Simple Summary
Laboratory diagnostics of the presence of Melissococcus plutonius is necessary for the confirmation of European foulbrood (EFB) in honey bee colonies. Seeking EFB positive colonies is based on inspections performed by beekeepers or authorized inspectors. Bee brood, adult bees, or honey are usually sampled and confirmed by PCR. Here, we tested a new concept of searching suspected colonies by simple and effective collecting hive debris. Conventional PCR detection of M. plutonius in hive debris gives comparable sensitivity and specificity as for adults or honey. The main advantage of testing hive debris combines its effectiveness and costs compared to the sampling of adult bees or honey. Easy and fast collection of hive debris samples in areas with EFB outbreaks could fasten disease early detection and elimination.
Abstract
European foulbrood (EFB) is an infectious disease of honey bees caused by the bacterium Melissococcus plutonius. A method for DNA isolation and conventional PCR diagnosis was developed using hive debris, which was non-invasively collected on paper sheets placed on the bottom boards of hives. Field trials utilized 23 honey bee colonies with clinically positive symptoms and 21 colonies without symptoms. Bayes statistics were applied to calculate the comparable parameters for EFB diagnostics when using honey, hive debris, or samples of adult bees. The reliability of the conventional PCR was 100% at 6.7 × 103 Colony Forming Unit of M. plutonius in 1 g of debris. The sensitivity of the method for the sampled honey, hive debris, and adult bees was 0.867, 0.714, and 1.000, respectively. The specificity for the tested matrices was 0.842, 0.800, and 0.833. The predictive values for the positive tests from selected populations with 52% prevalence were 0.813, 0.833, and 0.842, and the real accuracies were 0.853, 0.750, and 0.912, for the honey, hive debris, and adult bees, respectively. It was concluded that hive debris can effectively be utilized to non-invasively monitor EFB in honey bee colonies.
1. Introduction
Honey bees suffer from many pests and pathogens that can weaken or lead to the collapse of their colonies [1]. American foulbrood (AFB, infectious agent Paenibacillus larvae) and European foulbrood (EFB, infectious agent Gram-positive bacterium Melissococcus plutonius) are the main bacterial infections, and they affect honey bee health by impairing their larval development [2,3]. Both diseases can be detected from the clinical symptoms in infected colonies [4,5]. AFB typically results in a brownish semi-fluid “ropy-mass” [6], while younger EFB-infected larvae are mostly transparent, with visible trachea, and the older larvae die twisted around the cell walls [2]. European foulbrood (EFB) is a globally distributed and economically important disease of honey bees [7]. The bacterium only affects bee larvae shortly after they have hatched via contaminated food. Adult honey bees are not affected but transfer vegetative bacteria, and thus, spread the disease [2,8]. The bacterium multiplies in the digestive tract of the infected larvae after ingestion [9], and they may die at any time between the 4th day and pupation [6]. Whether or not the infection is lethal to all larvae depends on its extent [4,9,10] and how virulent the bacterial strain is [11,12,13,14]. The molecular mechanisms of EFB infection are not currently understood [2,15]. Weakened or dead larvae are attacked by secondary accompanying bacterial invaders [6,10,16].
Outbreaks of EFB have been observed in the Czech Republic, United Kingdom, Switzerland, Norway, and other European countries [17,18]. However, EFB is not limited to Europe as it has also been detected in Asia, America, and Africa [17]. Although the disease is spread worldwide, information about M. plutonius strains and their distribution is limited. At present, three different clonal complex (CC) groups that contain strains with several different sequence types have been identified: CC3, CC12, and CC13 [15]. They differ in biochemical characteristics, virulence, and prevalence [19].
One of the basic diagnostic methods for EFB is microscopic detection of M. plutonius from carbol fuchsin stained smears of the dead or suspected larvae, but it does not discriminate the secondary invaders [4,20]. Microbial cultivation techniques for M. plutonius are slow with variable results because of the contaminating bacteria [20]. More recently, the MALDI-TOF diagnosis technique has been used for AFB [21] and EFB, but it requires culturing on a medium beforehand, which makes it a less attractive option for routine diagnostics (Dainat B. & Schäfer M., personal communication).
