Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
2.2. Sample Collection
2.3. Experimental Design
2.4. Sample Processing
2.5. Data Analysis and Bioinformatics
2.6. Indicator Species Analysis
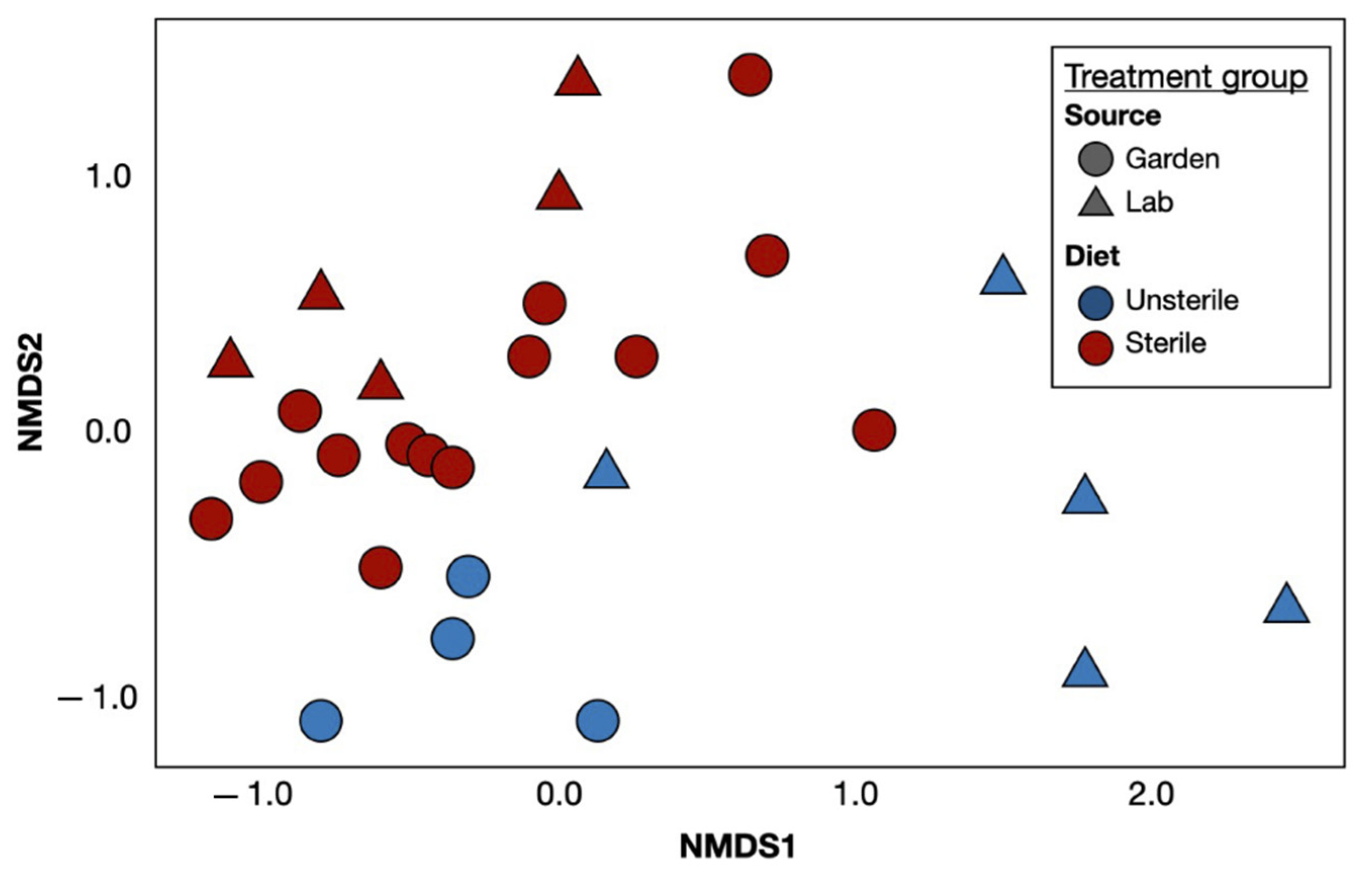
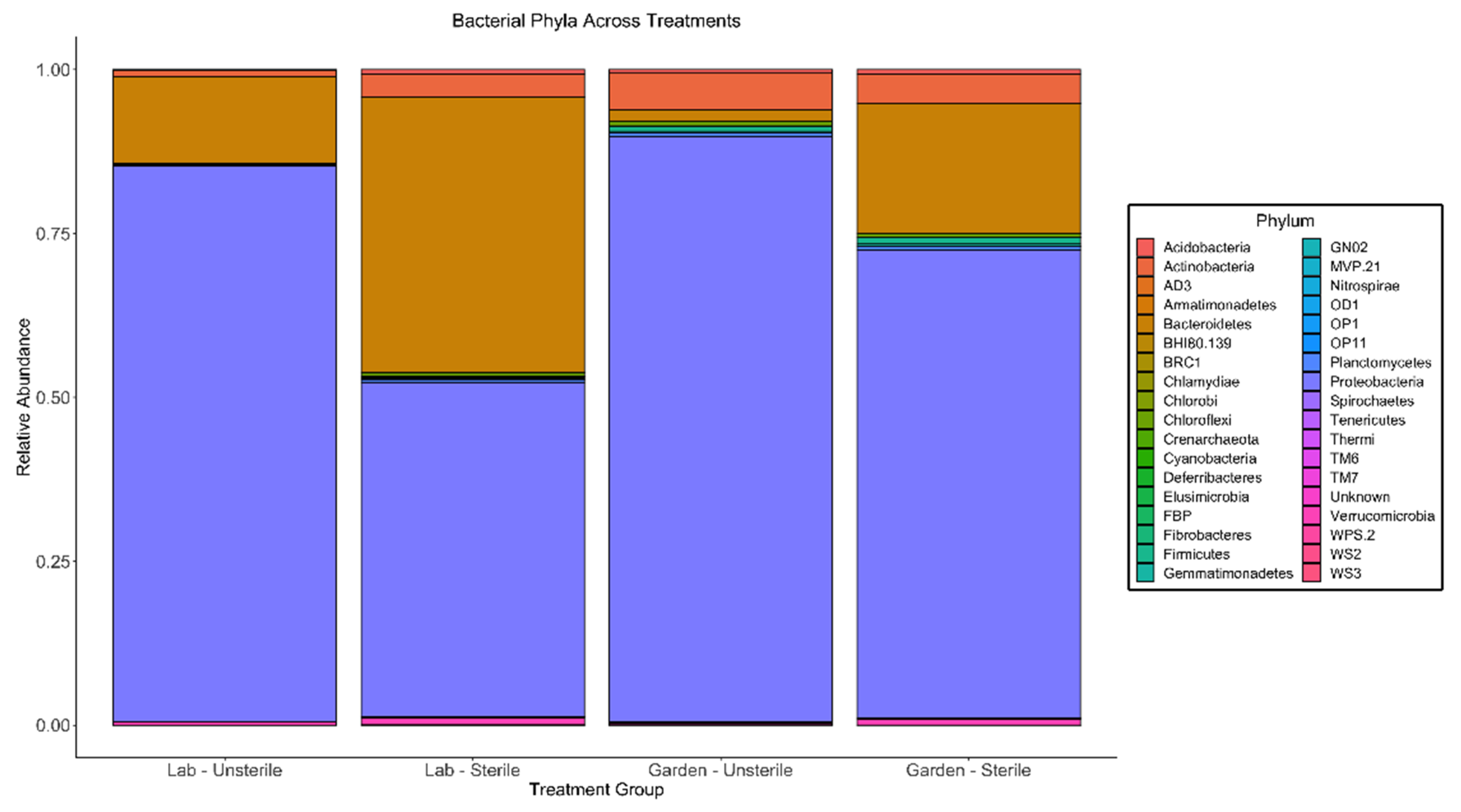
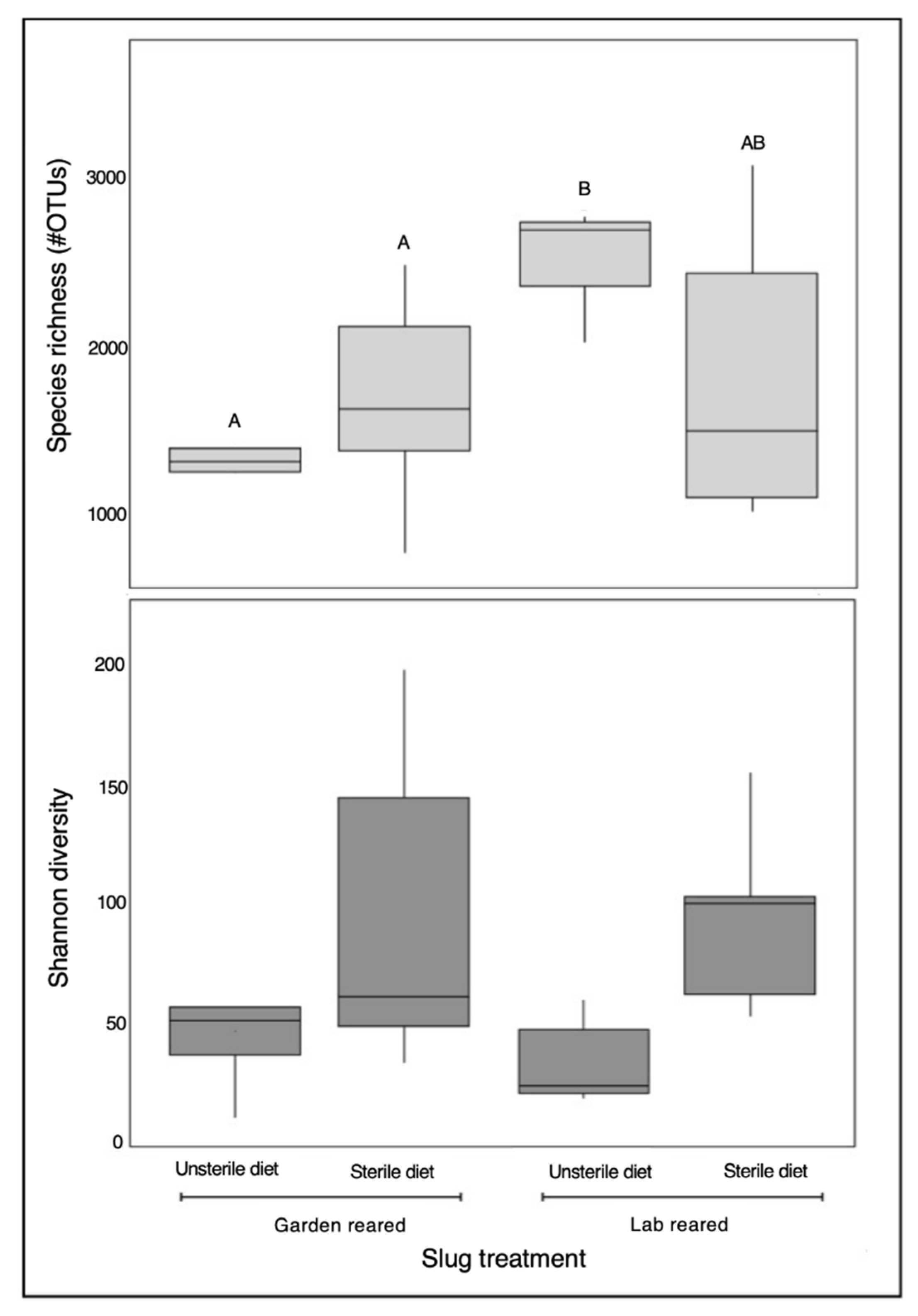
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bahrndorff, S.; Alemu, T.; Alemneh, T.; Nielsen, J.L. The Microbiome of Animals: Implications for Conservation Biology. Int. J. Genom. 2016, 2016, 5304028. [Google Scholar] [CrossRef]
- Douglas, A.E. Multiorganismal Insects: Diversity and Function of Resident Microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Newton, I.L.; Sheehan, K.B.; Lee, F.J.; Horton, M.A.; Hicks, R.D. Invertebrate systems for hypothesis-driven microbiome research. Microbiome Sci. Med. 2013, 1, 1–9. [Google Scholar] [CrossRef]
- Howlett, S.A. Terrestrial slug problems: Classical biological control and beyond. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2012, 7. [Google Scholar] [CrossRef]
- Bees, M.A.; Angulo, O.; López-Marcos, J.C.; Schley, D. Dynamics of a structured slug population model in the absence of seasonal variation. Math. Models Methods Appl. Sci. 2006, 16, 1961–1985. [Google Scholar] [CrossRef]
- Beyer, N.W.; Saari, D.M. Activity and Ecological Distribution of the Slug, Arion subfuscus (Draparnaud) (Stylom-matophora, Arionidae). Am. Midl. Nat. 1978, 100, 359–367. [Google Scholar] [CrossRef]
- Capinera, J.L.; Rodrigues, C.G. Biology and Control of the Leatherleaf Slug Leidyula floridana (Mollusca: Gastropoda: Veronicellidae). Fla. Entomol. 2015, 98, 243–253. [Google Scholar] [CrossRef]
- Jordaens, K.; Pinceel, J.; Backeljau, T. Life-history variation in selfing multilocus genotypes of the land slug Deroceras laeve (Pulmonata: Agriolimacidae). J. Molluscan Stud. 2006, 72, 229–233. [Google Scholar] [CrossRef]
- Shirley, M.; Howlett, S.; Port, G. Not All Slugs Are the Same: Variation in Growth and Development of the Slug Deroceras reticulatum. Insects 2020, 11, 742. [Google Scholar] [CrossRef]
- Russo, J.; Madec, L. Dual Strategy for Immune Defense in the Land Snail Cornu aspersum (Gastropoda, Pulmonata). Physiol. Biochem. Zool. 2011, 84, 212–221. [Google Scholar] [CrossRef]
