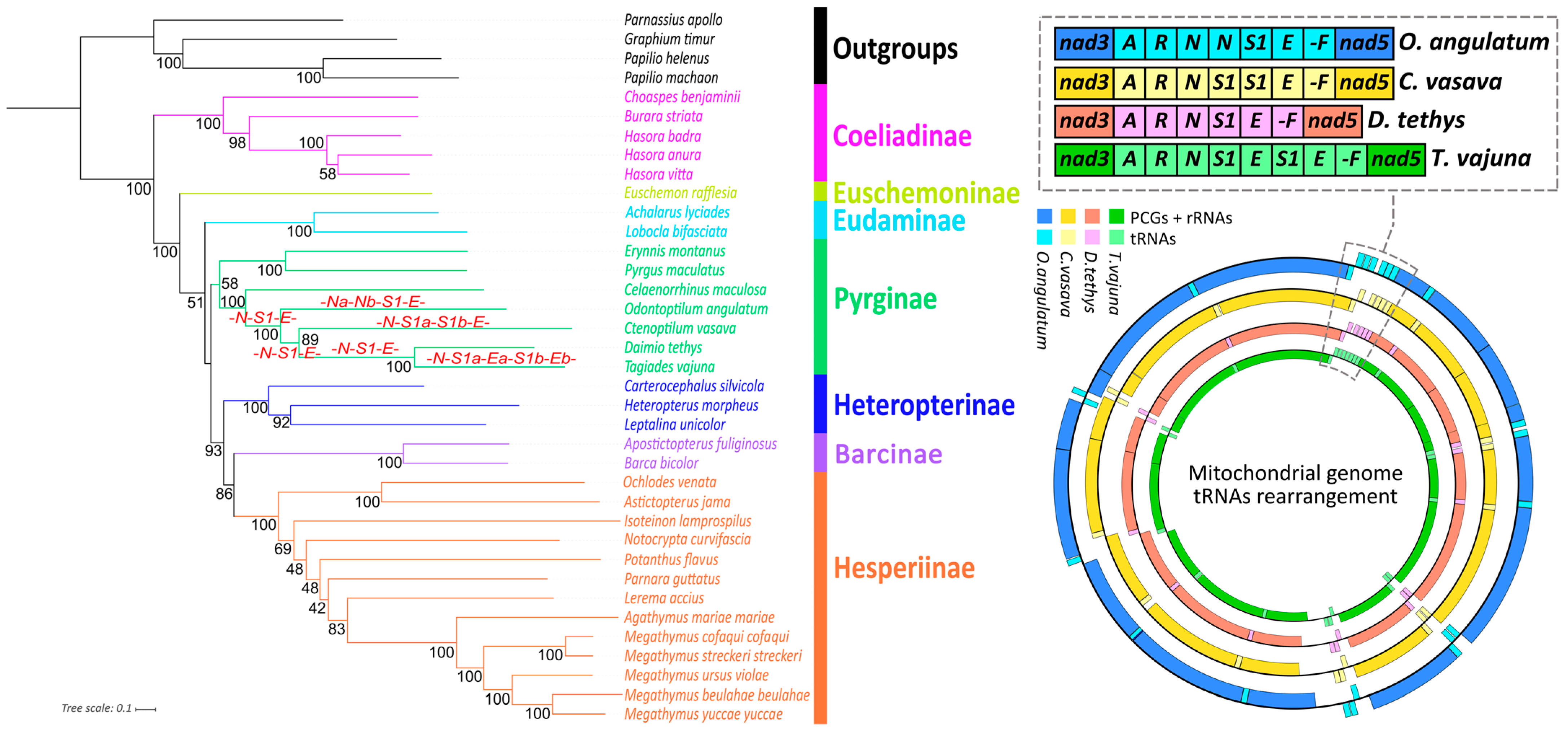
Unique Duplication of trnN in Odontoptilum angulatum (Lepidoptera: Pyrginae) and Phylogeny within Hesperiidae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sequencing
2.3. Phylogenetic Analysis
3. Results and Discussion
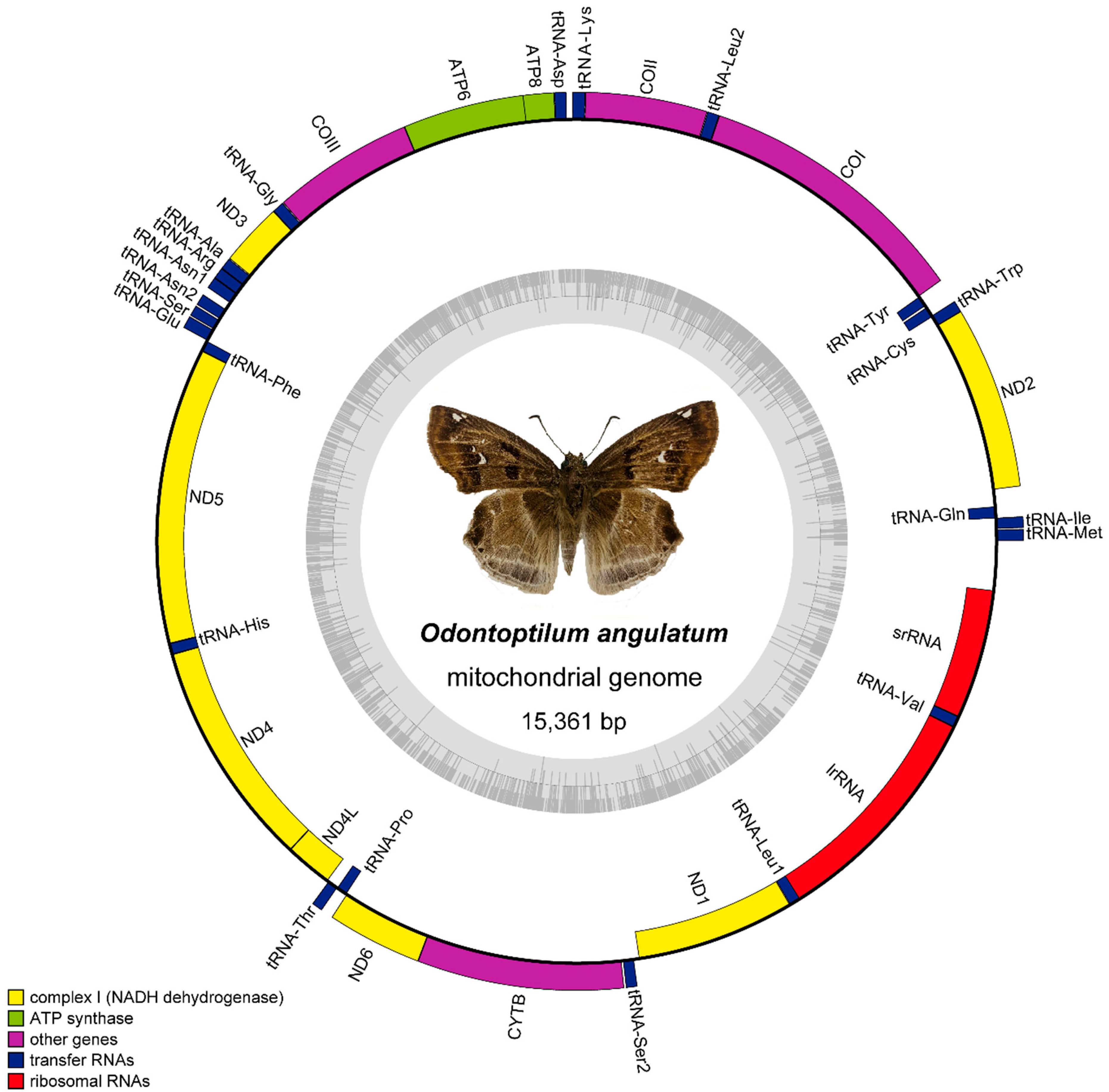
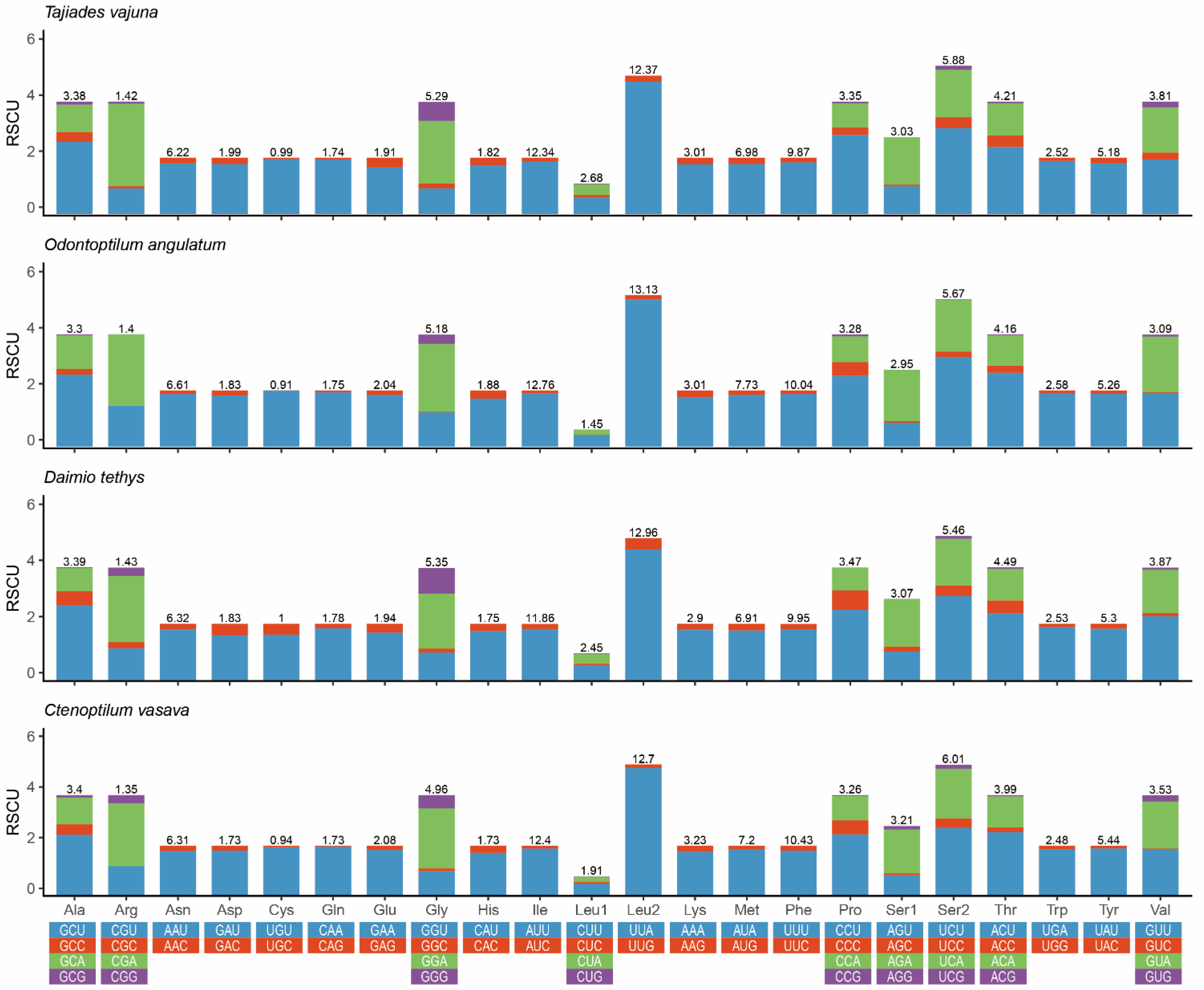
3.1. Mitochondrial Genome Organization
3.2. Non-Coding Regions (NCR) and a Pseudo Gene
3.3. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barr, C.M.; Neiman, M.; Taylor, D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Bae, J.S.; Kim, I.; Sohn, H.D.; Jin, B.R. The mitochondrial genome of the firefly, Pyrocoelia rufa: Complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol. Phylogenet. Evol. 2004, 32, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kang, A.R.; Jeong, H.C.; Kim, K.-G.; Kim, I. Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol. Phylogenet. Evol. 2011, 61, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Song, W.; Shi, S.L.; Liu, Y.Q.; Pan, M.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. Mitochondrial genome nucleotide substitution pattern between domesticated silkmoth, Bombyx mori, and its wild ancestors, Chinese Bombyx mandarina and Japanese Bombyx mandarina. Genet. Mol. Biol. 2010, 33, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Clayton, D.A. Replication of animal mitochondrial DNA. Cell 1982, 28, 693–705. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Yu, D.W.; Zhou, X.; Vogler, A.P. Mitochondrial metagenomics: Letting the genes out of the bottle. GigaScience 2016, 5, 15. [Google Scholar] [CrossRef]
- Gillett, C.P.; Crampton-Platt, A.; Timmermans, M.J.; Jordal, B.H.; Emerson, B.C.; Vogler, A.P. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol. Biol. Evol. 2014, 31, 2223–2237. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.Q.; Ma, C.; Chen, J.Y.; Yang, D.R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: The ancestral gene arrangement in Lepidoptera. BMC Genom. 2012, 13, 276. [Google Scholar] [CrossRef]
