Antibiotic Treatment Decrease the Fitness of Honeybee (Apis mellifera) Larvae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Honeybee
2.2. Antibiotic Treatment of A. mellifera Larvae
2.3. RNA Isolation and cDNA Synthesis
2.4. Quantitative Real-Time PCR (qPCR)
2.5. Statistical Analysis
3. Results
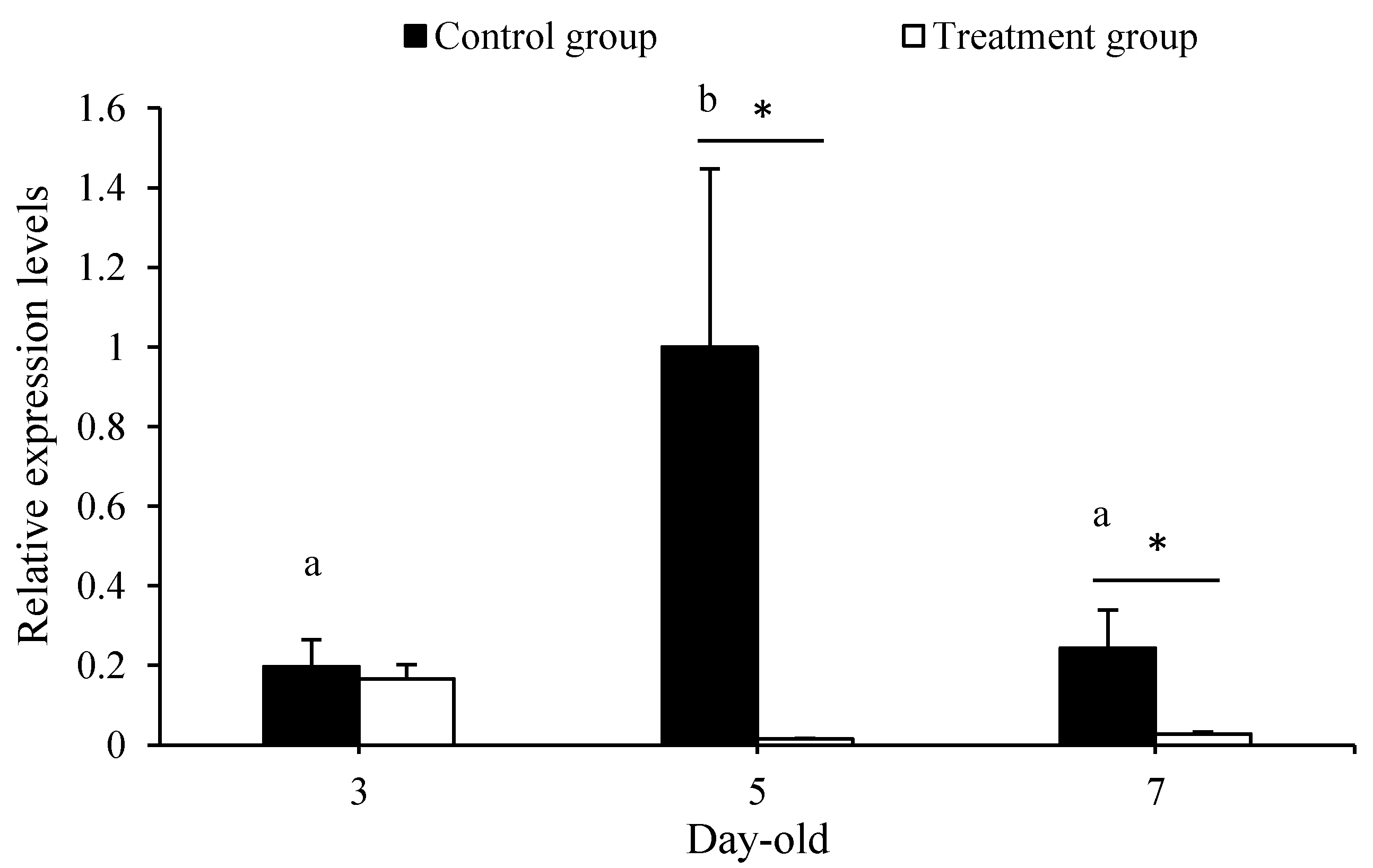
3.1. Penicillin–Streptomycin Inhibited Symbiotic Bacteria
3.2. Antibiotics Treatment Extended Eclosion Time and Decreased Body Weight
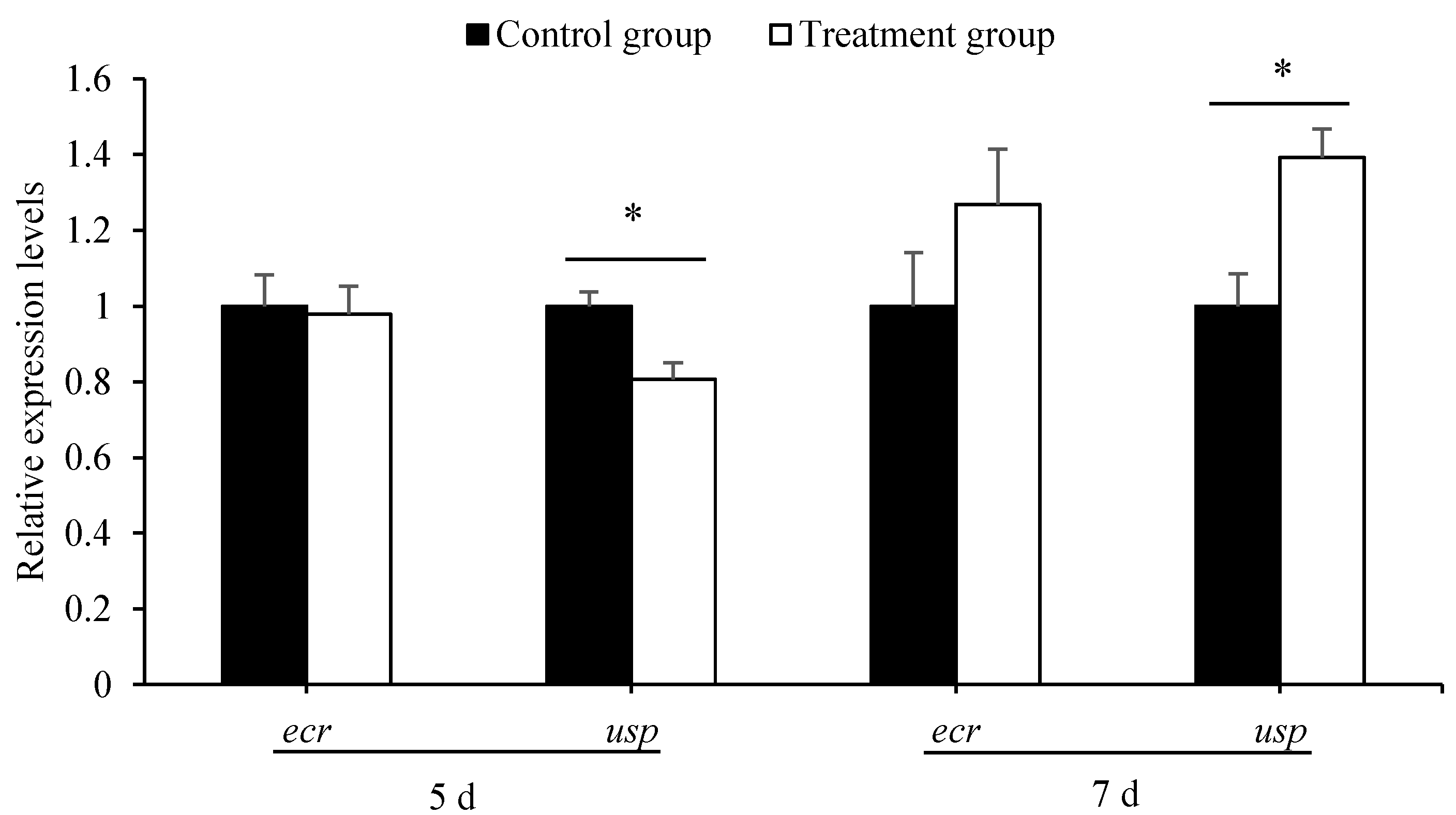
3.3. Antibiotics Treatment Disturbed the Expression of Development-Regulation Genes
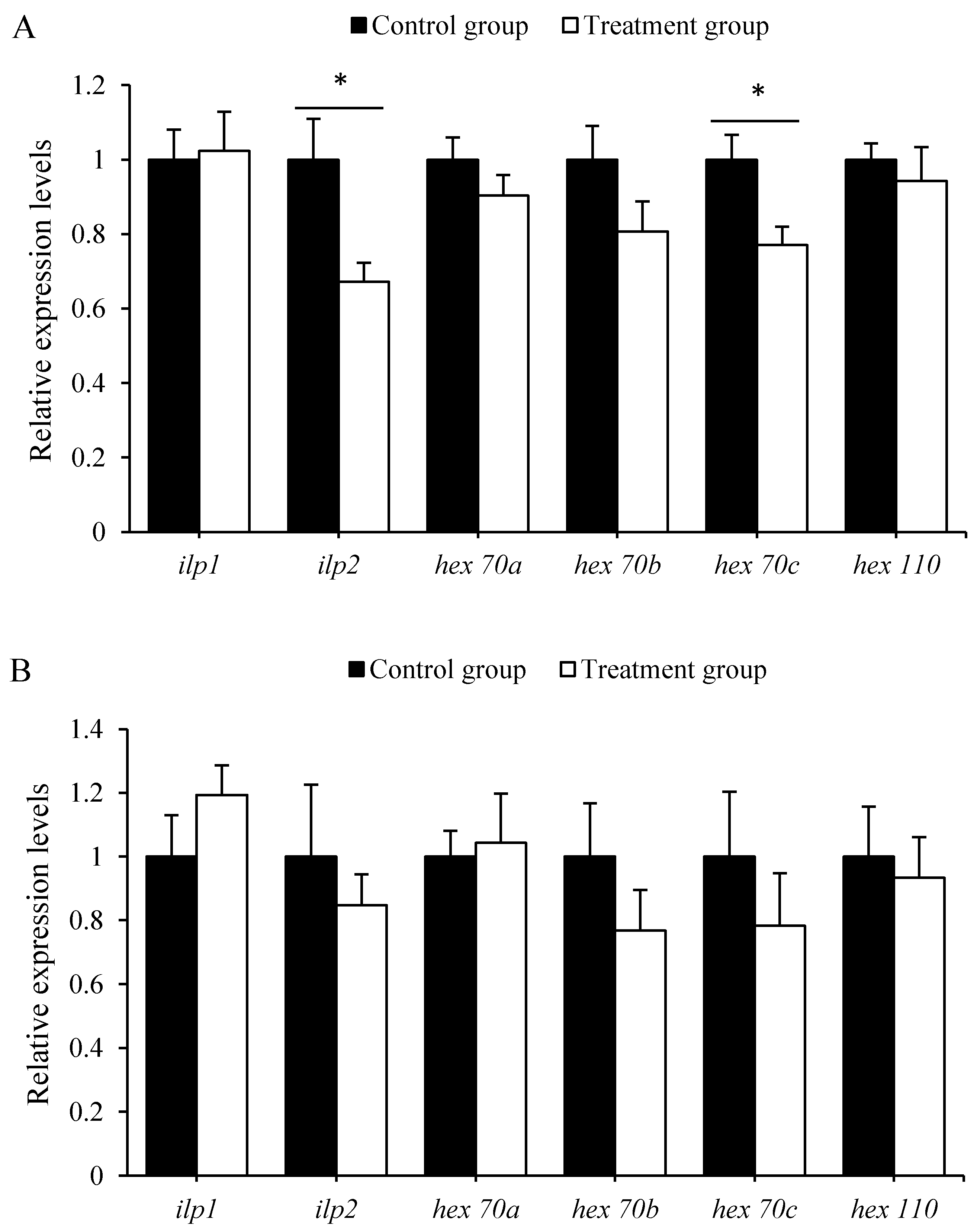
3.4. Antibiotics Treatment Down-Regulated the Expression of Nutrient Metabolism Genes
3.5. Antibiotics Treatment Decreased Immunity Genes Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gallai, N.; Salles, J.M.; Settele, J.; Vaissiere, B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Champetier, A.; Sumner, D.A.; Wilen, J.E. The Bioeconomics of Honey Bees and Pollination. Environ. Resour. Econ. 2015, 60, 143–164. [Google Scholar] [CrossRef]
- Zheng, B.Y.; Zhao, B.A.; Jin, X.; Duan, X.L.; Huang, S.K.; Li, J.H. Effect of Sacbrood virus infection on nutritional and immune responses of Apis cerana cerana (Hymenoptera: Apidae). Acta Entomol. Sin. 2019, 62, 1054–1064. [Google Scholar]
- Van Der Zee, R.; Pisa, L.; Andonov, S.; Brodschneider, R.; Charriere, J.D.; Chlebo, R.; Coffey, M.F.; Crailsheim, K.; Dahle, B.; Gajda, A.; et al. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J. Apic. Res. 2012, 51, 100–114. [Google Scholar] [CrossRef]
- Van Dooremalen, C.; Cornelissen, B.; Poleij-Hok-Ahin, C.; Blacquière, T. Single and interactive effects of Varroa destructor, Nosema spp., and imidacloprid on honey bee colonies (Apis mellifera). Ecosphere 2018, 9, e02378. [Google Scholar] [CrossRef]