Polymerase chain reaction (PCR) based diagnostics sample adult bee tissues, larvae, honey, and pollen with primer pairs specific to M. plutonius [20,22]. Enzyme Linked Immunosorbent Assays (ELISA) are used in laboratories to detect antibodies, antigens, and proteins. Polachova et al. [23] recently developed a highly sensitive up-conversion-linked immunosorbent assay (ULISA) that employs photon-up-conversion nanoparticles (UCNPs) for the detection of M. plutonius using polyclonal antibodies made in house. Biosensors have also recently been established in laboratory diagnostics [24,25]. The routine use of immune-based techniques is limited, however, because they require EFB specific antibodies. To the best of our knowledge, there are no commercial antibodies targeted to M. plutonius epitopes that are currently available (December 2020).
Lateral flow immunoassays are commercially available for field EFB confirmation at apiaries [26]. They function similarly to at-home pregnancy tests, and are useful for distinguishing between EFB symptoms and other diseases with commutable clinical signs, such as AFB, viruses, chalkbrood, and non-pathogenic brood sepsis.
The efficient management of honey bee pests and pathogens is integral for beekeeping practices to help limit the spread of disease by migratory beekeepers, and for sustainable pollination service [27,28]. Antibiotic treatment is banned in many countries; thus, the main control methods are stamping out diseased colonies or the shook swarm method [29]. The spread of honey bee diseases is increased if the colony density is high in particular areas. This seems to be a factor for the transmission of EFB via adult bees that rob or drift between colonies [30,31]. The immediate reaction after finding a new outbreak is important to avoid the spread of the pathogen. In countries where EFB is classified as a notifiable disease, the following is a recommended practice when EFB is encountered in a colony. All colonies at the same apiary and at neighboring apiaries must be inspected: colonies are opened, the brood nest is checked and brood samples with suspected larvae are moved into laboratories for further confirmation. This is time consuming, demanding on human resources, and increases the costs for the prevention of honey bee diseases. Hive debris has been successfully used for AFB spore detection using cultivation [32] and PCR techniques [33,34,35], and for small hive beetle (Aethina tumida) using PCR [36,37].
Here we tested the hypothesis that hive debris was suitable for EFB diagnostics using conventional PCR, which has not been done previously. Hive debris can be easily collected over several days (summer) or weeks (winter) using hive bottom boards, which are then sent to a laboratory for M. plutonius diagnosis. We optimized a protocol for DNA extraction from hive debris and tested our approach on Swiss EFB positive and negative colonies. This investigation aimed to increase the speed of early EFB diagnosis in wide areas, where hive debris could be easily collected by beekeepers. Furthermore, this would enable prophylactic measures to be implemented earlier, and consequently, earlier eradication could reduce the economic impacts on beekeepers.
2. Materials and Methods
2.1. Cultivation of Bacterial Isolates
A reference strain of M. plutonius CCM 3707T was obtained from Czech Collection of Microorganisms (Masaryk University, Brno, Czech Republic). The strain was inoculated on Basal medium [20] and incubated under anaerobic condition using Genbox anaer (Biomérieux, Marcy I’Etoile, France) at 37 °C for 7 days. Identity of grown colonies was confirmed by PCR [38]. This strain was used for validation of PCR method (see below).
2.2. Spiked Debris and Honey for PCR Method Validation
The bacterial colonies of the reference strain M. plutonius (CCM 3707T) grown up on Basal medium [20] were re-suspended in 0.9% NaCl to an optical density 1 McFarland (3 × 108 CFU/mL). A dilution series of four concentrations of was prepared in 0.9% NaCl 3 × 107 CFU/mL, 3 × 106 CFU/mL, 3 × 105 CFU/mL, or 3 × 104 CFU/mL. A 0.5 mL aliquot of each bacterial suspension was used for dissolving in 4.5 g of debris or honey. Both matrices were tested for M. plutonius absence prior to spiking by culturing. Samples were stored at −30 °C until use. Then 1 g of spiked debris or honey was processed for DNA isolation.