- Tian, L.L.; Sun, X.Y.; Chen, M.; Gai, Y.H.; Hao, J.S.; Yang, Q. Complete mitochondrial genome of the Five-dot Sergeant Parathyma sulpitia(Nymphalidae: Limenitidinae) and its phylogenetic implications. Zool. Res. 2012, 33, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kilpert, F.; Podsiadlowski, L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 2006, 7, 241. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Barker, S.C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): Convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003, 20, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.J.; Hodson, C.N.; Hamilton, P.T.; Opit, G.P.; Gowen, B.E. Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, 10162–10168. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Song, F.; Gu, W.; Feng, J.; Cai, W.; Shao, R. Novel insights into mitochondrial gene rearrangement in thrips (Insecta: Thysanoptera) from the grass thrips, Anaphothrips obscurus. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Shao, R.; Zhu, X.Q.; Barker, S.C.; Herd, K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol. Evol. 2012, 4, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Li, H.; Liu, G.-H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.; Cai, W.; Shao, R. Mitochondrial genome fragmentation unites the parasitic lice of eutherian mammals. Syst. Biol. 2019, 68, 430–440. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, Y.; Yang, C.; Gu, X.; Wilson, J.J.; Li, H.; Cai, W.; Yang, H.; Song, F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int. J. Biol. Macromol. 2020, 164, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Dickey, A.M.; Kumar, V.; Morgan, J.K.; Jara-Cavieres, A.; Shatters, R.G.; McKenzie, C.L.; Osborne, L.S. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): Extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genom. 2015, 16, 439. [Google Scholar] [CrossRef]
- Moritz, C.; Brown, W.M. Tandem duplications in animal mitochondrial DNAs: Variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. USA 1987, 84, 7183–7187. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Poulton, J.; Deadman, M.E.; Bindoff, L.; Morten, K.; Land, J.; Brown, G. Families of mtDNA re-arrangements can be detected in patients with mtDNA deletions: Duplications may be a transient intermediate form. Hum. Mol. Genet. 1993, 2, 23–30. [Google Scholar] [CrossRef]
- Lunt, D.H.; Hyman, B.C. Animal mitochondrial DNA recombination. Nature 1997, 387, 247. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Campbell, N.J. Intramitochondrial recombination–is it why some mitochondrial genes sleep around? Trends Ecol. Evol. 2001, 16, 269–271. [Google Scholar] [CrossRef]