- Russo, J.; Madec, L. Linking Immune Patterns and Life History Shows Two Distinct Defense Strategies in Land Snails (Gastropoda, Pulmonata). Physiol. Biochem. Zool. 2013, 86, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Koleva, Z.; Dedov, I.; Kizheva, J.; Lipovanska, R.; Moncheva, P.; Hristova, P. Lactic acid microflora of the gut of snail Cornu aspersum. Biotechnol. Biotechnol. Equip. 2014, 28, 627–634. [Google Scholar] [CrossRef]
- Van Horn, D.J.; Garcia, J.R.; Loker, E.S.; Mitchell, K.R.; Mkoji, G.M.; Adema, C.M.; Takacs-Vesbach, C.D. Complex intestinal bacterial communities in three species of planorbid snails. J. Molluscan Stud. 2011, 78, 74–80. [Google Scholar] [CrossRef]
- Charrier, M.; Combet-Blanc, Y.; Ollivier, B. Bacterial flora in the gut of Helix aspersa (Gastropoda Pulmonata): Evidence for a permanent population with a dominant homolactic intestinal bacterium, Enterococcus casseliflavus. Can. J. Microbiol. 1998, 44, 20–27. [Google Scholar] [CrossRef]
- Reich, I.; Ijaz, U.Z.; Gormally, M.; Smith, C.J. 16S rRNA sequencing reveals likely beneficial core microbes within faecal samples of the EU protected slug Geomalacus maculosus. Sci. Rep. 2018, 8, 10402. [Google Scholar] [CrossRef]
- Cardoso, A.M.; Cavalcante, J.J.; Cantão, M.E.; Thompson, C.E.; Flatschart, R.B.; Glogauer, A.; Scapin, S.M.N.; Sade, Y.B.; Beltrão, P.J.M.S.I.; Gerber, A.L.; et al. Metagenomic Analysis of the Microbiota from the Crop of an Invasive Snail Reveals a Rich Reservoir of Novel Genes. PLoS ONE 2012, 7, e48505. [Google Scholar] [CrossRef]
- Joynson, R.; Pritchard, L.; Osemwekha, E.; Ferry, N. Metagenomic Analysis of the Gut Microbiome of the Common Black Slug Arion ater in Search of Novel Lignocellulose Degrading Enzymes. Front. Microbiol. 2017, 8, 2181. [Google Scholar] [CrossRef]
- Cardoso, A.M.; Cavalcante, J.J.; Vieira, R.P.; Lima, J.L.; Grieco, M.A.B.; Clementino, M.M.; Vasconcelos, A.T.R.; Garcia, E.S.; de Souza, W.; Albano, R.M.; et al. Gut Bacterial Communities in the Giant Land Snail Achatina fulica and Their Modification by Sugarcane-Based Diet. PLoS ONE 2012, 7, e33440. [Google Scholar] [CrossRef]
- O’Rorke, R.; Cobian, G.M.; Holland, B.S.; Price, M.R.; Costello, V.; Amend, A.S. Dining local: The microbial diet of a snail that grazes microbial communities is geographically structured. Environ. Microbiol. 2014, 17, 1753–1764. [Google Scholar] [CrossRef]
- Pinheiro, G.L.; Correa, R.F.; Cunha, R.S.; Cardoso, A.M.; Chaia, C.; Clementino, M.M.; Garcia, E.S.; de Souza, W.; Frasés, S. Isolation of aerobic cultivable cellulolytic bacteria from different regions of the gastrointestinal tract of giant land snail Achatina fulica. Front. Microbiol. 2015, 6, 860. [Google Scholar] [CrossRef]
