First Complete Mitochondrial Genome of Melyridae(Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
2.2. DNA Sequencing and Assembly
2.3. Sequence Annotation and Analyses
2.4. Phylogenetic Analysis
3. Results and Discussion
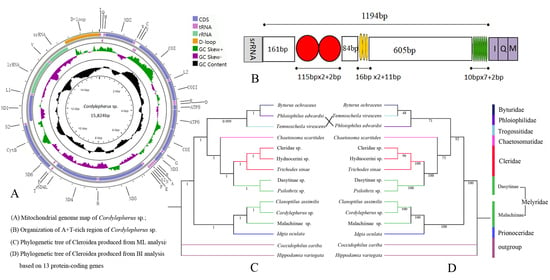
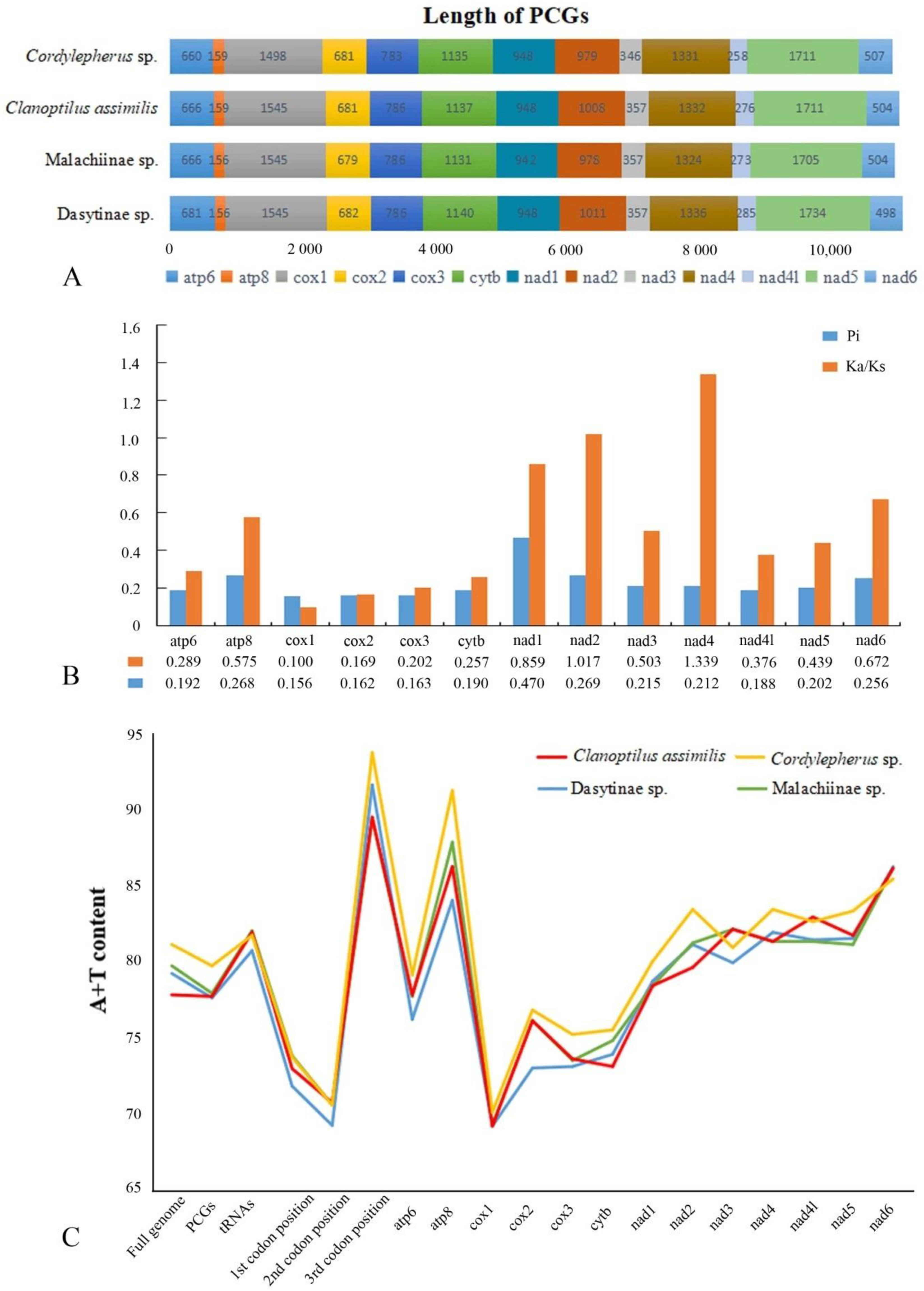
3.1. Mitogenome Organization and Base Composition
3.2. Protein-CodingGenes
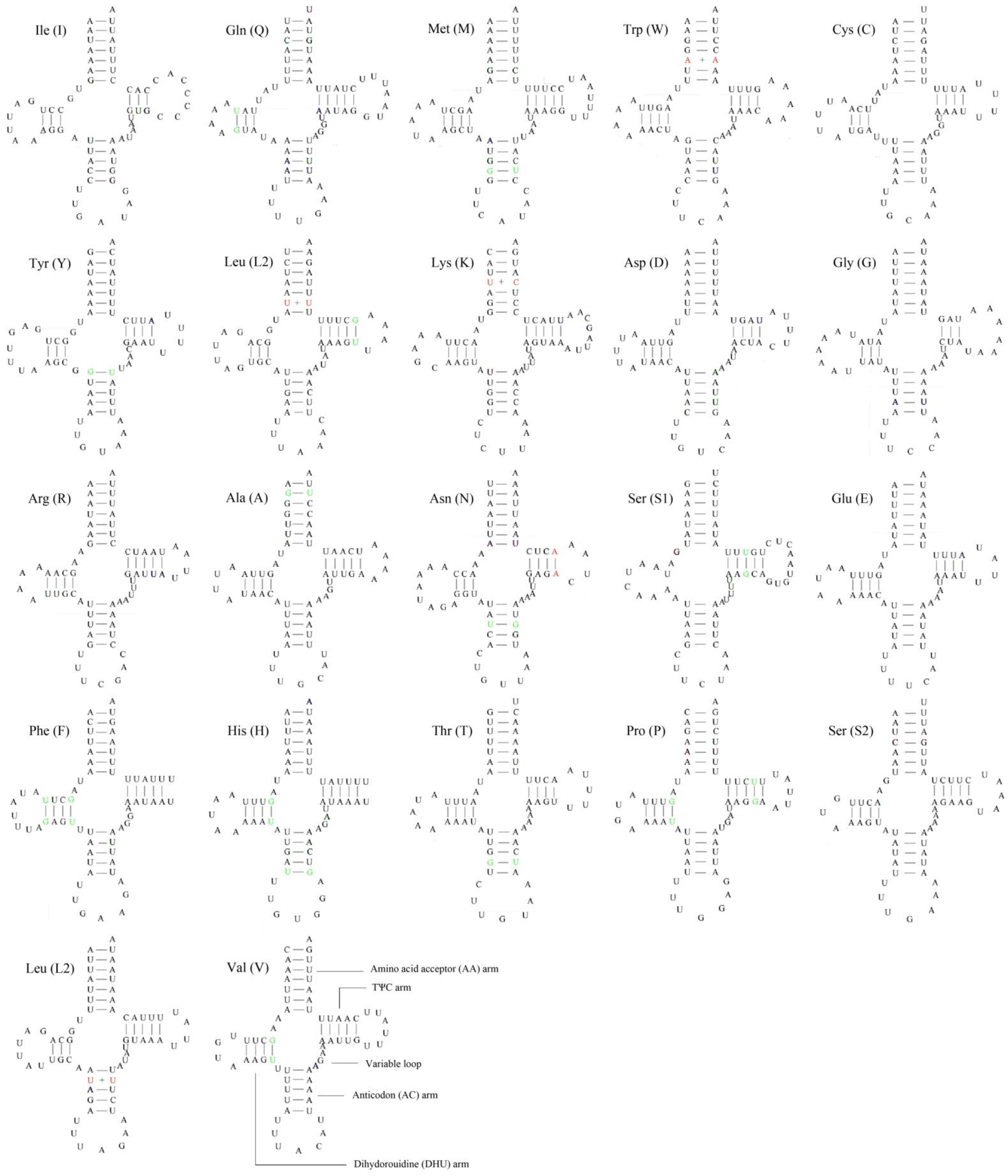
3.3. Transfer and Ribosomal RNA Genes
3.4. A + T-RichRegion
3.5. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bocakova, M.; Constantin, R.; Bocak, L. Molecular phylogenetics of the melyrid lineage (Coleoptera: Cleroidea). Cladistics 2011, 28, 117–129. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Leschen, R.A.B. 9.11. Melyridae Leach, 1815. In Coleoptera, Beetles: Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformiapartim); Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; W. DeGruyter: Berlin, Germany, 2010; Volume 2, pp. 273–280. [Google Scholar]
- Böving, A.G.; Craighead, F.C. An illustrated synopsis ofthe principal larval forms of the order Coleoptera. Entomol. Am. 1931, 11, 1–351. [Google Scholar]
- Crowson, R.A. The Phylogeny of Coleoptera. Annu. Rev. Entomol. 1960, 5, 111–134. [Google Scholar] [CrossRef]
- Crowson, R.A. A review of the classification of Cleroidea (Coleoptera), with descriptions of two new genera of Peltidae and of several new larval types. Ecol. Entomol. 2009, 116, 275–327. [Google Scholar] [CrossRef]
- Majer, K. A review of the classification of the Melyridae and related families (Coleoptera, Cleroidea). Entomol. Basil. 1994, 17, 319–390. [Google Scholar] [CrossRef]
- Mayor, A. Melyridae, Dasytidae, Malachiidae. In Catalogue of Palaearctic Coleoptera; Löbl, I., Smetana, A., Eds.; Apollo Books: Stenstrup, Denmark, 2007; Volume 4, pp. 386–454. [Google Scholar]
- Lawrence, J.F.; Newton, A.F. Families and subfamilies of Coleoptera (with selected genera, notes, references and data on family-group names). In Biology, Phylogeny, and Classification of Coleoptera; Lawrence, J., Pakaluk, J.F., Slípínski, S.A., Eds.; Museum i Instytut Zoologii PAN: Warzawa, Poland, 1995; pp. 779–1092. [Google Scholar]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Ślipiński, S.A.; et al. Family-Group Names In Coleoptera (Insecta). ZooKeys 2011, 88, 1–972. [Google Scholar] [CrossRef] [Green Version]
- Gimmel, M.L.; Bocakova, M.; Gunter, N.; Leschen, R.A.B. Comprehensive phylogeny of the Cleroidea (Coleoptera: Cucujiformia). Syst. Entomol. 2019, 44, 527–558. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lambkin, C.L.; Barker, S.C.; Whiting, M.F. A mitochondrial genome of Diptera: Whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007, 32, 40–59. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [Green Version]