- Cantatore, P.; Gadaleta, M.; Roberti, M.; Saccone, C.; Wilson, A. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature 1987, 329, 853–855. [Google Scholar] [CrossRef] [PubMed]
- San Mauro, D.; Gower, D.J.; Zardoya, R.; Wilkinson, M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006, 23, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Johnson, K.P.; Sweet, A.D.; Yao, I.; Ferreira, R.L.; Cameron, S.L. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol. Phylogenet. Evol. 2018, 119, 118–127. [Google Scholar] [CrossRef]
- Rand, D.M. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol. Evol. 1994, 9, 125–131. [Google Scholar] [CrossRef]
- Warren, A.D.; Ogawa, J.R.; Brower, A.V. Phylogenetic relationships of subfamilies and circumscription of tribes in the family Hesperiidae (Lepidoptera: Hesperioidea). Cladistics 2008, 24, 642–676. [Google Scholar] [CrossRef]
- Warren, A.D.; Grishin, N.V. A new species of Oxynetra from Mexico (Hesperiidae, Pyrginae, Pyrrhopygini). ZooKeys 2017, 667, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Grishin, N.V.; Janzen, D.H.; Hallwachs, W. A new species of Eracon (Hesperiidae: Pyrginae) substantiated by a number of traits, including female genitalia. J. Lepid. Soc. 2014, 68, 149–161. [Google Scholar] [CrossRef]
- Grishin, N.V.; Burns, J.M.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Oxynetra: Facies and DNA barcodes point to a new species from Costa Rica (Hesperiidae: Pyrginae: Pyrrhopygini). J. Lepid. Soc. 2013, 67, 1–14. [Google Scholar] [CrossRef]
- Grishin, N.V. Adding to the rich fauna of the Chocó region in Ecuador, a new species of Potamanaxas (Hesperiidae: Pyrginae: Erynnini). Trop. Lepid. Res. 2013, 23, I–III. [Google Scholar]
- Ferrer-Paris, J.R.; Sánchez-Mercado, A.; Viloria, A.L.; Donaldson, J. Congruence and diversity of butterfly-host plant associations at higher taxonomic levels. PLoS ONE 2013, 8, e63570. [Google Scholar] [CrossRef]
- Yuan, F.; Yuan, X.Q.; Xue, G.X. Fauna Sinica (Insecta: Lepidoptera: Hesperiidae); Science Press: Beijing, China, 2015. [Google Scholar]
- Hao, J.S.; Sun, Q.Q.; Zhao, H.B.; Sun, X.Y.; Gai, Y.H.; Yang, Q. The complete mitochondrial genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and its phylogenetic implication. Comp. Funct. Genom. 2012, 2012, 328049. [Google Scholar] [CrossRef]