- Hu, Z.; Tong, Q.; Chang, J.; Yu, J.; Li, S.; Niu, H.; Ma, D. Gut bacterial communities in the freshwater snail Planorbella trivolvis and their modification by a non-herbivorous diet. PeerJ 2021, 9, e10716. [Google Scholar] [CrossRef] [PubMed]
- Rossing, J.S.; Hietpas, N.J. Isolation of Anaerobic Microbiota of Slug Intestinal Tract; UW-L Journal of Undergraduate Research X: La Crosse, WI, USA, 2007; pp. 2–5. [Google Scholar]
- Pawar, K.D.; Banskar, S.; Rane, S.D.; Charan, S.S.; Kulkarni, G.J.; Sawant, S.S.; Ghate, H.V.; Patole, M.S.; Shouche, Y.S. Bacterial diversity in different regions of gastrointestinal tract of Giant African Snail (Achatina fulica). Microbiologyopen 2012, 1, 415–426. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, X.; Chang, J.; Yu, J.; Tong, Q.; Li, S.; Niu, H. Compositional and predicted functional analysis of the gut microbiota of Radix auricularia (Linnaeus) via high-throughput Illumina sequencing. PeerJ 2018, 6, e5537. [Google Scholar] [CrossRef] [PubMed]
- Pawar, K.D.; Dar, M.A.; Rajput, B.P.; Kulkarni, G.J. Enrichment and Identification of Cellulolytic Bacteria from the Gastrointestinal Tract of Giant African Snail, Achatina fulica. Appl. Biochem. Biotechnol. 2014, 175, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Nicolai, A.; Rouland-LeFèvre, C.; Ansart, A.; Filser, J.; Lenz, R.; Pando, A.; Charrier, M. Inter-Population Differences and Seasonal Dynamic of the Bacterial Gut Community in the Endangered Land Snail Helix pomatia (Gastropoda: Helicidae). Malacologia 2015, 59, 177–190. [Google Scholar] [CrossRef]
- Ottaviani, E. Molluscan immunorecognition. Invertebr. Surviv. J. 2006, 3, 50–63. [Google Scholar]
- Ottaviani, E. Immunorecognition in the gastropod molluscs with particular reference to the freshwater snail Planorbarius corneus (L.) (Gastropoda, Pulmonata). Bolletino Zool. 1992, 59, 129–139. [Google Scholar] [CrossRef][Green Version]
- Loker, E.S. Gastropod Immunobiology. In Invertebrate Immunity; Söderhäll, K., Ed.; Springer: Austin, TX, USA, 2010; pp. 17–43. [Google Scholar]
- El-Ghany, A.M.A.; El-Ghany, N.M.A. Molluscicidal activity of Bacillus thuringiensis strains against Biomphalaria alexandrina snails. Beni Suef Univ. J. Basic Appl. Sci. 2017, 6, 391–393. [Google Scholar] [CrossRef]
- Chattopadhyay, P. Characterization of Bacterial Isolates as natural Biocontrol Agents of Bollworm from an Epizootic Pest (Heliothis armigera). Pest Technol. 2011, 5, 81–85. [Google Scholar]
