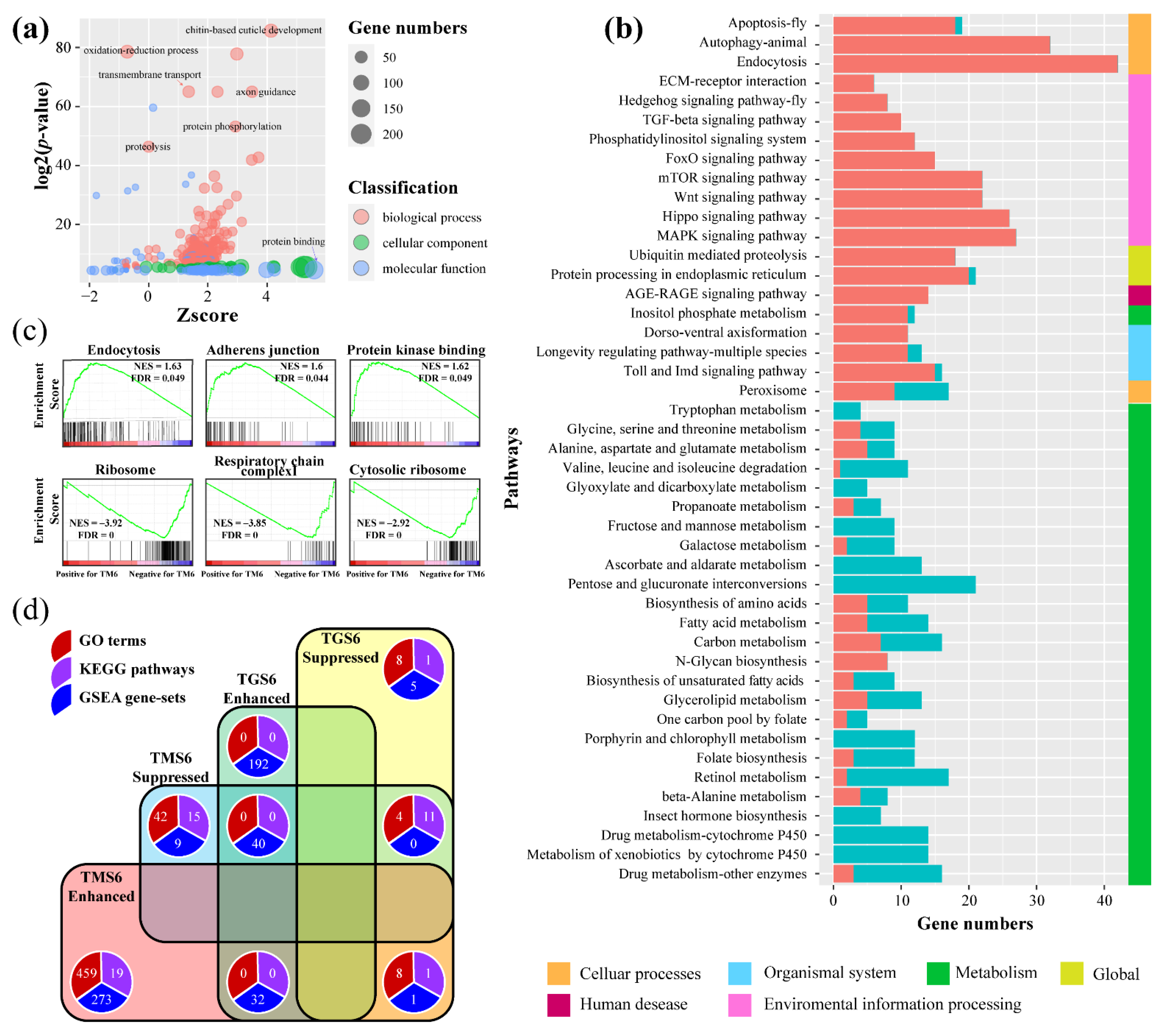
3.2.3. Molecular Responses of T. xiaojinensis Larvae When Caged with H. armigera of Similar Size
3.3. WGCNA Analysis for T. xiaojinensis
3.3.1. Construction of Gene Co-Expression Network for T. xiaojinensis
3.3.2. Interaction Analysis of Co-Expression Module
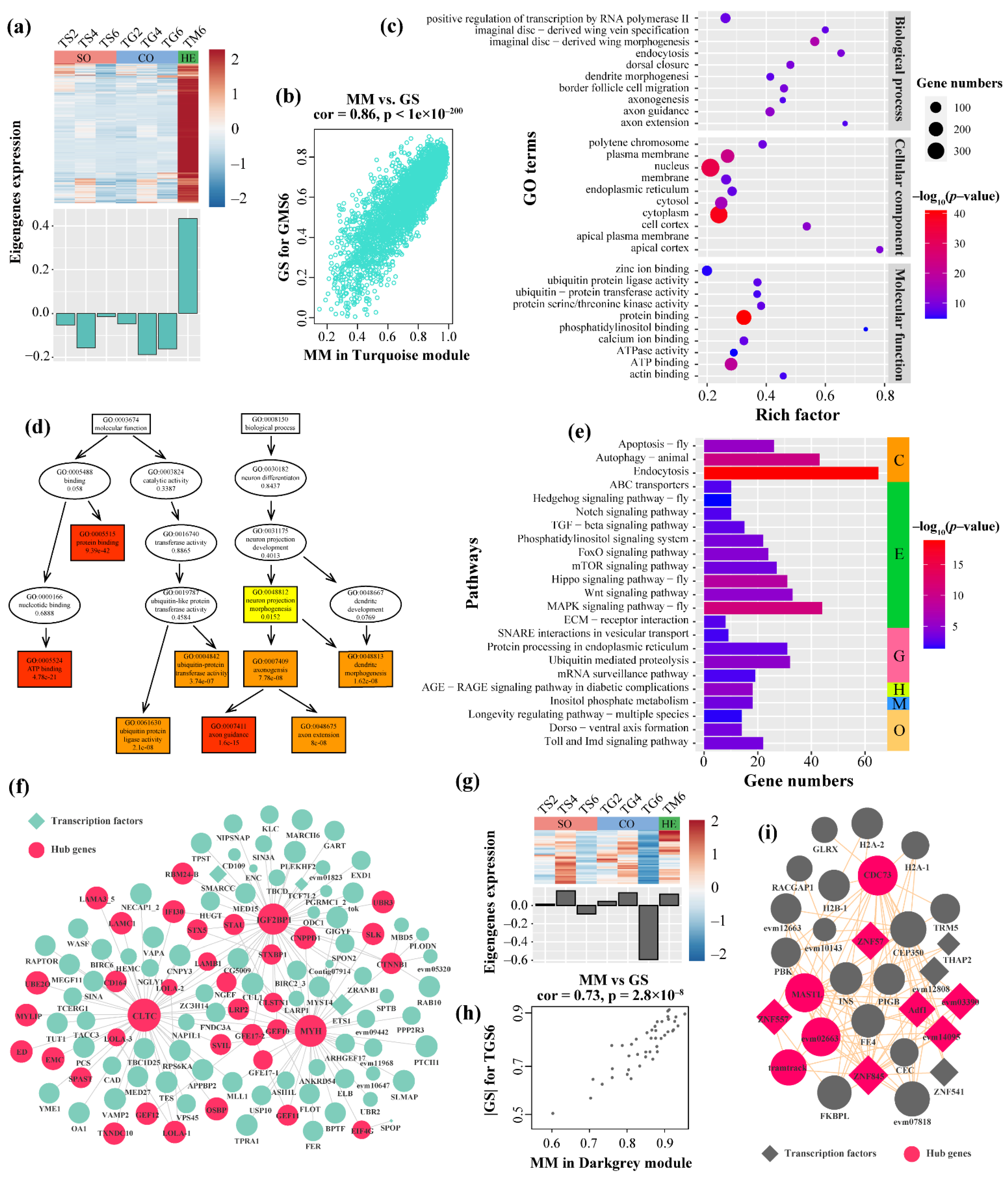
3.3.3. Functional Enrichment Analysis and Hub Genes Identification in Turquoise Module
3.3.4. Functional Enrichment Analysis and Hub Genes Identification in the DarkGrey Module
3.4. Experimental Validation
4. Discussion
4.1. Influences of Food on Aggressive Behavior
4.2. Transcriptional Response When T. xiaojinensis Showed Aggressiveness toward Conspecifics
4.3. Transcriptional Response When T. xiaojinensis Showed Aggressiveness toward Heterospecific
4.4. Hub Genes Modulating Aggressive Behavior
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Takahashi, A.; Miczek, K.A. Neurogenetics of Aggressive Behavior: Studies in Rodents. Curr. Top. Behav. Neurosci. 2014, 17, 3–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, X.; Mehren, J.; Hiroi, M.; Billeter, J.-C.; Miyamoto, T.; Amrein, H.; Levine, J.D.; Anderson, D.J. Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat. Neurosci. 2011, 14, 757–762. [Google Scholar] [CrossRef]
- Andrews, J.C.; Fernández, M.P.; Yu, Q.; Leary, G.P.; Leung, A.K.W.; Kavanaugh, M.P.; Kravitz, E.A.; Certel, S.J. Octopamine neuro-modulation regulates Gr32a-linked aggression and courtship pathways in Drosophila males. PLoS Genet. 2014, 10, e1004356. [Google Scholar] [CrossRef]
- Koganezawa, M.; Kimura, K.-I.; Yamamoto, D. The Neural Circuitry that Functions as a Switch for Courtship versus Aggression in Drosophila Males. Curr. Biol. 2016, 26, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Hoopfer, E.D.; Jung, Y.; Inagaki, H.K.; Rubin, G.M.; Anderson, D.J. P1 interneurons promote a persistent internal state that en-hances inter-male aggression in Drosophila. eLife 2015, 4, e11346. [Google Scholar] [CrossRef] [PubMed]
- Asahina, K.; Watanabe, K.; Duistermars, B.J.; Hoopfer, E.; González, C.R.; Eyjólfsdóttir, E.A.; Perona, P.; Anderson, D.J. Tachykin-in-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 2014, 156, 221–235. [Google Scholar] [CrossRef]
- Alekseyenko, O.V.; Lee, C.; Kravitz, E.A. Targeted Manipulation of Serotonergic Neurotransmission Affects the Escalation of Aggression in Adult Male Drosophila melanogaster. PLoS ONE 2010, 5, e10806. [Google Scholar] [CrossRef]
- Hoyer, S.C.; Eckart, A.; Herrel, A.; Zars, T.; Fischer, S.A.; Hardie, S.L.; Heisenberg, M. Octopamine in Male Aggression of Drosophila. Curr. Biol. 2008, 18, 159–167. [Google Scholar] [CrossRef]