- Liu, F.F.; Li, Y.P.; Jakovlić, I.; Yuan, X.Q. Tandem duplication of two tRNA genes in the mitochondrial genome of Tagiades vajuna (Lepidoptera: Hesperiidae). Eur. J. Entomol. 2017, 114, 407–415. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Res. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Shen, J.; Wang, R.; Grishin, N.V. The complete mitochondrial genome of a skipper Burara striata (Lepidoptera: Hesperiidae). Mitochondrial DNA Part B 2017, 2, 145–147. [Google Scholar] [CrossRef]
- Kim, M.J.; Wang, A.R.; Park, J.S.; Kim, I. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene 2014, 549, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; James John, Y.; Xuan, S.; Cao, T.; Yuan, X. The complete mitochondrial genome of the butterfly Hasora anura (Lepidoptera: Hesperiidae). Mitochondrial DNA Part A 2015, 27, 4401–4402. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.M.; Wang, J.P.; James, J.Y.; Yau, S.M.; Yuan, X.Q.; Liu, J.P.; Cao, T.W. The complete mitochondrial genome of Hasora vitta (Butler, 1870) (Lepidoptera: Hesperiidae). Mitochondrial DNA Part A 2016, 27, 3020–3021. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Shen, J.; Fan, X.L.; Wang, M.; Grishin, N.V. The complete mitogenome of Euschemon rafflesia (Lepidoptera: Hesperiidae). Mitochondrial DNA Part B 2017, 2, 136–138. [Google Scholar] [CrossRef]
- Wang, K.; Hao, J.; Zhao, H. Characterization of complete mitochondrial genome of the skipper butterfly, Celaenorrhinus maculosus (Lepidoptera: Hesperiidae). Mitochondrial DNA 2015, 26, 690. [Google Scholar] [CrossRef] [PubMed]
- Zuo, N.; Gan, S.; Chen, Y.; Hao, J. The complete mitochondrial genome of the Daimio tethys (Lepidoptera: Hesperoidea: Hesperiidae). Mitochondrial DNA 2014, 27, 1099–1100. [Google Scholar] [CrossRef]
- Wang, A.R.; Jeong, H.C.; Han, Y.S.; Kim, I. The complete mitochondrial genome of the mountainous duskywing, Erynnis montanus (Lepidoptera: Hesperiidae): A new gene arrangement in Lepidoptera. Mitochondrial DNA 2014, 25, 93–94. [Google Scholar] [CrossRef]
- Shen, J.H.; Cong, Q.; Grishin, N.V. The complete mitogenome of Achalarus lyciades (Lepidoptera: Hesperiidae). Mitochondrial DNA Part B 2016, 1, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.Y.; Kim, M.J.; Jeong, N.R.; Kim, I. Complete mitochondrial genome of the silver stripped skipper, Leptalina unicolor (Lepidoptera: Hesperiidae). Mitochondrial DNA Part B 2019, 4, 3418–3420. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Huang, Z.; Tang, J.; Chiba, H.; Fan, X. The complete mitochondrial genomes of two skipper genera (Lepidoptera: Hesperiidae) and their associated phylogenetic analysis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Qian, C.; Grishin, N.V. The complete mitochondrial genome of Lerema accius and its phylogenetic implications. PeerJ 2016, 4, e1546. [Google Scholar]