- Jaki, B.; Orjala, J.; Bürgi, H.R.; Sticher, O. Biological screening of cyanobacteria for antimicrobial and molluscicidal activity, brine shrimp lethality, and cytotoxicity. Pharm. Biol. 1999, 37, 138–143. [Google Scholar] [CrossRef]
- Lamovšek, J.; Urek, G.; Trdan, S. Biological control of root-knot nematodes (Meloidogyne spp.): Microbes against the pests. Acta Agric. Slov. 2013, 101, 263–275. [Google Scholar] [CrossRef]
- Cakici, F.O.; Sevim, A.; Demirbağ, Z.; Demir, I. Investigating internal bacteria of Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae) larvae and some Bacillus strains as biocontrol agents. Turk. J. Agric. For. 2014, 38, 99–110. [Google Scholar] [CrossRef]
- Sicuia, O.; Dinu, S.; Dinu, M.; Fătu, C.; Valimareanu, D.; Mincea, C.; Constantinescu, F. Pests and diseases management using compatible biocontrol bacteria and entomopathogenic fungal strains. Sci. Bull. Ser. F Biotechnol. 2014, 18, 66–72. [Google Scholar]
- Singer, S.F.; van Fleet, A.L.; Viel, J.J.; Genevese, E.E. Biological control of the zebra mussel Dreissena polymorpha and the snail Biomphalaria glabrata, using Gramicidin S and D and molluscicidal strains of Bacillus. J. Ind. Microbiol. Biotechnol. 1997, 18, 226–231. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, N.; Xie, S.; Zhang, X.; He, J.; Muhammad, A.; Sun, C.; Lu, X.; Shao, Y. Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 2020, 143, 105886. [Google Scholar] [CrossRef]
- Paddock, K.J.; Pereira, A.E.; Finke, D.L.; Ericsson, A.C.; Hibbard, B.E.; Shelby, K.S. Host resistance to Bacillus thuringiensis is linked to altered bacterial community within a specialist insect herbivore. Mol. Ecol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, X.; Zhang, F. New Insights into Cockroach Control: Using Functional Diversity of Blattella germanica Symbionts. Insects 2020, 11, 696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sun, X.X.; Zhang, X.C.; Zhang, S.; Lu, J.; Xia, Y.M.; Huang, Y.H.; Wang, X.J. The interactions between gut microbiota and entomopathogenic fungi: A potential approach for biological control of Blattella germanica (L.). Pest Manag. Sci. 2018, 74, 438–447. [Google Scholar] [CrossRef]
- Oliver, K.; Smith, A.; Russell, J.A. Defensive symbiosis in the real world-advancing ecological studies of heritble, protective bacteria in aphids and beyond. Funct. Ecol. 2014, 28, 341–355. [Google Scholar] [CrossRef]