- Calatayud-Vernich, P.; Calatayud, F.; Simó, E.; Aguilar, J.A.P.; Picó, Y. A two-year monitoring of pesticide hazard in-hive: High honey bee mortality rates during insecticide poisoning episodes in apiaries located near agricultural settings. Chemosphere 2019, 232, 471–480. [Google Scholar] [CrossRef]
- Kablaua, A.; Eckertb, J.H.; Pistoriusb, J.; Sharbatia, S.; Einspanier, R. Effects of selected insecticidal substances on mRNA transcriptome in larvae of Apis mellifera. Pestic. Biochemi. Phys. 2020, 170, 104703. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.L.; Xiong, M.Q.; Liu, W.B.; Zhao, B.A.; Huang, S.K.; Li, J.H. Effects of three fungicides on the activities of protective enzymes and detoxifying enzymes in Apis mellifera. Acta Pratac. Sin. 2020, 29, 74–82. [Google Scholar]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Nancy, A.M. Genomics of the honey bee microbiome. Curr. Opin. Insect Sci. 2015, 10, 22–28. [Google Scholar]
- Li, C.Y.; Zhou, X.; Zheng, H. Diversity and function of insect gut microbiome. Acta Microbiol. Sin. 2018, 58, 1016–1024. [Google Scholar]
- Cilia, G.; Fratini, F.; Tafi, E.; Mancini, S.; Turchi, B.; Mancini, S.; Sagona, S.; Nanetti, A.; Cerri, D.; Felicioli, A. Microbial Profile of the Ventriculum of Honey Bee (Apis mellifera ligustica Spinola, 1806) Fed with Veterinary Drugs, Dietary Supplements and Non-Protein Amino Acids. Vet. Sci. 2020, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Fratini, F.; Tafi, E.; Mancini, S.; Turchi, B.; Sagona, S.; Cerri, D.; Felicioli, A.; Nanetti, A. Changes of Western honey bee Apis mellifera ligustica (Spinola, 1806) ventriculus microbial profile related to their in-hive tasks. J. Apic. Res. 2020, 60, 198–202. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef]
- Ament, S.A.; Wang, Y.; Robinson, G.E. Nutritional regulation of division of labor in honey bees: Toward a systems biology perspective. WIREs Syst. Biol. Med. 2010, 2, 566–576. [Google Scholar] [CrossRef]