- Edwards, A.C.; Zwarts, L.; Yamamoto, A.; Callaerts, P.; Mackay, T.F. Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol. 2009, 7, 29. [Google Scholar] [CrossRef]
- Davis, S.M.; Thomas, A.L.; Liu, L.; Campbell, I.M.; Dierick, H.A. Isolation of Aggressive Behavior Mutants in Drosophila Using a Screen for Wing Damage. Genetics 2018, 208, 273–282. [Google Scholar] [CrossRef]
- Wang, L.; Anderson, D.J. Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nat. Cell Biol. 2010, 463, 227–231. [Google Scholar] [CrossRef]
- Liu, W.; Liang, X.; Gong, J.; Yang, Z.; Zhang, Y.-H.; Zhang, J.-X.; Rao, Y. Social regulation of aggression by pheromonal activation of Or65a olfactory neurons in Drosophila. Nat. Neurosci. 2011, 14, 896–902. [Google Scholar] [CrossRef]
- Hoffmann, A.A. The influence of age and experience with conspecifics on territorial behavior in Drosophila melanogaster. J. Insect Behav. 1990, 3, 1–12. [Google Scholar] [CrossRef]
- Svetec, N.; Ferveur, J.-F. Social experience and pheromonal perception can change male–male interactions in Drosophila melanogaster. J. Exp. Biol. 2005, 208, 891. [Google Scholar] [CrossRef]
- Shorter, J.; Couch, C.; Huang, W.; Carbone, M.A.; Peiffer, J.; Anholt, R.R.H.; Mackay, T.F.C. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 2015, 112, E3555–E3563. [Google Scholar] [CrossRef] [PubMed]
- Zampolini, M.; Zaccaria, B.; Tolli, V.; Frustaci, A.; Franceschini, M. Rehabilitation of traumatic brain injury in Italy: A multicentred study. Brain Injury 2012, 26, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Alia-Klein, N.; Goldstein, R.Z.; Kriplani, A.; Logan, J.; Tomasi, D.; Williams, B.; Telang, F.; Shumay, E.; Biegon, A.; Craig, I.W.; et al. Brain monoamine oxidase A activity predicts trait aggression. J. Neurosci. 2008, 28, 5099–5104. [Google Scholar] [CrossRef]
- De Almeida, R.M.; Ferrari, P.F.; Parmigiani, S.; Miczek, K.A. Escalated aggressive behavior: Dopamine, serotonin and GABA. Eur. J. Pharmacol. 2005, 526, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.H.; A Kravitz, E. Serotonin, social status and aggression. Curr. Opin. Neurobiol. 1997, 7, 812–819. [Google Scholar] [CrossRef]
- Jacobs, M.E. Influence of beta-alanine on mating and territorialism in Drosophila melanogaster. Behav. Genet. 1978, 8, 487. [Google Scholar] [CrossRef]
- Certel, S.J.; Savella, M.G.; Schlegel, D.C.F.; Kravitz, E.A. Modulation of Drosophila male behavioral choice. Proc. Natl. Acad. Sci. USA 2007, 104, 4706–4711. [Google Scholar] [CrossRef]