- Oliver, K.M.; Russell, J.A.; Moran, N.A.; Hunter, M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 2003, 100, 1803–1807. [Google Scholar] [CrossRef]
- Frago, E.; Mala, M.; Weldegergis, B.T.; Yang, C.; McLean, A.; Godfray, H.C.J.; Gols, R.; Dicke, M. Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 2017, 8, 1860. [Google Scholar] [CrossRef]
- Meriweather, M.; Matthews, S.; Rio, R.; Baucom, R.S. A 454 Survey Reveals the Community Composition and Core Microbiome of the Common Bed Bug (Cimex lectularius) across an Urban Landscape. PLoS ONE 2013, 8, e61465. [Google Scholar] [CrossRef]
- Broderick, N.; Raffa, K.F.; Goodman, R.M.; Handelsman, J. Census of the Bacterial Community of the Gypsy Moth Larval Midgut by Using Culturing and Culture-Independent Methods. Appl. Environ. Microbiol. 2004, 70, 293–300. [Google Scholar] [CrossRef]
- Mc Donnell, R.J.; Paine, T.D.; Gormally, M.J. Slugs: A Guide to the Invasive and Native Fauna of California; UCANR Press: Davis, CA, USA, 2009; pp. 1–21. [Google Scholar]
- Douglas, M.R.; Tooker, J.F. Slug (Mollusca: Agriolimacidae, Arionidae) Ecology and Management in No-Till Field Crops, with an Emphasis on the mid-Atlantic Region. J. Integr. Pest Manag. 2012, 3, C1–C9. [Google Scholar] [CrossRef]
- Capinera, J.L. Culture of Terrestrial Slugs and Snails (Gastropoda): Acceptance and Suitability of Synthetic Insect Diets. Fla. Entomol. 2012, 95, 1077–1085. [Google Scholar] [CrossRef]
- Jensen, K.; Engelke, S.; Simpson, S.J.; Mayntz, D.; Hunt, J. Balancing of specific nutrients and subsequent growth and body composition in the slug Arion lusitanicus. Physiol. Behav. 2013, 122, 84–92. [Google Scholar] [CrossRef][Green Version]
- Rueda, A.A.; Slansky, F., Jr.; Wheeler, G.S. Compensatory Feeding Response of the Slug Sarasinula plebeia to Dietary Dilution. Oecologia 1991, 88, 181–188. [Google Scholar] [CrossRef]
- Udaka, H.; Goto, S.G.; Numata, H. Effects of photoperiod and acclimation temperature on heat and cold tolerance in the terrestrial slug, Lehmannia valentiana (Pulmonata: Limacidae). Appl. Entomol. Zool. 2008, 43, 547–551. [Google Scholar] [CrossRef]
- Udaka, H.; Mori, M.; Goto, S.G.; Numata, H. Seasonal reproductive cycle in relation to tolerance to high temperatures in the terrestrial slug Lehmannia valentiana. Invertebr. Biol. 2007, 126, 154–162. [Google Scholar] [CrossRef]
- Faberi, A.J.; López, A.N.; Manetti, P.L.; Clemente, N.L.; Álvarez Castillo, H.A. Growth and reproduction of the slug Deroceras laeve (Müller) (Pulmonata: Stylommatophora) under controlled conditions. Span. J. Agric. Res. 2006, 4, 345–350. [Google Scholar] [CrossRef]