2.3. Collecting Honey, Debris, and Bees from Colonies
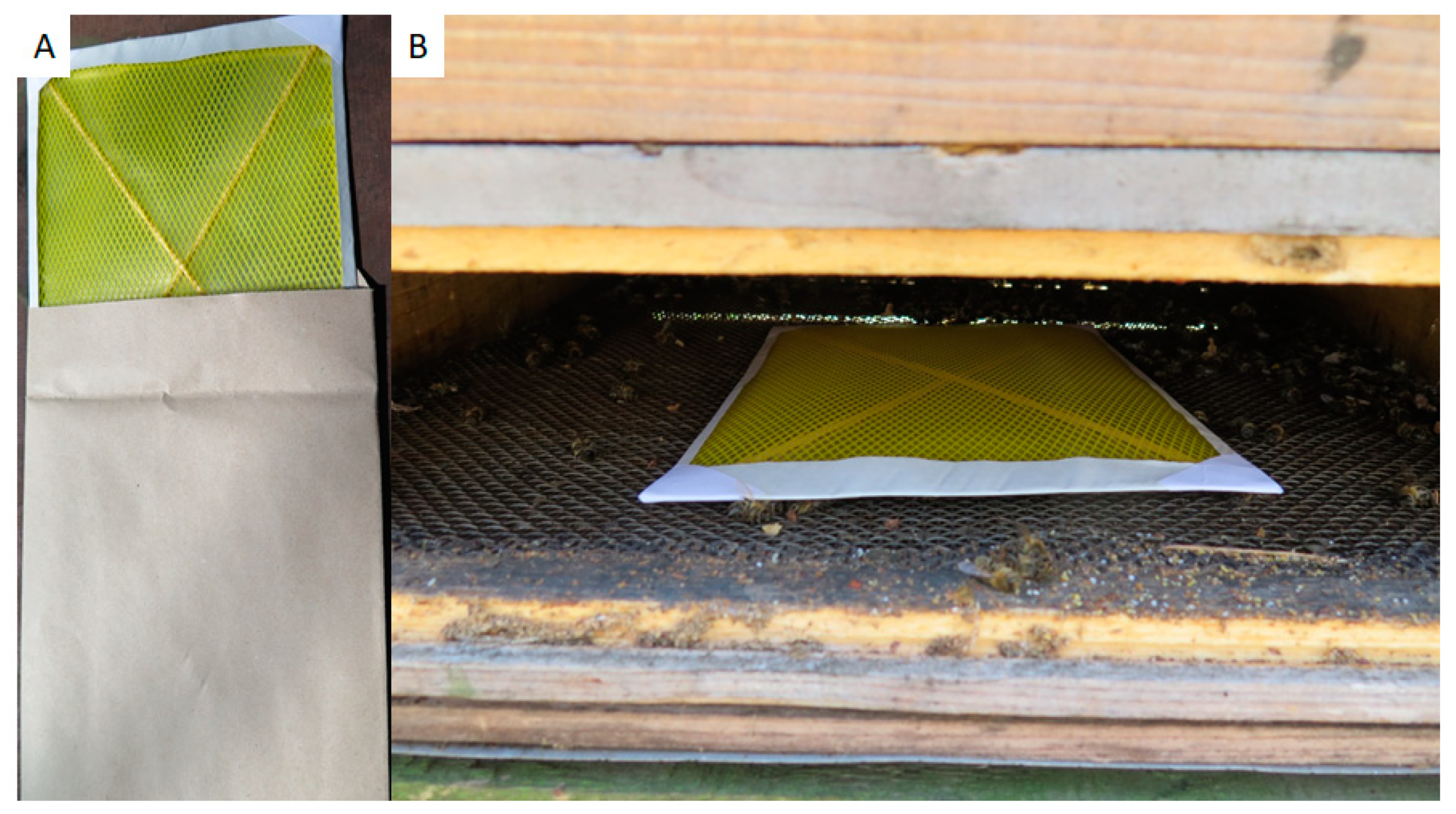
Field samples were collected in Switzerland, near Bern, in May–June 2018 and 2019. All colonies involved in this study were clinically inspected by trained bee inspectors. Honey was collected from individual colonies directly from the honey chamber. A pool of approx. 200 bees was sampled from a brood chamber of each tested colony, transported to laboratory, and frozen as soon as possible. Hive debris was gathered on bottom boards from individual colonies after 7 days of collection. For easier collection, paper sheets covered with a plastic sieve (common A4 format, supplier Bee Research Institute Dol, Czech Republic) were inserted to the hive bottom for one week (Figure 1). Dry debris was collected and stored in a freezer at −20 °C prior to use. Debris samples with molds are unusable for DNA isolation and M. plutonius detection by culture and, therefore, were wasted.