- Nie, R.; Andújar, C.; Gómez-Rodríguez, C.; Bai, M.; Xue, H.; Tang, M.; Yang, C.; Tang, P.; Yang, X.; Vogler, A.P. The phylogeny of leaf beetles (Chrysomelidae) inferred from mitochondrial genomes. Syst. Entomol. 2019, 45, 188–204. [Google Scholar] [CrossRef]
- Liu, Y.; Song, F.; Jiang, P.; Wilson, J.-J.; Cai, W.; Li, H. Compositional heterogeneity in true bug mitochondrial phylogenomics. Mol. Phylogenet. Evol. 2018, 118, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Nie, R.; Vogler, A.P.; Yang, X.-K.; Lin, M. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst. Entomol. 2020. [Google Scholar] [CrossRef]
- Li, H.; Shao, R.; Song, N.; Song, F.; Jiang, P.; Li, Z.; Cai, W. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015, 5, 8527. [Google Scholar] [CrossRef] [Green Version]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Liu, S.; Yang, Q.; Su, X.; Zhou, L.; Tang, M.; Fu, R.; Li, J.; Huang, Q. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. GigaScience 2013, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Gärtner, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Wu, L.; Nie, R.E.; Bai, M.; Yang, Y.X. The complete mitochondrial genome of Idgia oculata (Coleoptera: Cleroidea: Prionoceridae) and a related phylogenetic analysis of Cleroidea. Mitochondrial DNA Part B 2019, 4, 491–493. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE.A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CG View Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sitesof animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. Mafft Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usa-bility. Mol. Biol. Evol. 2013, 4, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Ahrens, D.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-Level Sampling of Mitochondrial Genomes in Coleoptera: Compositional Heterogeneity and Phyloge-netics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Linard, B.; Crampton-Platt, A.; Moriniere, J.; Timmermans, M.J.T.N.; Andújar, C.; Arribas, P.; Miller, K.E.; Lipecki, J.; Favreau, E.; Hunter, A. The contribution of mitochondrial metagenomics to large-scale data mining and phylogenetic analysis of Coleoptera. Mol. Phylogenet. Evol. 2018, 128, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, P.A.; Feng, R.Q.; Zhang, L.; Wang, J.; Wang, X.T.; Zhang, L.J.; Yuan, M.L. Mitochondrial genomes of three Bostrichiformia species and phylogenetic analysis of Polyphaga (Insecta, Coleoptera). Genomics 2020, 112, 2970–2977. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.J.; Gimmel, M.L.; Kutty, S.N.; Cockerill, T.D.; Khen, C.V.; Vogler, A.P. Soup to Tree: The Phylogeny of Beetles Inferred by Mitochondrial Metagenomics of a Bornean Rainforest Sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. A Comparative Analysis of Mitochondrial Genomes in Coleoptera (Arthropoda: Insecta) and Genome Descriptions of Six New Beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nattier, R.; Salazar, K. Next-generation sequencing yields mitochondrial genome of Coccidophilus cariba Gordon (Coleoptera: Coccinellidae) from museum specimen. Mitochondrial DNA Part B 2019, 4, 3780–3781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.-N.; Liu, C.-Z.; Sun, Y.-X. The complete mitochondrial genome of the Adonis ladybird, Hippodamia variegata (Coleoptera: Coccinellidae). Mitochondrial DNA Part B 2019, 4, 1087–1088. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba nucleotide sequence, gene organiza-tion, and genetic code. J. Mol. Phylogenet. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal Mitochondrial DNA: Structure and Evolution. Adv. Virus Res. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nat. Cell Biol. 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demari-Silva, B.; Foster, P.G.; Oliveira-de, T.M.P.; Bergo, E.S.; Sanabani, S.S.; Pessôa, R.; Sallum, M.A.M. Mitochondrial ge-nomes and comparative analyses of Culex camposi, Culex coronator, Culex usquatus and Culex usquatissimus (Diptera: Culicidae), members of the Coronator group. BMC Genom. 2015, 16, 831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Dietrich, C.H.; Huang, M. Characterization of the complete mitochondrial genomes of two species with preliminary investigation on phylogenetic status of Zyginellini (Hemioptera: Cicadellidae: Typhlocybinae). Insects 2020, 11, 684. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; He, S.; Song, X.; Liao, Q.; Zhang, X.; Yue, B. The complete mitochondrial genome of Epicauta chinensis (Coleoptera: Meloidae) and phylogenetic analysis among Coleopteran insects. Gene 2016, 578, 274–280. [Google Scholar] [CrossRef]
- Du, C.; Zhang, L.; Lu, T.; Ma, J.; Zeng, C.; Yue, B.; Zhang, X. Mitochondrial genomes of blister beetles (Coleoptera, Meloidae) and two large intergenic spacers in Hycleus genera. BMC Genom. 2017, 18, 698. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, G. The mitochondrial genomes of two walnut pests, Gastrolina depressa depressa and G. depressa thoracica (Coleoptera: Chrysomelidae), and phylogenetic analyses. PeerJ 2018, 6, e4919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballard, J.W.O. Comparative Genomics of Mitochondrial DNA in Members of the Drosophila melanogaster Subgroup. J. Mol. Evol. 2000, 51, 48–63. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Robertson, J.A.; Ślipiński, A.; Moulton, M.; Shockley, F.W.; Giorgi, A.; Lord, N.P.; Mckenna, D.D.; Tomaszewska, W.; Forrester, J.; Miller, K.B.; et al. Phylogeny and classification of Cucujoidea and the recognition of a new super-family Coccinelloidea (Coleoptera: Cucujiformia). Syst. Entomol. 2015, 40, 745–778. [Google Scholar] [CrossRef]



| Superfamily | Family/Subfamily | Species | GenBankNo. | References |
|---|---|---|---|---|
| Cleroidea | Phloiophilidae | Phloiophilus edwardsi | JX412815.1 | Unpublished |
| (Ingroup) | Melyridae/ | Malachiinae sp. | JX412799.1 | Unpublished |
| Malachiinae | Clanoptilus assimilis | JX412833.1 | Unpublished | |
| Cordylepherus sp. | MW365444 | This study | ||
| Melyridae/ | Psilothrix sp. | JX412801.1 | [33] | |
| Dasytinae | Dasytinae sp. | JX412765.1 | Unpublished | |
| Trogossitidae | Temnoscheila virescens | JX412752.1 | Unpublished | |
| Prionoceridae | Idgia oculata | NC_044896.1 | [23] | |
| Byturidae | Byturus ochraceus | NC_036267.1 | [34] | |
| Cleridae | Hydnocerini sp. | KX035157.1 | Unpublished | |
| Trichodes sinae | NC_033340.1 | [35] | ||
| Cleridae sp. | MH789728.1 | [36] | ||
| Chaetosomatidae | Chaetosomas caritides | NC_011324.1 | [37] | |
| Cucujoidea | Coccinellidae | Coccidophilus cariba | MN447521.1 | [38] |
| (Outgroup) | Hippodamia variegata | MK334129.1 | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.; Ge, X.; Xie, G.; Liu, H.; Yang, Y. First Complete Mitochondrial Genome of Melyridae(Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications. Insects 2021, 12, 87. https://doi.org/10.3390/insects12020087
Yuan L, Ge X, Xie G, Liu H, Yang Y. First Complete Mitochondrial Genome of Melyridae(Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications. Insects. 2021; 12(2):87. https://doi.org/10.3390/insects12020087
Chicago/Turabian StyleYuan, Lilan, Xueying Ge, Guanglin Xie, Haoyu Liu, and Yuxia Yang. 2021. "First Complete Mitochondrial Genome of Melyridae(Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications" Insects 12, no. 2: 87. https://doi.org/10.3390/insects12020087
APA StyleYuan, L., Ge, X., Xie, G., Liu, H., & Yang, Y. (2021). First Complete Mitochondrial Genome of Melyridae(Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications. Insects, 12(2), 87. https://doi.org/10.3390/insects12020087