- Zotin, A.A. Patterns of individual growth of gray garden slug Deroceras reticulatum. Biol. Bull. 2007, 34, 457–462. [Google Scholar] [CrossRef]
- Rae, R.G.; Robertson, J.F.; Wilson, M.J. Susceptibility and immune response of Deroceras reticulatum, Milax gagates and Limax pseudoflavus exposed to the slug parasitic nematode Phasmarhabditis hermaphrodita. J. Invertebr. Pathol. 2008, 97, 61–69. [Google Scholar] [CrossRef]
- Wilson, M.; Robertson, J.F.; Rae, R.G. The chemotactic response of Phasmarhabditis hermaphrodita (Nematoda: Rhabditida) to cues of Deroceras reticulatum (Mollusca: Gastropoda). Nematology 2006, 8, 197–200. [Google Scholar] [CrossRef]
- Kaya, H.; Mitani, D. Molluscicidal nematodes for the biological control of pest slugs. Slosson Rep. 2000, 14, 1–5. [Google Scholar]
- Grewal, S.K.; Grewal, P.S.; Hammond, R.B. Susceptibility of North American Native and Non-native Slugs (Mollusca: Gastropoda) to Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae). Biocontrol Sci. Technol. 2003, 13, 119–125. [Google Scholar] [CrossRef]
- Pechova, H.; Foltan, P. The parasitic nematode Phasmarhabditis hermaphrodita defends its slug host from being predated or scavenged by manipulating host spatial behaviour. Behav. Processes 2008, 78, 416–420. [Google Scholar] [CrossRef]
- Rae, R.G.; Robertson, J.F.; Wilson, M.J. Chemoattraction and Host Preference of the Gastropod Parasitic Nematode Phasmarhabditis hermaphrodita. J. Parasitol. 2009, 95, 517–526. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Correspondence QIIME allows analysis of high-throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Publ. Gr. 2010, 7, 335–336. [Google Scholar]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-liggett, C.; Knight, R.; Gordon, J.I. The human microbiome project: Exploring ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Vegan: Community Ecology Package. R Package Version 2.0-2. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 16 June 2021).
- Kassambara, A. Machine Learning Essentials: Practical Guide in R. Available online: http://www.sthda.com (accessed on 16 June 2021).
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Maltz, M.; Jackson, D.; Freund, H.; Aronson, E. Dryad Dataset: Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California. 2021. Available online: https://doi.org/10.6086/D1W39K (accessed on 16 June 2021).
