Enhanced Myc Expression in Silkworm Silk Gland Promotes DNA Replication and Silk Production
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Strain
2.2. Construction of Recombinant Plasmid
2.3. DNA Quantification
2.4. Immunostaining
2.5. RNA Extraction and Quantitative Real-Time RT-PCR (RT-qPCR)
2.6. Statistical Methods
3. Results
3.1. Construction of Transgenic Silkworm with PSG-Specific Myc Overexpression
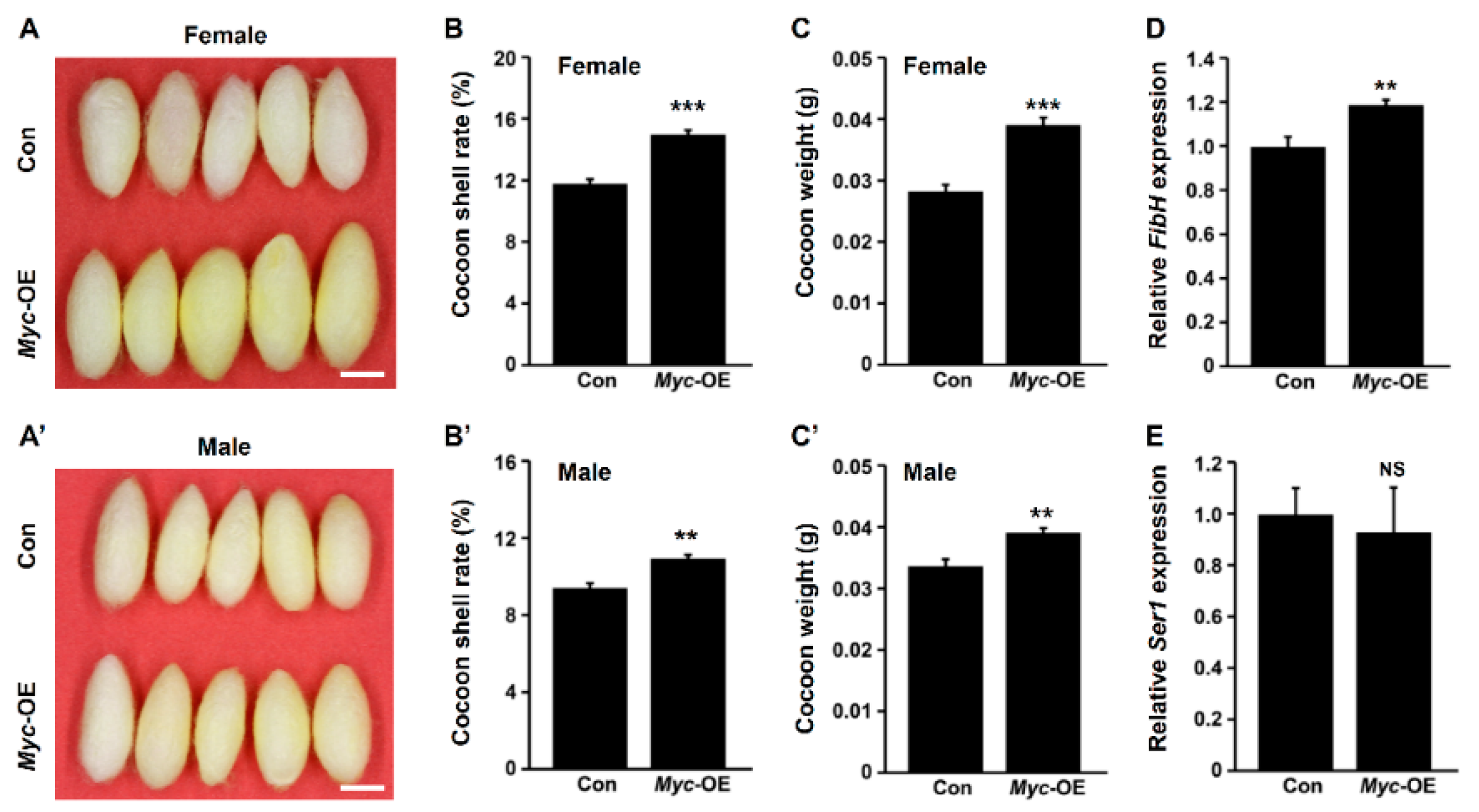
3.2. PSG-Specific Myc Overexpression Improves Silk Yield
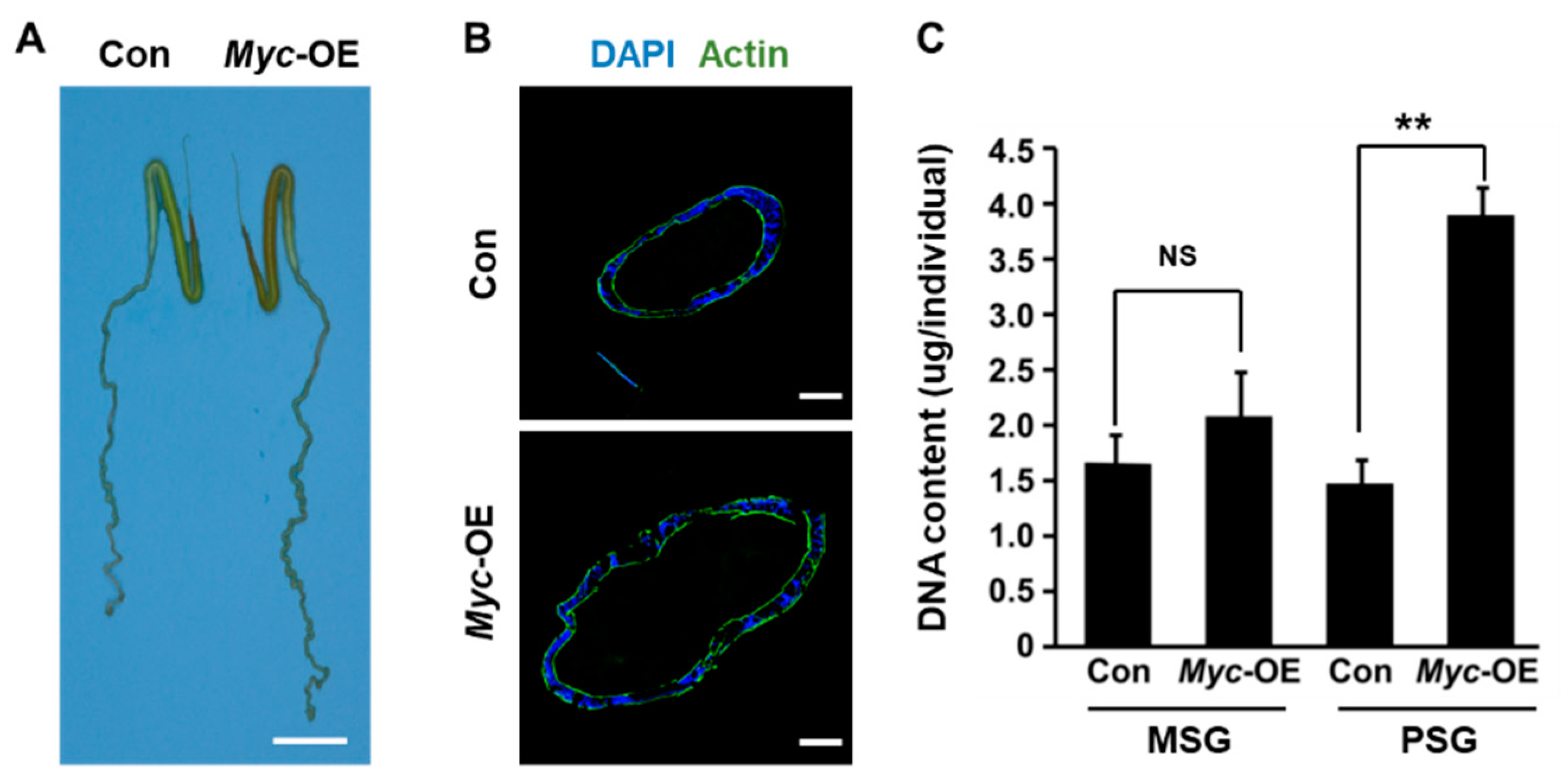
3.3. PSG-specific Myc Overexpression Promotes DNA Replication
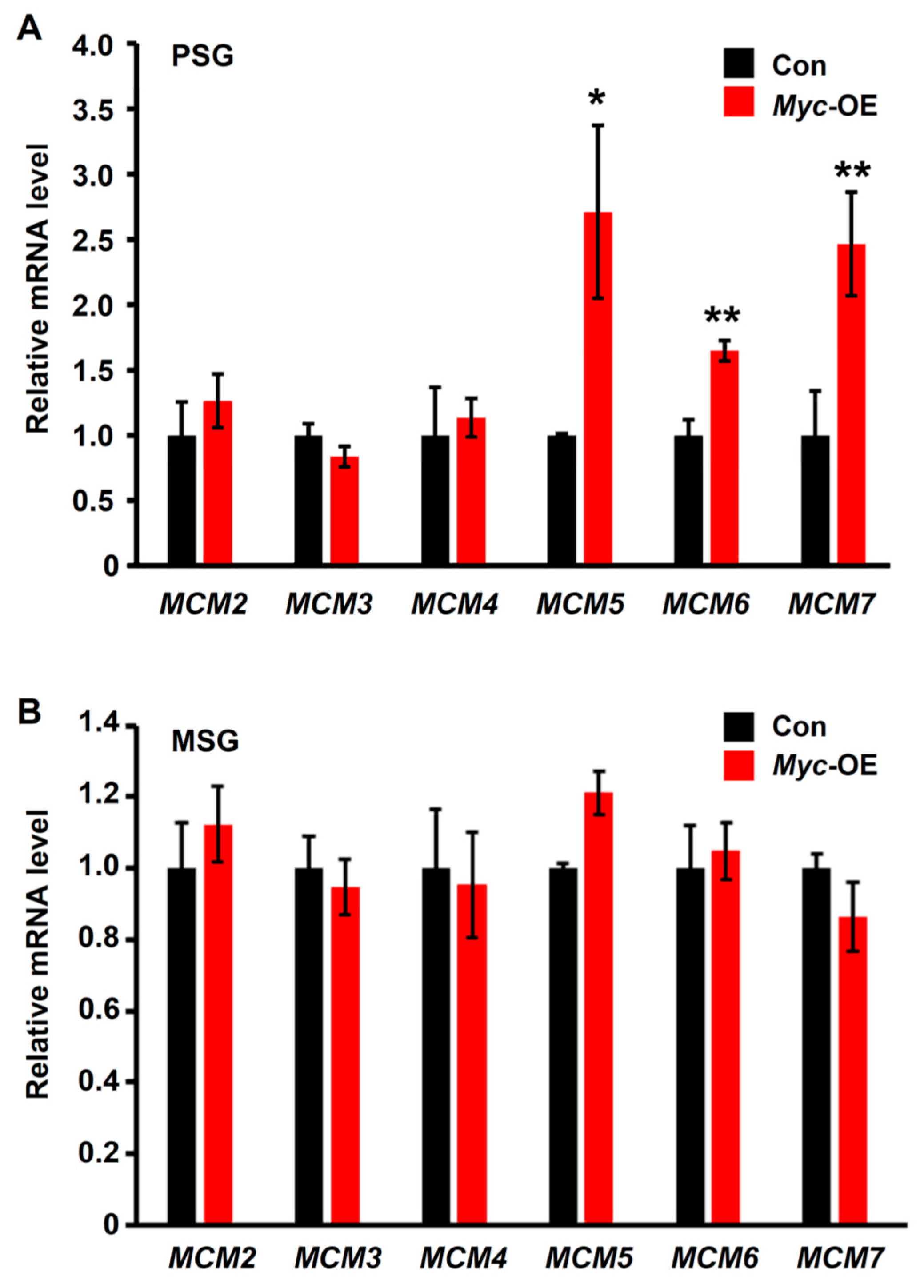
3.4. Myc Positively Regulates the Transcription of the MCM Genes Involving in DNA Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Perdrix-Gillot, S. DNA synthesis and endomitoses in the giant nuclei of the silkgland of Bombyx mori. Biochimie 1979, 61, 171–204. [Google Scholar] [CrossRef]
- Henderson, S.C.; Locke, M. The development of branched silk gland nuclei. Tissue Cell 1991, 23, 867–880. [Google Scholar] [CrossRef]
- Niranjanakumari, S.; Gopinathan, K.P. DNA polymerase Delta from the silk glands of Bombyx mori. J. Biol. Chem. 1992, 267, 17531–17539. [Google Scholar] [CrossRef]
- Dhawan, S.; Gopinathan, K.P. Cell cycle events during the development of the silk glands in the mulberry silkworm Bombyx mori. Dev. Genes Evol. 2003, 213, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell. Biol. 2014, 15, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Zielke, N.; Edgar, B.A.; De Pamphilis, M.L. Endoreplication. Cold Spring Harb. Perspect. Biol. 2013, 5, a012948. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, S.J.; Lehner, C.F. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 1997, 90, 671–681. [Google Scholar] [CrossRef]