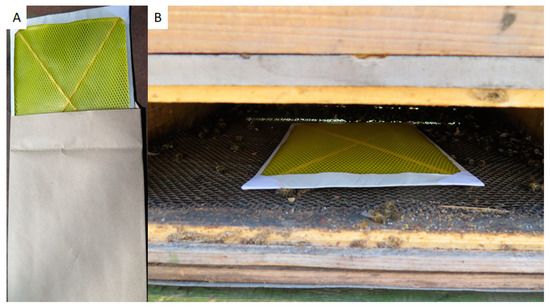
Figure 1.
Paper sheets for hive debris collection (A); installed in the hive bottom board (B).
2.4. Isolation of DNA from Hive Debris
An amount of 1 g of hive debris was extracted to GITC buffer (5.25 M guanidine isothiocyanate, 50 mM TRIS-HCl, pH 6.4, 20 mM EDTA, 1.3% Triton X-100; autoclaved 120 °C for 20 min, then added β-mercaptoethanol to final 1% concentration) [39]. The mixture was vortexed 3 times for 50 s and then centrifuged at 10,000× g for 3 min at room temperature. The supernatant was used for DNA isolation using the DNeasy Plant Mini Kit (Qiagen) according to instruction with minor modifications. Briefly: volume of 400 µL of a debris supernatant was mixed with 130 µL of AP2 buffer. RNase A was not added to the reaction. The samples were mixed and incubated for 5 min on ice. After that, the samples were centrifuged for 5 min at 20,000× g and the lysates were pipetted into QIAshredder spin columns. They were centrifuged for 2 min at 20,000× g and the flow-through was gently mixed with 1.5 amount of AP3/E buffer. The 650 µL of samples were put into DNeasy Mini spin columns and the columns were centrifuged for 1 min at 6000× g. The step was repeated until the whole volume of samples was used. The columns were washed twice with 500 µL of AW buffer and then centrifuged at the highest speed (2 × 2 min at 20,000× g). The columns were transferred into new tubes and 50 µL of AE buffer was added. After 5 min of incubation, the DNA was eluted with centrifugation at room temperature (1 min at 6000× g).
2.5. Isolation of DNA from Honey
An amount of 1 g of honey was fully dissolved with 2.25 mL of sterile distilled water and centrifuged at 16,000× g for 30 min at room temperature. The supernatant was discarded and the pellet was gently (by pipetting) dissolved with 1 mL of sterile distilled water and centrifuged again at 16,000× g for 30 min at room temperature. The supernatant was discarded and the pellet was used for the DNA isolation. The DNA was isolated from the pellets using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer instructions. Purified DNA was eluted to 50 µL of AE buffer (supplied within the kit).
2.6. Isolation of DNA from Honey Bee Workers
The DNA extraction was done according to Roetschi et al. [40]. Bees (12 g = ∼100 workers) were placed in a bag with 50 mL sterile water and crushed twice for 2 min in a stomacher apparatus at high speed. The homogenate was poured into a 50 mL plastic tube and centrifuged for 15 min at 1150× g. The supernatant (1.5 mL) was transferred to a 2 mL tube and centrifuged at 20,000× g for 2 min. The supernatant was then discarded and the pellet was resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1.2% Triton X-100, Sigma-Aldrich, St. Louis, MO, USA), amended with 20 mg/mL lysozyme and incubated for one hour at 37 °C. DNA was extracted from the lysate with the EZ1 DNA Tissue kit and a BioRobot EZ1 workstation (Qiagen, Hilden, Germany). Two μL of the extracted DNA was used as the template for real-time PCR.
2.7. Polymerase Chain Reaction (PCR)
Prior to PCR, the concentration and quality of isolated DNA from samples were quantified by a Synergy HT microplate reader (BioTek, Bad Friedrichshall, Germany). FastStart Taq DNA Polymerase (Roche, Basel, Switzerland) was used for PCR in a final volume of 25 µL; for the details, see Supplementary Material File S1. When a PCR product of 16S rRNA was detected on the gel, the reaction was judged as positive results. The length of the EFB-specific PCR product was in accordance with the previously published data: 831 bp [38]. The amplified fragments were analyzed by Sanger sequencing using commercial service and subsequently queried in BLAST (NCBI) to confirm identity and coverage of the PCR product and the template sequence of the M. plutonius 16S rRNA gene. BioEdit software [41] was used as a tool for visualization of the Sanger sequencing results.