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef] [PubMed]
- Landry, M.; Comeau, A.M.; Derome, N.; Cusson, M.; Levesque, R.C. Composition of the Spruce Budworm (Choristoneura fumiferana) Midgut Microbiota as Affected by Rearing Conditions. PLoS ONE 2015, 10, e0144077. [Google Scholar] [CrossRef]
- Bonnet, M.; Lagier, J.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2020, 34, 100622. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208–236. [Google Scholar] [CrossRef]
- Bing, X.; Gerlach, J.; Loeb, G.; Buchon, N. Nutrient-Dependent Impact of Microbes on Drosophila suzukii Development. mBio 2018, 9, e02199-17. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, P. Evolutionary Relationships of Primary Prokaryotic Endosymbionts of Whiteflies and Their Hosts. Appl. Environ. Microbiol. 2004, 70, 3401–3406. [Google Scholar] [CrossRef]
- Russell, J.A.; Moreau, C.S.; Goldman-Huertas, B.; Fujiwara, M.; Lohman, D.J.; Pierce, N.E. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. USA 2009, 106, 21236–21241. [Google Scholar] [CrossRef]
- Jones, R.T.; Sanchez, L.G.; Fierer, N. A Cross-Taxon Analysis of Insect-Associated Bacterial Diversity. PLoS ONE 2013, 8, e61218. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D. The Slug Microbiome: Elucidating the Influences of an Invasive Slug on Soil Bacterial Communities. Ph.D. Thesis, The University of California, Riverside, CA, USA, 2021. [Google Scholar]
- Fokin, S.I.; Giamberini, L.; Molloy, D.P.; de Vaate, A.B. Bacterial endocytobionts within endosymbiotic ciliates in Dreissena polymorpha (Lamellibranchia: Mollusca). Acta Protozool. 2003, 42, 31–39. [Google Scholar]
- Latorre, A.; Moya, A.; Gil, R.; Latorre, A.; Moya, A. Bacterial endosymbionts of insects: Insights from comparative genomics. Environ. Microbiol. 2004, 6, 1109–1122. [Google Scholar]
- Heddi, A.; Gross, R. Proteobacteria as Primary Endosymbionts of Arthropods. In Manipulative Tenants: Bacteria Associated with Arthropods; CRC Press: Boca Raton, FL, USA, 2018; pp. 39–66. [Google Scholar]
- Ip, J.C.-H.; Xu, T.; Sun, J.; Li, R.; Chen, C.; Lan, Y.; Han, Z.; Zhang, H.; Wei, J.; Wang, H.; et al. Host-Endosymbiont Genome Integration in a Deep-Sea Chemosymbiotic Clam. Mol. Biol. Evol. 2021, 38, 502–518. [Google Scholar] [CrossRef] [PubMed]
- Thao, M.L.; Gullan, P.J.; Baumann, P. Secondary (γ-Proteobacteria ) Endosymbionts Infect the Primary (β-Proteobacteria ) Endosymbionts of Mealybugs Multiple Times and Coevolve with Their Hosts. Appl. Environ. Microbiol. 2002, 68, 3190–3197. [Google Scholar] [CrossRef]
- Hammer, T.J.; Bowers, M.D. Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia 2015, 179, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Dubreuil, G.; Giron, D.; Simon, J.-C. Plant-insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 2015, 66, 467–478. [Google Scholar] [CrossRef]
- Brossi, M.J.D.L.; Jiménez, D.J.; Cortes-Tolalpa, L.; van Elsas, J.D. Soil-Derived Microbial Consortia Enriched with Different Plant Biomass Reveal Distinct Players Acting in Lignocellulose Degradation. Microb. Ecol. 2016, 71, 616–627. [Google Scholar] [CrossRef]
- Wilkinson, P.G. Characterisation of the Bacterial Flora Associated with the Grey Field Slug Deroceras reticulatum and Assessment of Its Suitability as a Target for Biological Control. Ph.D. Thesis, The University of Edinburgh, Edinburgh, UK, 2010. [Google Scholar]
- Umunna, O.E.; Austin, A.A. An Overview of Characterization and Identification of Soft Rot Bacterium Erwinia in Some Vegetable Crops. Green. J. Biol. Sci. 2016, 6, 046–055. [Google Scholar]
- Putnam, M.L.; Miller, M.L. Rhodococcus fascians in Herbaceous Perennials. Plant Dis. 2007, 91, 1064–1076. [Google Scholar] [CrossRef] [PubMed]
- Heydari, A.; Khodakaramian, G.; Zafari, D. Characterization of Pseudomonas viridiflava Causing Alfalfa Root Rot Disease in Hamedan Province of Iran. Plant Pathol. Microbiol. 2012, 3, 135. [Google Scholar] [CrossRef]
- Lim, K.C.; Yusoff, F.M.; Shariff, M.; Kamarudin, M.S. Astaxanthin as feed supplement in aquatic animals. Rev. Aquac. 2018, 10, 738–773. [Google Scholar] [CrossRef]