- A Miczek, K.; Fish, E.W.; De Bold, J.F. Neurosteroids, GABAA receptors, and escalated aggressive behavior. Horm. Behav. 2003, 44, 242–257. [Google Scholar] [CrossRef]
- Nelson, R.J.; Demas, G.E.; Huang, P.L.; Fishman, M.C.; Dawson, V.L.; Dawson, T.M.; Snyder, S.H. Behavioural abnormalities in male mice lacking neuronal nitric oxide synthase. Nat. Cell Biol. 1995, 378, 383–386. [Google Scholar] [CrossRef]
- Rujescu, D.; Giegling, I.; Mandelli, L.; Schneider, B.; Hartmann, A.M.; Schnabel, A.; Maurer, K.; Möller, H.-J.; Serretti, A. NOS-I and -III gene variants are differentially associated with facets of suicidal behavior and aggression-related traits. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008, 147B, 42–48. [Google Scholar] [CrossRef]
- Gammie, S.C.; Auger, A.P.; Jessen, H.M.; Vanzo, R.J.; Awad, T.A.; Stevenson, S.A. Altered gene expression in mice selected for high maternal aggression. Genes Brain Behav. 2010, 6, 432–443. [Google Scholar] [CrossRef]
- Karl, T.; Herzog, H. Behavioral profiling of NPY in aggression and neuropsychiatric diseases. Peptides 2007, 28, 326–333. [Google Scholar] [CrossRef] [PubMed]
- A Dierick, H.; Greenspan, R.J. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat. Genet. 2007, 39, 678–682. [Google Scholar] [CrossRef]
- A Dierick, H.; Greenspan, R.J. Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 2006, 38, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.C.; Rollmann, S.M.; Morgan, T.J.; Mackay, T.F. Quantitative genomics of aggressive behavior in Drosophila melano-gaster. PLoS Genet. 2006, 2, e154. [Google Scholar] [CrossRef] [PubMed]
- Rollmann, S.M.; Zwarts, L.; Edwards, A.C.; Yamamoto, A.; Callaerts, P.; Norga, K.; Mackay, T.F.C.; Anholt, R.R.H. Pleiotropic effects of Drosophila neuralized on complex behaviors and brain structure. Genetics 2008, 179, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, E.G.; Plowes, R.M.; Folgarait, P.J.; Bollazzi, M.; Gilbert, L.E. Ritualized aggressive behavior reveals distinct social structures in native and introduced range tawny crazy ants. PLoS ONE 2019, 14, e0225597. [Google Scholar] [CrossRef]
- Yadava, R.P.; Smith, M.V. Aggressive behavior of Apis mellifera L. workers towards introduced queens. II. Role of the man-dibular gland contents of the queen in releasing aggressive behavior. Can. J. Zool. 1971, 49, 1179–1183. [Google Scholar] [CrossRef]
- Sakura, M.; Aonuma, H. Aggressive behavior in the antennectomized male cricket Gryllus bimaculatus. J. Exp. Biol. 2013, 216, 2221–2228. [Google Scholar] [CrossRef]