- Shao, L.L.; Sun, Q.Q.; Hao, J.S. The complete mitochondrial genome of Parara guttata (Lepidoptera: Hesperiidae). Mitochondrial DNA 2015, 26, 724–725. [Google Scholar] [CrossRef]
- Ma, L.; Liu, F.; Chiba, H.; Yuan, X.Q. The mitochondrial genomes of three skippers: Insights into the evolution of the family Hesperiidae (Lepidoptera). Genomics 2019, 112, 432–441. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Fan, X.; Wang, R.; Wang, M.; Grishin, N.V. Mitogenomes of giant-skipper butterflies reveal an ancient split between deep and shallow root feeders. F1000Research 2017, 6, 222. [Google Scholar] [CrossRef]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X.; Liu, S.; Song, W.; Li, Y.; Wu, Q.; Zhang, A. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef]
- Chen, Y.; Gan, S.; Shao, L.; Cheng, C.; Hao, J. The complete mitochondrial genome of the Pazala timur (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA Part A 2016, 27, 533–534. [Google Scholar] [CrossRef]
- Chen, Y.H.; Huang, D.Y.; Wang, Y.L.; Zhu, C.D.; Hao, J.S. The complete mitochondrial genome of the endangered Apollo butterfly, Parnassius apollo (Lepidoptera: Papilionidae) and its comparison to other Papilionidae species. J. Asia-Pac. Entomol. 2014, 17, 663–671. [Google Scholar] [CrossRef]
- Zhang, D. MitoTool Software. Available online: https://github.com/dongzhang0725/MitoTool (accessed on 13 April 2021).
- Zhang, D. BioSuite Software. Available online: https://github.com/dongzhang0725/BioSuite (accessed on 13 April 2021).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Meth. 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Whiting, M.F. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 2008, 408, 112–123. [Google Scholar] [CrossRef]
- Kim, M.I.; Baek, J.Y.; Kim, M.J.; Jeong, H.C.; Kim, K.-G.; Bae, C.H.; Han, Y.S.; Jin, B.R.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol. Cells 2009, 28, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Z.M.; Gu, X.S.; Wang, M.; Huang, G.H.; Zwick, A. Phylogenetic relationships among Bombycidae sl (Lepidoptera) based on analyses of complete mitochondrial genomes. Syst. Entomol. 2019, 44, 490–498. [Google Scholar] [CrossRef]