- Davinelli, S.; Nielsen, M.E.; Scapagnini, G. Astaxanthin in Skin Health, Repair, and Disease: A Comprehensive Review. Nutrients 2018, 10, 522. [Google Scholar] [CrossRef]
- Harker, M.; Hirschberg, J.; Oren, A. Paracoccus marcusii sp. nov., an orange gram-negative coccus. Int. J. Syst. Bacteriol. 1998, 48 Pt 2, 543–548. [Google Scholar] [CrossRef]
- Sharma, A.; Thakur, D.R.; Kanwar, S.; Chandla, V.K. Diversity of entomopathogenic bacteria associated with the white grub, Brahmina coriacea. J. Pest Sci. 2013, 86, 261–273. [Google Scholar] [CrossRef]
- Hutcheson, J.; Balaji, U.; Porembka, M.R.; Wachsmann, M.B.; McCue, P.A.; Knudsen, E.S.; Witkiewicz, A.K. Immunological and Metabolic Features of Pancreatic Ductal Adenocarcinoma Define Prognostic Subtypes of Disease. Clin. Cancer Res. 2016, 22, 3606–3617. [Google Scholar] [CrossRef]
- Proença, D.; Fonseca, L.; Powers, T.O.; Abrantes, I.; Morais, P.V. Diversity of Bacteria Carried by Pinewood Nematode in USA and Phylogenetic Comparison with Isolates from Other Countries. PLoS ONE 2014, 9, e105190. [Google Scholar] [CrossRef]
- Baquiran, J.-P.; Thater, B.; Sedky, S.; de Ley, P.; Crowley, D.; Orwin, P.M. Culture-Independent Investigation of the Microbiome Associated with the Nematode Acrobeloides maximus. PLoS ONE 2013, 8, e67425. [Google Scholar] [CrossRef]
- Hu, G.; Zhang, L.; Yun, Y.; Peng, Y. Taking insight into the gut microbiota of three spider species: No characteristic symbiont was found corresponding to the special feeding style of spiders. Ecol. Evol. 2019, 9, 8146–8156. [Google Scholar] [CrossRef]
- Monteiro, C.C.; Villegas, L.E.M.; Campolina, T.B.; Pires, A.C.M.A.; Miranda, J.C.; Pimenta, P.F.P.; Secundino, N.F.C. Bacterial diversity of the American sand fly Lutzomyia intermedia using high-throughput metagenomic sequencing. Parasites Vectors 2016, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, H. The Impact of Environmental Heterogeneity and Life Stage on the Hindgut Microbiota of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae). PLoS ONE 2013, 8, e57169. [Google Scholar] [CrossRef]
- Reid, N.M.; Addison, S.L.; West, M.A.; Lloyd-Jones, G. The bacterial microbiota of Stolotermes ruficeps (Stolotermitidae), a phylogenetically basal termite endemic to New Zealand. FEMS Microbiol. Ecol. 2014, 90, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Bredon, M.; Dittmer, J.; Noël, C.; Moumen, B.; Bouchon, D. Lignocellulose degradation at the holobiont level: Teamwork in a keystone soil invertebrate. Microbiome 2018, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Wang, H.; Xiang, X.; Wang, R.; Xu, Y. Structural Variations of Bacterial Community Driven by Sphagnum Microhabitat Differentiation in a Subalpine Peatland. Front. Microbiol. 2019, 10, 1661. [Google Scholar] [CrossRef]
- Tláskal, V.; Zrůstová, P.; Vrška, T.; Baldrian, P. Bacteria associated with decomposing dead wood in a natural temperate forest. FEMS Microbiol. Ecol. 2017, 93, fix157. [Google Scholar] [CrossRef] [PubMed]
| Bacterial Genus | Bacterial Family |
|---|---|
| Citrobacter | Enterobacteriaceae |
| Delftia | Comamonadaceae |
| Erwinia | Enterobacteriaceae |
| Arthrobacter | Micrococcaceae |
| Stenotrophomonas | Xanthomonadaceae |
| Pseudomonas | Pseudomonadaceae |
| Rhodococcus | Nocardiaceae |
| Bacillus | Bacillaceae |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, D.; Maltz, M.R.; Freund, H.L.; Borneman, J.; Aronson, E. Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California. Insects 2021, 12, 575. https://doi.org/10.3390/insects12070575
Jackson D, Maltz MR, Freund HL, Borneman J, Aronson E. Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California. Insects. 2021; 12(7):575. https://doi.org/10.3390/insects12070575
Chicago/Turabian StyleJackson, Denise, Mia R. Maltz, Hannah L. Freund, James Borneman, and Emma Aronson. 2021. "Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California" Insects 12, no. 7: 575. https://doi.org/10.3390/insects12070575
APA StyleJackson, D., Maltz, M. R., Freund, H. L., Borneman, J., & Aronson, E. (2021). Environment and Diet Influence the Bacterial Microbiome of Ambigolimax valentianus, an Invasive Slug in California. Insects, 12(7), 575. https://doi.org/10.3390/insects12070575