2.8. qPCR from Adult Honey Bees
To determine the presence of M. plutonius in the extract of adult workers, we used the triplex qPCR described by Dainat et al. [42]. This triplex real-time PCR allows us to analyze the following target organisms: M. plutonius, P. larvae, and Apis mellifera. The PCR was set up in a 20 μL volume; for details, see Supplementary Material File S1.
2.9. Statistics
The sensitivity is the ability to recognize a truly positive samples (laboratory result is positive and the colony shows clinical symptoms). Specificity means the ability to recognize a truly negative sample (laboratory result is negative and the colony shows no clinical symptoms). Predictive value of a positive test shows the probability of the presence of disease if the laboratory test is positive. Predictive value of a negative test is the probability that the colony is healthy if the laboratory test is negative. Predictive value of positive and negative tests involves the prevalence of the disease in the studied population, where prevalence is the ratio of clinically positive items from the total population [43,44].
We adopted the concept of combined sensitivity and specificity [45,46] according to the standard equation:
where true positives are colonies showing EFB symptoms that tested positive for the disease, true negatives are healthy colonies that tested negative, false negatives are colonies showing EFB symptoms that tested negative, and false positives are healthy colonies that tested positive for the EFB disease.
To better understand missing data values in Real Accuracy, we calculate Pessimistic Accuracy that represents a “worst-case scenario” that also incorporates colonies with unknown symptoms and/or unknown EFB test information. The worst-case scenario accuracy is calculated according to the following equation:
In this work, we use Average Accuracy to compare varying quality and size data sets and Combined Accuracy Pessimistic to evaluate missing info value. An important measure of disease spread in the tested population is EFB prevalence that was calculated based on the ratio of positive and negative colonies inspected for EFB.
3. Results
3.1. Primer Specificity
The primers used to amplify the M. plutonius template sequence were verified using the PCR conditions described in Material and Methods. We confirmed 100% identity and 100% coverage of the 16S rRNA gene from the M. plutonius NCDO 2443 (synonymous of LME 9003 strain) strain, which is in accordance with previous research [38]. Supplementary Material File S1 describes the identity of the sequences and further compares the 16S rRNA gene among other M. plutonius strains. It should be noted that the primers can be utilized to detect all known M. plutonius strains as they cover highly conserved regions where the strains are 100% identical. We then tested whether these primers could also amplify the 16S rRNA gene of the other microbes, mainly the second infection candidates of the genus Enterococcus and Lactobacillus [6,46], when using the same PCR conditions. The results are presented in Supplementary Material File S1.
3.2. Reliability of PCR Detection from Honey and Hive Debris
The reliability of the method was tested using six sets of honey and debris samples with four different CFUs of M. plutonius (6.7 × 102; 103; 104, and 105 CFU per 1 g of honey or debris). Detection success with 6.7 × 102 CFU of M. plutonius in 1 g of hive debris and 1 g of honey was 50% and 80%, respectively. When there were more than 6.7 × 103 CFUs, there was 100% reliability of the M. plutonius detection using conventional PCR (Table 1).

Table 1.
Reliability of the analysis using Colony Forming Units (CFUs) of M. plutonius in spiked samples.
3.3. Comparison of PCR Confirmations Using Different Sample Types
Inspected colonies were assumed to be EFB positive when clinical symptoms were present. Positivity of the result was based on a positive PCR or qPCR reaction and the presence of clinical symptoms. The results were positive for EFB in 38.2%, 41.7%, and 47.1% of the honey, debris, and adult bee samples, respectively (Table 2). In total, 10 honey samples, 8 debris samples, and 8 bee samples were missing or degraded and 2 of the adult bee samples tested using qPCR were confirmed as unsure, because the Ct value was over 30, and detailed results are listed in Supplementary Material File S2.

Table 2.
Overview of the PCR results obtained for each sample matrix normalized to the clinical inspections.