- Tao, Z.; Cao, L.; Zhang, Y.; Ye, Y.; Han, R. Laboratory rearing of Thitarodes armoricanus and Thitarodes jianchuanensis (Lepidoptera: Hepialidae), hosts of the Chinese medicinal fungus Ophiocordyceps sinensis (Hypocreales: Ophiocordycipitaceae). J. Econ. Entomol. 2015, 109, 176–181. [Google Scholar] [CrossRef]
- Jun-Feng, L.I.; Zou, Z.W.; Liu, X.; Zhang, G.R. Biology of Thitarodes pui (Lepidoptera, Hepialidae), a host species of Ophiocordyceps sinensis. J. Environ. Entomol. 2011, 33, 195–202. [Google Scholar]
- Vergara, F.; Shino, A.; Kikuchi, J. Cannibalism affects core metabolic processes in Helicoverpa armigera Larvae—A 2D NMR Metabolomics Study. Int. J. Mol. Sci. 2016, 17, 1470. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Xing, Y.M.; Guo, S.X. Molecular cloning and characterization of two kind of heat-shock protein gene from Polyporus umbellatus. China J. Chin. Mater. Med. 2016, 41, 4550–4555. [Google Scholar]
- Borgan, R. Kaplan-Meier Estimator; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Kaplan, E.S. Non-parametric estimations from incomplete observations. J. Am. Stat. Assoc. 1958, 34. [Google Scholar]
- Breslow, N.E. Analysis of Survival Data under the Proportional Hazards Model. Int. Stat. Rev. 1975, 43, 45. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.J.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visual-ization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. TopGO: Enrichment Analysis for Gene Ontology. R Package Version 2.40.40. 2020. Available online: https://bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 8 June 2021).
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fuller, T.F.; Ghazalpour, A.; Aten, J.E.; Drake, T.A.; Lusis, A.J.; Horvath, S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 2007, 18, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.B.; Wang, S.; Yang, B.; Jiang, Z.; Lenahan, C.; Wang, J.; Zhang, J.; Shao, A. Transcriptome analyses reveal molecular mechanisms underlying phenotypic differences among transcriptional subtypes of glioblastoma. J. Cell. Mol. Med. 2020, 24, 3901–3916. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Yao, Y.-J.; Wang, X.-L. Host insect species of Ophiocordyceps sinensis: A review. ZooKeys 2011, 43. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Chen, X.X.; Zheng, S.C.; Chen, P.; Mo, M.H. The mechanisms of pharmacological activities of Ophiocordyceps sinensis fungi. Phytother. Res. 2016, 30, 1572–1583. [Google Scholar] [CrossRef]
- Umbers, K.D.L.; Tatarnic, N.J.; Holwell, G.I.; Herberstein, M.E. Ferocious Fighting between Male Grasshoppers. PLoS ONE 2012, 7, e49600. [Google Scholar] [CrossRef][Green Version]