- Schaeffer, V.; Althauser, C.; Shcherbata, H.R.; Deng, W.M.; Ruohola-Baker, H. Notch-dependent Fizzy-related/Hec1/Cdh1 expression is required for the mitotic-to-endocycle transition in Drosophila follicle cells. Curr. Biol. 2004, 14, 630–636. [Google Scholar] [CrossRef]
- Shcherbata, H.R.; Althauser, C.; Findley, S.D.; Ruohola-Baker, H. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 2004, 131, 3169–3181. [Google Scholar] [CrossRef]
- Qian, W.; Li, Z.; Song, W.; Zhao, T.; Wang, W.; Peng, J.; Wei, L.; Xia, Q.; Cheng, D. A novel transcriptional cascade is involved in Fzr-mediated endoreplication. Nucl. Acids Res. 2020, 48, 4214–4229. [Google Scholar] [CrossRef]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell. Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef]
- Costa, A.; Hood, I.V.; Berger, J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013, 82, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef]
- Eward, K.L.; Obermann, E.C.; Shreeram, S.; Loddo, M.; Fanshawe, T.; Williams, C.; Jung, H.I.; Prevost, A.T.; Blow, J.J.; Stoeber, K.; et al. DNA replication licensing in somatic and germ cells. J. Cell Sci. 2004, 117, 5875–5886. [Google Scholar] [CrossRef]
- Ma, L.; Xu, H.; Zhu, J.; Ma, S.; Liu, Y.; Jiang, R.J.; Xia, Q.; Li, S. Ras1(CA) overexpression in the posterior silk gland improves silk yield. Cell Res. 2011, 21, 934–943. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Tang, X.; Zhang, C.; Wang, L.; Chen, P.; Pan, M.; Lu, C. DNA synthesis during endomitosis is stimulated by insulin via the PI3K/Akt and TOR signaling pathways in the silk gland cells of Bombyx mori. Int. J. Mol. Sci. 2015, 16, 6266–6280. [Google Scholar] [CrossRef]