- Li, W.F.; Chen, Y.P.; Cook, S.C. Chronic Nosema ceranae infection inflicts comprehensive and persistent immuno-suppression and accelerated lipid loss in host Apis mellifera honey bees. Int. J. Parasitol. 2018, 48, 433–444. [Google Scholar] [CrossRef]
- Antonio, D.S.M.; Guidugli-Lazzarini, K.R.; do Nascimento, A.M.; Simoes, Z.L.P.; Hartfelder, K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 2008, 95, 953–961. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Ortiz-Alvarado, Y.; Clark, D.R.; Vega-Melendez, C.J.; Flores-Cruz, Z.; Domingez-Bello, M.G.; Giray, T. Antibiotics in hives and their effects on honey bee physiology and behavioral development. Biol. Open 2020, 9, bio053884. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Armstrong, T.N. Inhibition of the American foulbrood bacterium, Paenibacillus larvae larvae, by bacteria isolated from honey bees. J. Apic. Res. 2005, 44, 168–171. [Google Scholar] [CrossRef]
- McFrederick, Q.; Wcislo, W.T.; Taylor, D.R.; Ishak, H.D.; Dowd, S.E.; Mueller, U. Environment or kin: Whence do bees obtain acidophilic bacteria? Mol. Ecol. 2012, 21, 1754–1768. [Google Scholar] [CrossRef]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef]
- Vojvodic, S.; Rehan, S.M.; Anderson, K.E. Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PLoS ONE 2013, 8, e72106. [Google Scholar] [CrossRef] [PubMed]
- Forsgren, E.; Olofsson, T.C.; Váasquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2010, 41, 99–108. [Google Scholar] [CrossRef]
- Evans, J.D.; Lopez, D.L. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 2004, 97, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Janashia, I.; Alaux, C. Specific immune stimulationby endogenous bacteria in honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 2016, 109, 1474–1477. [Google Scholar] [CrossRef]