3.4. Sensitivity and Specificity
Based on our results from hypothetical populations with varying disease prevalence, we performed a comparative analysis of the statistical parameters on a testing population with 52% prevalence of the disease (Table 3). The specificity of the tests was comparable for both methods and all three samples were analyzed, whereas sensitivity was 100% only for the adult bees with the qPCR. Although the sensitivity of the conventional PCR when the hive debris was used as the template was only 70% in the testing population, predictive values for the positive tests were comparable to the other two sample groups.

Table 3.
Comparative statistical parameters in a model population with 52% prevalence of the M. plutonius in honey, hive debris and adult bees.
3.5. Predictive Values of the Positive and Negative Tests Based on the Prevalence of M. plutonius in Hypothetical Populations
When there was a high prevalence of M. plutonius in a population, the predictive value of the positive test was relatively high when compared to the populations with a low prevalence of the disease, regardless of the sample type.
The highest predictive value for the positive test was 0.934 for the adults when the M. plutonius prevalence was 30%, and 0.006 when prevalence was 0.1%. In contrast, the predictive value of the negative test increased with the decreasing prevalence of M. plutonius in the tested population; however, this value remains high even for populations with high levels of prevalence for M. plutonius (Table 4).

Table 4.
Predictive values for the positive and negative tests based on the prevalence of M. plutonius in honey, hive debris, and adult bees.
4. Discussion
Hive debris may indicate the health status of honey bee colonies and can be easily collected by beekeepers [32,33,34,35,36]. It is common and an easy task to sample honey, suspected broods, or adult bees [20]; however, this is time consuming, dependent on weather conditions, requires opening colonies, and increases the risk of losing the queen during the sampling process and requires significant logistics for sending the samples to the laboratory.
The optimized method for DNA isolation presented here allows for large amounts of hive debris to be processed using an in-house made GITC buffer with DNaeasy Plant Mini Kits (Qiagen). Ryba, Titera, Haklova, and Stopka [33] reported that PCR could confirm the presence of P. larvae spores (AFB) from hive debris at levels of 102 spores per 1 g of debris. The reliability of the PCR results in this investigation were 50% at 6.7 × 102 CFU of M. plutonius per 1 g of debris. Reports from Alippi et al. [47] and Ryba, Titera, Haklova, and Stopka [33] also showed that there were detection limits of 283 and 105 spores of P. larvae per 1 g of honey, respectively. This study detected 80% reliability for the detection of M. plutonius at 6.7 × 102 CFU per 1 g honey, and this is comparable to the results for the P. larvae spores. For comparison, periods of debris collection on the bottom board [34], the place the honey was sampled from [48], and the extraction methods could all influence the reliability of the PCR confirmation of AFB from hive debris or honey, as previously described by Forsgren and Laugen [49].
To evaluate the detection of EFB when using conventional or qPCR methods on three of the most easily accessible hive samples, we selected several statistical measures of the test performance. Here, sensitivity and specificity were calculated based on the clinical symptoms detected by the bee inspector, which is considered a “golden standard”. Predictive values of positive and negative tests that were based on the prevalence of a disease among different geographically isolated populations were also an important measure of disease spread. Besides the predictive values, real accuracy was a strict measure of test performance and, thus, more appropriate for the comparison of data sets. Pessimistic accuracy counts occurred for the missing data in cases where a test underperformed, i.e., pessimistic accuracy shows uncertainty. There are undoubtedly many other statistical parameters that could be used to compare the testing methods [50,51]; here, we suggest routine calculations of the above-mentioned statistical parameters.
Bee inspectors collected samples from the suspected colonies, but all sample types were not always available, because of beekeeper’s intervention or technical reasons.