- Li, Y.F.; Chen, X.Y.; Zhang, C.D.; Tang, X.F.; Wang, L.; Liu, T.H.; Pan, M.H.; Lu, C. Effects of starvation and hormones on DNA synthesis in silk gland cells of the silkworm, Bombyx mori. Insect Sci. 2016, 23, 569–578. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, S.; Song, H.S.; Zhang, G.; Jia, Q.; Li, S. Yorkie (CA) overexpression in the posterior silk gland improves silk yield in Bombyx mori. J. Insect Physiol. 2017, 100, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, S.; Chang, J.; Zhang, T.; Wang, X.; Shi, R.; Zhang, J.; Lu, W.; Liu, Y.; Xia, Q. Tissue-specific genome editing of laminA/C in the posterior silk glands of Bombyx mori. J. Genet. Genom. 2017, 44, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Roussel, M.; Saule, S.; Lagrou, C.; Rommens, C.; Beug, H.; Graf, T.; Stehelin, D. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature 1979, 281, 452–455. [Google Scholar] [CrossRef]
- Fulco, C.P.; Munschauer, M.; Anyoha, R.; Munson, G.; Grossman, S.R.; Perez, E.M.; Kane, M.; Cleary, B.; Lander, E.S.; Engreitz, J.M. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 2016, 354, 769–773. [Google Scholar] [CrossRef]
- Scognamiglio, R.; Cabezas-Wallscheid, N.; Thier, M.C.; Altamura, S.; Reyes, A.; Prendergast, A.M.; Baumgartner, D.; Carnevalli, L.S.; Atzberger, A.; Haas, S.; et al. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell 2016, 164, 668–680. [Google Scholar] [CrossRef]
- Fan, L.; Peng, G.; Sahgal, N.; Fazli, L.; Gleave, M.; Zhang, Y.; Hussain, A.; Qi, J. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene 2016, 35, 2441–2452. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Tanaka, T.; Ogonuki, N.; Ogura, A.; Morimoto, H.; Cheng, P.F.; Eisenman, R.N.; Trumpp, A.; Shinohara, T. Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 2016, 30, 2637–2648. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 427–437. [Google Scholar] [CrossRef]