- Mutti, N.S.; Dolezal, A.G.; Wolschin, F.; Mutti, J.S.; Gill, K.S.; Amdam, G.V. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 2011, 214, 3977–3984. [Google Scholar] [CrossRef]
- Jensen, A.B.; Pedersen, B.V.; Eilenberg, J. Differential susceptibility across honey bee colonies in larval chalk-brood resistance. Apidologie 2009, 40, 524–534. [Google Scholar] [CrossRef]
- Vojvodic, S.; Jensen, A.B.; James, R.R.; Boomsma, J.J.; Eilenberg, J. Temperature dependent virulence of obligate and facultative fungal pathogens of honeybee brood. Vet. Microbiol. 2011, 149, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.Y.; Wang, X.Y.; Gao, Y.; Lin, Y.; Zhu, Y.N.; Yang, D.L.; Li, Z.G.; Su, S.K. Changes in the gene expression of chalkbrood resistance in Apis mellifera larvae infected by Ascosphaera apis. Apidologie 2020, 51, 35–47. [Google Scholar] [CrossRef]
- Simone, M.; Evans, J.D.; Spivak, M. Resin collection and social immunity in honey bees. Evolution 2009, 63, 3016–3022. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.Z.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef]
- Liu, C.D.; Wang, Q.; Yu, H.; Zhang, Z.B.; Zheng, B.Y.; Li, J.H. Expression and functional analysis of early-response factors ecr and usp of ecdysteroid in different castes of Apis mellifera. Genom. Appl. Biol. 2018, 37, 686–692. [Google Scholar]
- De Azevedo, S.V.; Hartfelder, K. The insulin signaling pathway in honey bee (Apis mellifera) caste development-differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 2008, 54, 1064–1071. [Google Scholar] [CrossRef]
- Zhang, G.D. Transcription Analysis of Hexamerin in Different Development Stage of Honeybee (Apis mellifera). Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2014. [Google Scholar]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Raymann, K.; Bobay, L.M.; Moran, N.A. Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol. 2018, 27, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Al-Ghamdi, A.A.; Ghramh, H.A.; Ansari, M.J.; Ali, H.; Alamri, S.A.; Kahtani, S.N.A.; Adgaba, N.; Qasim, M.; Hafeez, M. Structural diversity and functional variability of gut microbial communities associated with honey bees. Microb. Pathog. 2019, 138, 103793. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Romero, S.; Nastasa, A.; Chapman, A.; Kwong, W.K.; Foster, L.J. The honey bee gut microbiota: Strategies for study and characterization. Insect Mol. Biol. 2019, 28, 455–472. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.S.; Shah, A.; Brockmann, A. Honey bee foraging induces upregulation of early growth response protein 1, hormone receptor 38 and candidate downstream genes of the ecdysteroid signalling pathway. Insect Mol. Biol. 2017, 27, 90–98. [Google Scholar] [CrossRef]
- Lu, C.Y.; Huang, P.J.; Hsu, C.Y. The cholesterol-hydroxyecdysone-vitellogenin pathway is involved in the longevity of trophocytes and oenocytes of queen honey bees (Apis mellifera). Apidologie 2018, 49, 721–733. [Google Scholar] [CrossRef]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; De Grandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- Flatt, T.; Tu, M.P.; Tatar, M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. BioEssays 2005, 27, 999–1010. [Google Scholar] [CrossRef]
- Martins, J.R.; Nunes Francis, M.F.; Cristino, A.S.; Simes Zilá, L.P.; Bitondi Márcia, M.G. The four hexamerin genes in the honey bee: Structure, molecular evolution and function deduced from expression patterns in queens, workers and drones. BMC Mol. Bio. 2010, 11, 23. [Google Scholar] [CrossRef]
- Kanost, M.R. Encyclopedia of Insects, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2009; Chapter 17; pp. 446–449. ISBN 978-0-12-374144-8. [Google Scholar]