Here, the positive hive debris and honey samples were detected by either conventional PCR or qPCR and the results compared. The methods differed in the time required for analysis, price, and sensitivity, but both are used in clinical routines [52,53]. Colonies with positive adult bees that were detected using qPCR were not detected as positive from conventional PCR from the honey (1 case), hive debris (4 cases), or bee inspector (3 cases; for further details see Supplementary Material File S2). Adult bees can carry and disperse M. plutonius. Colonies in apiaries without EFB positive colonies that were close to EFB positive apiaries also had 30% positive detections for M. plutonius [54]. Social immunity plays an important role in removing diseased or dead brood from the hive nest [55,56]. Colonies with high levels of hygienic behavior could decrease the infectious pressure by removing the diseased broods [57,58,59], and thus, EFB would remain undetected by the bee inspector. This might be the cause of the missing clinical symptoms of colonies 27, 29, and 34 (see Supplementary Material File S2). Those colonies were confirmed in the laboratory as positive from debris, honey, and bee samples. The colonies were not tested or inspected later in this study. False-positive samples are also informative from a practical point of view. Those colonies might be sources for the agents (infection) and should be monitored, put under regimented controls (no exchange of frames between colonies, no moving), or preventive prophylactic measures should be performed (shook swarm method or replace old frames; [29]. In general, a colony diagnosed in the laboratory as positive but without clinical symptoms could be at risk of future disease in the apiary.
Paper sheets allow for the collection of hive debris without the need for further processing at the apiary. A beekeeper simply places the paper on the bottom board and collects hive debris for the recommended time. Here, we used 1 week during early spring time. Exposure time could be extended during the cold weather period because there is reduced activity in the colonies. The paper sheets could then be sent to the laboratory to confirm a colony at an apiary level or as a pooled sample. Pooled samples were not tested in this study, as they could be validated in another assay. All these steps could be done by beekeeper; hence, apiaries from a wide region could be sampled in a very short time with very low costs. Bee inspectors could then visit apiaries in a more targeted manner at a later date, based on the positive results from the laboratory analysis, thus reducing the time and expense required for the visual inspection. Saving time and spotting infected apiaries early in the season is important to quickly prevent the spread of disease. An example of a hypothetical schedule for sampling and testing in suspected regions is as follows: beekeepers collect hive debris, the laboratory make confirmation of hive debris by PCR, and then at an early stage of infection, a bee inspector could inspect the suspected apiary or colonies. Honey and bee samples appeared to provide more reliable results. In contrast to previous findings, the honey collected from the brood chambers of the EFB positive colonies were confirmed as positive in only 35% [48]. In general, the collection of honey, broods, or adults is more invasive and difficult to carry out early in the season and, therefore, more difficult to utilize in practice.
5. Conclusions
Based on our results, while the qPCR detection from the adult bees displayed higher sensitivity, the conventional PCR could be an appropriate alternative. Harvesting honey and sampling adult bees in brood nests is invasive for colonies and limited to the active season. Our findings support the idea that hive debris is a valuable matrix to utilize for M. plutonius detection with only a slight reduction in the sensitivity and specificity, in comparison with other tested materials. Sampling hive debris during the winter could be a preliminary check that could help to target the future in-person inspections.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/12/2/150/s1, Supplementary Material File S1: PCR and primer details; Supplementary Material File S2: Detailed results from individual colonies.
Author Contributions
J.B. (Jana Biová), J.D. and J.B. (Jaroslav Bzdil) performed the experiments; M.Š. and J.D. did the statistical analysis; J.D., J.B. (Jana Biová), S.D., M.Š. and M.P. wrote the manuscript; J.D., J.B. (Jana Biová), and J.-D.C. prepared the experimental design; all authors contributed to manuscript revision and interpretation of the results. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by Palacký University Internal Grant Agency (Project no. IGA_2020_013).
Data Availability Statement
Detailed data are available upon request.
Acknowledgments
We would like to thank Walter Gasser of the Bern Veterinary Service and the various beekeepers who made it possible to take samples from their apiaries. We would like to thank Nicolas Pradervand from Agroscope for the qPCR analyses.
Conflicts of Interest
None declared.