- Adhikary, S.; Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 2005, 6, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Gautier, J. MYC and the control of DNA replication. Cold Spring Harb. Perspect. Med. 2014, 4, a014423. [Google Scholar] [CrossRef]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Maines, J.Z.; Stevens, L.M.; Tong, X.; Stein, D. Drosophila dMyc is required for ovary cell growth and endoreplication. Development 2004, 131, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Osanai, K.; Ohyoshi, T.; Wang, H.B.; Iwanaga, M.; Kawasaki, H. Ecdysteroid promotes cell cycle progression in the Bombyx wing disc through activation of c-Myc. Insect Biochem. Mol. Biol. 2016, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mon, H.; Li, Z.; Kobayashi, I.; Tomita, S.; Lee, J.; Sezutsu, H.; Tamura, T.; Kusakabe, T. Soaking RNAi in Bombyx mori BmN4-SID1 cells arrests cell cycle progression. J. Insect Sci. 2013, 13, 155. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Yang, Y.; Wang, P.; Zhou, X.; Zhao, T.; Guo, M.; Meng, M.; Zhang, T.; Qian, W.; et al. Comparative transcriptome analysis provides novel insight into morphologic and metabolic changes in the fat body during silkworm metamorphosis. Int. J. Mol. Sci. 2018, 19, 3525. [Google Scholar] [CrossRef]
- Pierce, S.B.; Yost, C.; Britton, J.S.; Loo, L.W.; Flynn, E.M.; Edgar, B.A.; Eisenman, R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development 2004, 131, 2317–2327. [Google Scholar] [CrossRef]
- Frigola, J.; He, J.; Kinkelin, K.; Pye, V.E.; Renault, L.; Douglas, M.E.; Remus, D.; Cherepanov, P.; Costa, A.; Diffley, J.F.X. Cdt1 stabilizes an open MCM ring for helicase loading. Nat. Commun. 2017, 8, 15720. [Google Scholar] [CrossRef]
- Fernandez-Cid, A.; Riera, A.; Tognetti, S.; Herrera, M.C.; Samel, S.; Evrin, C.; Winkler, C.; Gardenal, E.; Uhle, S.; Speck, C. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol. Cell 2013, 50, 577–588. [Google Scholar] [CrossRef]