- Nilsen, K.A.; Ihle, K.E.; Frederick, K.; Fondrk, M.K.; Smedal, B.; Hartfelder, K.; Amdam, G.V. Insulin-like peptide genes in honey bee fat body respond differently to manipulation of social behavioral physiology. J. Exp. Biol. 2011, 214, 1488–1497. [Google Scholar] [CrossRef]
- Ryoo, H.D.; Baehrecke, E.H. Distinct death mechanisms in Drosophila development. Curr. Opin. Cell Biol. 2010, 22, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; Li, S. E93 predominantly transduces 20-hydroxyecdysone signaling to induce autophagy and caspase activity in Drosophila fat body. Insect Biochem. Mol. Biol. 2014, 45, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Danihlík, J.; Aronstein, K.; Petrivalsky, M. Antimicrobial peptides: A key component of honey bee innate immunity physiology, biochemistry, and chemical ecology. J. Apic. Res. 2015, 54, 123–136. [Google Scholar] [CrossRef]
- Bullet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]

| Genes | Primer Sequences (5′-3′) | Gene ID | Reference | |
|---|---|---|---|---|
| Reference gene | β-actin | F: TTGTATGCCAACACTGTCCTTT R: TGGCGCGATGATCTTAATTT | NM_001185145.1 | Simone et al. [33] |
| 16S rRNA gene | unibac | F: AGAGTTTGATCCTGGCTCAG R: CTGCTGCCTCCCGTAGGAGT | Schwarz et al. [34] | |
| Development-related genes | ecr | F: TGCCAACACTGTCCTTTCTG R: AAAGAGCCAGGCTGCGACAA | XM_016913298.2 | Liu et al. [35] Liu et al. [35] |
| usp | F: GCCAAGATGATGAAGAAGGAGA R: GGTTGATGAGGCTTGCTGTGTC | NM_001011634.2 | ||
| Nutrient metabolism-related genes | ilp1 | F: CGATAGTCCTGGTCGGTTTG R: CAAGCTGAGCATAGCTGCAC | XM_026442143.1 | De Azevedo et al. [36] |
| ilp2 | F: TTCCAGAAATGGAGATGGATG R: TAGGAGCGCAACTCCTCTGT | NM_001177903.1 | ||
| hex 70a | F: GCCTTCAGTTTGGTCGGTGC R: GCTGGTTGAGCGACGCGATA | NM_001110764.1 | Zhang [37] | |
| hex 70b | F: AACAGCCACGAATCCGTCTT R: CAGGCTTGTCCAGAGGGAAG | NM_001011600.1 | Zheng et al. [3] Zheng et al. [3] Zheng et al. [3] | |