- Chen, L.; Wahlberg, N.; Liao, C.-Q.; Wang, C.-B.; Ma, F.-Z.; Huang, G.-H. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics 2020, 112, 4435–4441. [Google Scholar] [CrossRef]
- Garey, J.R.; Wolstenholme, D.R. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989, 28, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y.; Nishida, M. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J. Mol. Evol. 1993, 37, 380–398. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Miura, S.; Yamada, C.; Hashiguchi, Y. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genom. 2014, 15, 930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Tamura, K.; Aotsuka, T. Replication origin of mitochondrial DNA in insects. Genetics 2005, 171, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In Comparative Genomics; Springer: Dordrecht, The Netherlands, 2000; pp. 133–147. [Google Scholar]
- Bernt, M.; Chen, K.Y.; Chen, M.C.; Chu, A.C.; Merkle, D.; Wang, H.L.; Chao, K.M.; Middendorf, M. Finding all sorting tandem duplication random loss operations. J. Discret. Algorithms 2011, 9, 32–48. [Google Scholar] [CrossRef][Green Version]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Lambkin, C.L.; Barker, S.C.; Whiting, M.F. A mitochondrial genome phylogeny of Diptera: Whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007, 32, 40–59. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Warren, A.D.; Wahlberg, N.; Brower, A.V.; Lukhtanov, V.A.; Kodandaramaiah, U. Ten genes and two topologies: An exploration of higher relationships in skipper butterflies (Hesperiidae). PeerJ 2016, 4, e2653. [Google Scholar] [CrossRef]
- Boore, J.L.; Lavrov, D.V.; Brown, W.M. Gene translocation links insects and crustaceans. Nature 1998, 392, 667–668. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]


| Taxon | Species | Accession Number | References |
|---|---|---|---|
| Hesperiidae | |||
| Coeliadinae | Burara striata | NC_034676 | [44] |
| Choaspes benjaminii | NC_024647 | [45] | |
| Hasora anura | KF881049 | [46] | |
| Hasora vitta | NC_027170 | [47] | |
| Hasora badra | NC_045249 | Unpublished | |
| Euschemoninae | Euschemon rafflesia | NC_034231 | [48] |
| Pyrginae | Celaenorrhinus maculosus | NC_022853 | [49] |
| Ctenoptilum vasava | JF713818 | [38] | |
| Tagiades (=Daimio) tethys | KJ813807 | [50] | |