- Zhao, X.M.; Liu, C.; Li, Q.Y.; Hu, W.B.; Zhou, M.T.; Nie, H.Y.; Zhang, Y.X.; Peng, Z.C.; Zhao, P.; Xia, Q.Y. Basic helix-loop-helix transcription factor Bmsage is involved in regulation of fibroin H-chain gene via interaction with SGF1 in Bombyx mori. PLoS ONE 2014, 9, e94091. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zha, X.; Shi, P.; Zhao, P.; Wang, H.; Zheng, R.; Xia, Q. Nuclear hormone receptor BmFTZ-F1 is involved in regulating the fibroin heavy chain gene in the silkworm, Bombyx mori. Biochim. Biophys. Acta 2016, 1860, 2529–2536. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Li, Y.; Guo, P.; Liu, C.; Li, Z.; Wang, F.; Zhao, P.; Xia, Q.; He, H. Insights into the repression of fibroin modulator binding protein-1 on the transcription of fibroin H-chain during molting in Bombyx mori. Insect Biochem. Mol. Biol. 2019, 104, 39–49. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, W.; Yang, Y.; Li, Z.; Wu, Y.; He, X.; Li, H.; Cheng, D. Enhanced Myc Expression in Silkworm Silk Gland Promotes DNA Replication and Silk Production. Insects 2021, 12, 361. https://doi.org/10.3390/insects12040361
Qian W, Yang Y, Li Z, Wu Y, He X, Li H, Cheng D. Enhanced Myc Expression in Silkworm Silk Gland Promotes DNA Replication and Silk Production. Insects. 2021; 12(4):361. https://doi.org/10.3390/insects12040361
Chicago/Turabian StyleQian, Wenliang, Yan Yang, Zheng Li, Yuting Wu, Xuechuan He, Hao Li, and Daojun Cheng. 2021. "Enhanced Myc Expression in Silkworm Silk Gland Promotes DNA Replication and Silk Production" Insects 12, no. 4: 361. https://doi.org/10.3390/insects12040361
APA StyleQian, W., Yang, Y., Li, Z., Wu, Y., He, X., Li, H., & Cheng, D. (2021). Enhanced Myc Expression in Silkworm Silk Gland Promotes DNA Replication and Silk Production. Insects, 12(4), 361. https://doi.org/10.3390/insects12040361