- Georgiev, A.V.; Klimczuk, A.; Traficonte, D.M.; Maestripieri, D. When violence pays: A cost-benefit analysis of aggressive be-havior in animals and humans. Evol. Psychol. 2013, 11, 147470491301100313. [Google Scholar] [CrossRef]
- Richardson, M.L.; Mitchell, R.F.; Reagel, P.F.; Hanks, L.M. Causes and consequences of cannibalism in noncarnivorous insects. Annu. Rev. Entomol. 2010, 55, 39–53. [Google Scholar] [CrossRef]
- Kakimoto, T.; Fujisaki, K.; Miyatake, T. Egg laying preference, larval dispersion, and cannibalism in Helicoverpa armigera (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 2003, 96, 793–798. [Google Scholar] [CrossRef]
- Nakamura, K.; Ohgushi, T. Studies on the population dynamics of a thistle-feeding lady beetle, Henosepilachna pustulosa (Kôno) in a cool temperature climax forest. Popul. Ecol. 1979, 20, 297–314. [Google Scholar] [CrossRef]
- Alaux, C.; Sinha, S.; Hasadsri, L.; Hunt, G.J.; Guzmán-Novoa, E.; DeGrandi-Hoffman, G.; Uribe-Rubio, J.L.; Southey, B.R.; Rodriguez-Zas, S.; Robinson, G.E. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 15400–15405. [Google Scholar] [CrossRef] [PubMed]
- Li-Byarlay, H.; Rittschof, C.C.; Massey, J.H.; Pittendrigh, B.R.; Robinson, G.E. Socially responsive effects of brain oxidative metabolism on aggression. Proc. Natl. Acad. Sci. USA 2014, 111, 12533–12537. [Google Scholar] [CrossRef]
- Miranda Mendonca, A.P.; Hoppe, L.Y.; Gaviraghi, A.; Araujo-Jorge, T.C.; de Oliveira, G.M.; Felippe, R.M.; Oliveira, M.F.; da Silva Fragoso, V.M. Highly aggressive behavior induced by social stress is associated to reduced cytochrome c oxidase activity in mice brain cortex. Neurochem. Int. 2019, 126, 210–217. [Google Scholar] [CrossRef]
- Nelson, R.J.; Chiavegatto, S. Molecular basis of aggression. Trends Neurosci. 2001, 24, 713–719. [Google Scholar] [CrossRef]
- Olivier, B.; Mos, J.; Van Oorschot, R.; Hen, R. Serotonin receptors and animal models of aggressive behavior. Pharmacopsychiatry 1995, 28, 80–90. [Google Scholar] [CrossRef]
- Miczek, K.A.; Barros, H.M.; Sakoda, L.; Weerts, E.M. Alcohol and heightened aggression in individual mice. Alcohol. Clin. Exp. Res. 1998, 22, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Fish, E.W.; Faccidomo, S.; Miczek, K.A. Aggression heightened by alcohol or social instigation in mice: Reduction by the 5-HT(1B) receptor agonist CP-94,253. Psychopharmacolohy 1999, 146, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Chiavegatto, S. Brain serotonin dysfunction accounts for aggression in male mice lacking neuronal nitric oxide synthase. Proc. Nat. Acad. Sci. USA 2001, 98, 1277–1281. [Google Scholar] [CrossRef] [PubMed]