| hex 70c | F: TAAGGCAGGCAGACTTGAGC R: AAACGCTGTGGTGAACATGC | NM_001098717.1 | ||
| hex 110 | F: ACGGACAATACCCGAACACC R: AGCATGCTGATGCCTCTGTT | NM_001101023.1 | ||
| Innate virus immunity genes | abaecin | F: CAGCATTCGCATACGTACCA R: GACCAGGAAACGTTGGAAAC | NM_001011617.1 | Simone et al. [33] Evans et al. [38] Evans et al. [38] |
| hymenoptaecin | F: CTCTTCTGTGCCGTTGCATA R: GCGTCTCCTGTCATTCCATT | NM_001011615.1 | ||
| defencin-1 | F: TGCGCTGCTAACTGTCTCAG R: AATGGCACTTAACCGAAACG | NM_001011616.2 | ||
| apidaecin | F: TTTTGCCTTAGCAATTCTTGTTG R: GTAGGTCGAGTAGGCGGATCT | NM_001011613.1 | Evans et al. [38] | |
| Treatment | Development Period (h) | Body Weight (mg) | ||
|---|---|---|---|---|
| Pupation | Eclosion | 5-Day-Old Larvae | Newly Emerged Adults | |
| Control group | 212.79 ± 0.90 | 406.94 ± 1.60 | 128.68 ± 2.95 | 113.46 ± 2.69 |
| Treatment group | 213.19 ± 0.72 | 413.69 ± 1.59 ** | 114.62 ± 3.39 ** | 102.38 ± 2.30 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, X.; Zhao, B.; Jin, X.; Cheng, X.; Huang, S.; Li, J. Antibiotic Treatment Decrease the Fitness of Honeybee (Apis mellifera) Larvae. Insects 2021, 12, 301. https://doi.org/10.3390/insects12040301
Duan X, Zhao B, Jin X, Cheng X, Huang S, Li J. Antibiotic Treatment Decrease the Fitness of Honeybee (Apis mellifera) Larvae. Insects. 2021; 12(4):301. https://doi.org/10.3390/insects12040301
Chicago/Turabian StyleDuan, Xinle, Bi’an Zhao, Xin Jin, Xuefen Cheng, Shaokang Huang, and Jianghong Li. 2021. "Antibiotic Treatment Decrease the Fitness of Honeybee (Apis mellifera) Larvae" Insects 12, no. 4: 301. https://doi.org/10.3390/insects12040301
APA StyleDuan, X., Zhao, B., Jin, X., Cheng, X., Huang, S., & Li, J. (2021). Antibiotic Treatment Decrease the Fitness of Honeybee (Apis mellifera) Larvae. Insects, 12(4), 301. https://doi.org/10.3390/insects12040301