| Erynnis montanus | NC_021427 | [51] | |
| Pyrgus maculatus | NC_030192 | Unpublished | |
| Tagiades vajuna | KX865091 | [39] | |
| Odontoptilum angulatum | MW381783 | This study | |
| Eudaminae | Achalarus lyciades | NC_030602 | [52] |
| Lobocla bifasciata | KJ629166 | [45] | |
| Heteropterinae | Carterocephalus silvicola | NC_024646 | [45] |
| Heteropterus morpheus | NC_028506 | Unpublished | |
| Leptalina unicolour | MK265705 | [53] | |
| Barcinae | Apostictopterus fuliginosus | NC_039946 | [54] |
| Barca bicolor | NC_039947 | [54] | |
| Hesperiinae | Lerema accius | NC_029826 | [55] |
| Ochlodes venata | HM243593 | Unpublished | |
| Parnara guttata | NC_029136 | [56] | |
| Potanthus flavus | KJ629167 | [45] | |
| Astictopterus jama | MH763663 | [57] | |
| Isoteinon lamprospilus | MH763664 | [57] | |
| Notocrypta curvifascia | MH763665 | [57] | |
| Agathymus mariae | KY630504 | [58] | |
| Megathymus beulahae | KY630505 | [58] | |
| Megathymus cofaqui | KY630503 | [58] | |
| Megathymus streckeri | KY630501 | [58] | |
| Megathymus ursus | KY630502 | [58] | |
| Megathymus yuccae | KY630500 | [58] | |
| Outgroup | |||
| Papilionidae | Papilio machaon | NC_018047 | Unpublished |
| Papilio helenus | NC_025757 | [59] | |
| Graphium timur | NC_024098 | [60] | |
| Parnassius apollo | NC_024727 | [61] |
| Position | Size (bp) | Intergenic Nucleotides | Codon | Strand | |||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| trnM | 1 | 66 | 66 | + | |||
| trnI | 72 | 135 | 64 | 5 | + | ||
| trnQ | 133 | 201 | 69 | −3 | - | ||
| nad2 | 300 | 1313 | 1014 | 98 | ATT | TAA | + |
| trnW | 1312 | 1378 | 67 | −2 | + | ||
| trnC | 1371 | 1435 | 65 | −8 | - | ||
| trnY | 1446 | 1510 | 65 | 10 | - | ||
| cox1 | 1513 | 3046 | 1534 | 2 | ATG | T | + |
| trnL2 | 3047 | 3113 | 67 | + | |||
| cox2 | 3115 | 3793 | 679 | 1 | ATG | T | + |
| trnK | 3794 | 3864 | 71 | + | |||
| trnD | 3898 | 3963 | 66 | 33 | + | ||
| atp8 | 3964 | 4137 | 174 | ATT | TAA | + | |
| atp6 | 4131 | 4808 | 678 | −7 | ATG | TAA | + |
| cox3 | 4808 | 5593 | 786 | −1 | ATG | TAA | + |
| trnG | 5596 | 5662 | 67 | 2 | + | ||
| nad3 | 5663 | 6016 | 354 | ATT | TAA | + | |
| trnA | 6019 | 6084 | 66 | 2 | + | ||
| trnR | 6085 | 6148 | 64 | + | |||
| trnN1 | 6149 | 6214 | 66 | + | |||
| trnN2 | 6259 | 6324 | 66 | 44 | + | ||
| trnS1 | 6332 | 6393 | 62 | 7 | + | ||
| trnE | 6399 | 6469 | 71 | 5 | + | ||