- Kravitz, E.A. Serotonin and aggression: Insights gained from a lobster model system and speculations on the role of amine neurons in a complex behavior. J. Comp. Physiol. A 2000, 186, 221–238. [Google Scholar] [CrossRef]
- Kravitz, E.A.; Huber, R. Aggression in invertebrates. Curr. Opin. Neurobiol. 2003, 13, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Manuck, S.B.; Flory, J.D.; Ferrell, R.E.; Dent, K.M.; Muldoon, M.F. Aggression and anger-related traits associated with a poly-morphism of the tryptophan hydroxylase gene. Biol. Psychiatry 1999, 45, 603–614. [Google Scholar] [CrossRef]
- Craig, D.; Hart, D.J.; Carson, R.; Mcilroy, S.P.; Passmore, A.P. Allelic variation at the A218C tryptophan hydroxylase polymor-phism influences agitation and aggression in Alzheimer’s disease. Neurosci. Lett. 2004, 363, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Frau, R.; Fanni, S.; Serra, V.; Simola, N.; Godar, S.C.; Traccis, F.; Devoto, P.; Bortolato, M.; Melis, M. Dysfunctional mesocortical dopamine circuit at pre-adolescence is associated to aggressive behavior in MAO-A hypomorphic mice exposed to early life stress. Neuropharmacology 2019, 159, 107517. [Google Scholar] [CrossRef]
- Van Erp, A.M.M.; Miczek, K.A. Aggressive behavior, increased accumbal dopamine, and decreased cortical serotonin in rats. J. Neurosci. 2000, 20, 9320–9325. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.-B.; Pang, R.; Li, W.-X.; Ojha, A.; Li, D.; Zhang, W.-Q. An Overview of Embryogenesis: External Morphology and Transcriptome Profiling in the Hemipteran Insect Nilaparvata lugens. Front. Physiol. 2020, 11, 106. [Google Scholar] [CrossRef]
- Tian, L.; Gao, X.; Zhang, S.; Zhang, Y.; Ma, D.; Cui, J. Dynamic changes of transcriptome of fifth-instar spodoptera litura larvae in response to insecticide. 3 Biotech 2021, 11, 98. [Google Scholar] [CrossRef]
- Alonen, A.; Finel, M.; Kostiainen, R. The human UDP-glucuronosyltransferase UGT1A3 is highly selective towards N2 in the tetrazole ring of losartan, candesartan, and zolarsartan. Biochem. Pharmacol. 2008, 76, 763–772. [Google Scholar] [CrossRef]
- Picard, N.; Ratanasavanh, D.; Prémaud, A.; Meur, Y.L.; Marquet, P. Identification of the UDP-Glucuronosyl transferase isoforms involved in mycophenolic acid phase II metabolism. Drug Metab. Dispos. Biol. Fate Chem. 2005, 33, 139. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.; An, J.; Yang, P.; Guo, L.; Li, Y.; Lv, J.; Yu, S. Transcriptome analyses and weighted gene coexpression network analysis reveal key pathways and genes involved in the rapid cold resistance of the Chinese white wax scale insect. Arch. Insect Biochem. Physiol. 2021, 107, e21781. [Google Scholar] [CrossRef]
- E Lloyd, T.; Verstreken, P.; Ostrin, E.J.; Phillippi, A.; Lichtarge, O.; Bellen, H.J. A Genome-Wide Search for Synaptic Vesicle Cycle Proteins in Drosophila. Neuron 2000, 26, 45–50. [Google Scholar] [CrossRef]
- Bashaw, G.J.; Hu, H.; Nobes, C.D.; Goodman, C.S. A novel Dbl family RhoGEF promotes Pho-dependent axon attraction to the central nervous system. J. Cell Biol. 2001, 155, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Lin, M.Z.; Goldberg, J.; Estrach, S.; Sahin, M.; Hu, L.; Bazalakova, M.; Neve, R.L.; Corfas, G.; Debant, A.; et al. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor Ephexin. Cell 2001, 105, 233–244. [Google Scholar] [CrossRef]
- Verhoeven, K.; Jonghe, P.D.; Putte, T.; Nelis, E.; An, Z.; Verpoorten, N.; Vriendt, E.D.; An, J.; Gerwen, V.V.; Francis, A. Slowed conduction and thin myelination of peripheral nerves associated with mutant rho Guanine-nucleotide exchange factor 10. Am. J. Hum. Genet. 2003, 73, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Lin, Y.M.; Kao, T.C.; Chuang, H.H.; Chen, R.H. PDZ-RhoGEF ubiquitination by Cullin3–KLHL20 controls neurotro-phin-induced neurite outgrowth. J. Cell Biol. 2011, 193, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Giniger, E.; Tietje, K.; Jan, L.Y.; Jan, Y.N. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 1994, 120, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, Z.; Cao, L.; Wu, H.; Han, R. Transcriptome Analyses Provide Insights into the Aggressive Behavior toward Conspecific and Heterospecific in Thitarodes xiaojinensis (Lepidoptera: Hepialidae). Insects 2021, 12, 577. https://doi.org/10.3390/insects12070577
Rao Z, Cao L, Wu H, Han R. Transcriptome Analyses Provide Insights into the Aggressive Behavior toward Conspecific and Heterospecific in Thitarodes xiaojinensis (Lepidoptera: Hepialidae). Insects. 2021; 12(7):577. https://doi.org/10.3390/insects12070577
Chicago/Turabian StyleRao, Zhongchen, Li Cao, Hua Wu, and Richou Han. 2021. "Transcriptome Analyses Provide Insights into the Aggressive Behavior toward Conspecific and Heterospecific in Thitarodes xiaojinensis (Lepidoptera: Hepialidae)" Insects 12, no. 7: 577. https://doi.org/10.3390/insects12070577
APA StyleRao, Z., Cao, L., Wu, H., & Han, R. (2021). Transcriptome Analyses Provide Insights into the Aggressive Behavior toward Conspecific and Heterospecific in Thitarodes xiaojinensis (Lepidoptera: Hepialidae). Insects, 12(7), 577. https://doi.org/10.3390/insects12070577