| trnF | 6473 | 6536 | 64 | 3 | + | ||
| nad5 | 6537 | 8280 | 1744 | - | |||
| trnH | 8281 | 8345 | 65 | ATA | T | - | |
| nad4 | 8346 | 9684 | 1339 | - | |||
| nad4L | 9685 | 9966 | 282 | ATG | T | - | |
| trnT | 9974 | 10,036 | 63 | 7 | ATG | TAA | - |
| trnP | 10,037 | 10,100 | 64 | + | |||
| nad6 | 10,103 | 10,633 | 531 | 2 | - | ||
| cytb | 10,633 | 11,781 | 1149 | −1 | ATT | TAA | + |
| trnS2 | 11,791 | 11,854 | 64 | 9 | ATG | TAA | + |
| nad1 | 11,885 | 12,823 | 939 | 30 | + | ||
| trnL1 | 12,825 | 12,891 | 67 | 1 | ATG | TAA | - |
| rrnL | 12,868 | 14,247 | 1380 | −24 | - | ||
| trnV | 14,248 | 14,313 | 66 | - | |||
| rrnS | 14,314 | 15,074 | 761 | - | |||
| A-T rich region | 15,075 | 15,361 | 287 | + | |||
| O. angulatum | ||||||||
|---|---|---|---|---|---|---|---|---|
| Regions | Size (bp) | T(U) | C | A | G | AT (%) | AT Skew | GC Skew |
| PCGs | 11199 | 46.1 | 9.9 | 33.7 | 10.2 | 79.8 | −0.155 | 0.014 |
| 1st codon position | 3733 | 37.7 | 9.7 | 37.1 | 15.4 | 74.8 | −0.008 | 0.226 |
| 2nd codon position | 3733 | 48.1 | 16.4 | 22.6 | 13 | 70.7 | −0.361 | −0.115 |
| 3rd codon position | 3733 | 52.5 | 3.8 | 41.5 | 2.3 | 94 | −0.117 | −0.239 |
| A + T rich region | 287 | 47 | 2.8 | 48.4 | 1.7 | 95.4 | 0.015 | −0.231 |
| tRNAs | 1515 | 39.4 | 7.7 | 42 | 10.9 | 81.4 | 0.032 | 0.174 |
| rRNAs | 2141 | 41.3 | 5.0 | 43.7 | 10 | 85 | 0.027 | 0.34 |
| Full genome | 15361 | 41.2 | 11.4 | 40 | 7.3 | 81.2 | −0.015 | −0.217 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Xiao, J.; Hao, X.; Yuan, X. Unique Duplication of trnN in Odontoptilum angulatum (Lepidoptera: Pyrginae) and Phylogeny within Hesperiidae. Insects 2021, 12, 348. https://doi.org/10.3390/insects12040348
Liu J, Xiao J, Hao X, Yuan X. Unique Duplication of trnN in Odontoptilum angulatum (Lepidoptera: Pyrginae) and Phylogeny within Hesperiidae. Insects. 2021; 12(4):348. https://doi.org/10.3390/insects12040348
Chicago/Turabian StyleLiu, Jiaqi, Jintian Xiao, Xiangyu Hao, and Xiangqun Yuan. 2021. "Unique Duplication of trnN in Odontoptilum angulatum (Lepidoptera: Pyrginae) and Phylogeny within Hesperiidae" Insects 12, no. 4: 348. https://doi.org/10.3390/insects12040348
APA StyleLiu, J., Xiao, J., Hao, X., & Yuan, X. (2021). Unique Duplication of trnN in Odontoptilum angulatum (Lepidoptera: Pyrginae) and Phylogeny within Hesperiidae. Insects, 12(4), 348. https://doi.org/10.3390/insects12040348