References
- Goulson, D.; Nicholls, E.; Botias, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Forsgren, E. European foulbrood in honey bees. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S5–S9. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Alippi, A.M.; Antúnez, K.; Aronstein, K.A.; Budge, G.; De Koker, D.; De Smet, L.; Dingman, D.W.; Evans, J.D.; Foster, L.J.; et al. Standard methods for american foulbrood research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. European foulbrood of honey bees (infection of honey bees with Melissococcus plutonius). In Oie Terrestrial Manual; WOAH: Paris, France, 2018. [Google Scholar]
- World Organisation for Animal Health. American foulbrood of honey bees (infection of honey bees with Paenibacillus larvae). In Oie Terrestrial Manual; WOAH: Paris, France, 2018. [Google Scholar]
- Forsgren, E.; Locke, B.; Sircoulomb, F.; Schäfer, M.O. Bacterial diseases in honeybees. Curr. Clin. Microbiol. Rep. 2018, 5, 18–25. [Google Scholar] [CrossRef]
- Boncristiani, H.; Ellis, J.D.; Bustamante, T.; Graham, J.; Jack, C.; Kimmel, C.B.; Mortensen, A.; Schmehl, D.R. World honey bee health: The global distribution of western honey bee (Apis mellifera L.) pests and pathogens. Bee World 2021, 98, 2–6. [Google Scholar] [CrossRef]
- Andrea Cecchini, P.H.; Dietemann, V.; Charrière, J.-D.; Grossar, D. The Influence of European Foulbrood on the Mortality of Adult Honeybees. In Proceedings of the 7th European Conference of Apidology, Cluj-Napoca, Romania, 7–9 September 2016. [Google Scholar]
- Bailey, L. Melissococcus pluton, the cause of European foulbrood of honey bees (Apis spp.). J. Appl. Bacteriol. 1983, 55, 65–69. [Google Scholar] [CrossRef]
- Alippi, A.M. Bacterial diseases of honey bees. In Bee Health and Veterinarians; Ritter, W., Ed.; World Organisation for Animal Health: Paris, France, 2014. [Google Scholar]
- Grossar, D.; Kilchenmann, V.; Forsgren, E.; Charriere, J.D.; Gauthier, L.; Chapuisat, M.; Dietemann, V. Putative determinants of virulence in Melissococcus plutonius, the bacterial agent causing european foulbrood in honey bees. Virulence 2020, 11, 554–567. [Google Scholar] [CrossRef]
- Arai, R.; Tominaga, K.; Wu, M.; Okura, M.; Ito, K.; Okamura, N.; Onishi, H.; Osaki, M.; Sugimura, Y.; Yoshiyama, M.; et al. Diversity of Melissococcus plutonius from honeybee larvae in japan and experimental reproduction of european foulbrood with cultured atypical isolates. PLoS ONE 2012, 7, e33708. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Okumura, K.; Harada, M.; Okamoto, M.; Okura, M.; Takamatsu, D. Different impacts of pmp19 on the virulence of Melissococcus plutonius strains with different genetic backgrounds. Environ. Microbiol. 2020, 22, 2756–2770. [Google Scholar] [CrossRef]
- Lewkowski, O.; Erler, S. Virulence of Melissococcus plutonius and secondary invaders associated with European foulbrood disease of the honey bee. Microbiol. Open 2019, 8, e00649. [Google Scholar] [CrossRef]
- Takamatsu, D.; Sato, M.; Yoshiyama, M. Infection of Melissococcus plutonius clonal complex 12 strain in european honeybee larvae is essentially confined to the digestive tract. J. Vet. Med. Sci. 2016, 78, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Erler, S.; Lewkowski, O.; Poehlein, A.; Forsgren, E. The curious case of achromobacter eurydice, a gram-variable pleomorphic bacterium associated with european foulbrood disease in honeybees. Microb. Ecol. 2017, 75, 1–6. [Google Scholar] [CrossRef]
- CABI. European Foulbrood. Available online: https://www.cabi.org/isc/datasheet/109547#tooverview (accessed on 12 January 2021).
- Pejchar, P. V Česku byl po 20 Letech Opět Prokázán Případ Hniloby Včelího Plodu; State Veterinary Administration: Prague, Czech Republic, 2015; Available online: https://www.svscr.cz/v_cesku_byl_po_20_letech_opet_prokazan_pripad/ (accessed on 12 January 2021).
- Nakamura, K.; Yamazaki, Y.; Shiraishi, A.; Kobayashi, S.; Harada, M.; Yoshiyama, M.; Osaki, M.; Okura, M.; Takamatsu, D. Virulence differences among Melissococcus plutonius strains with different genetic backgrounds in Apis mellifera larvae under an improved experimental condition. Sci. Rep. 2016, 6, 33329. [Google Scholar] [CrossRef]
- Forsgren, E.; Budge, G.E.; Charrière, J.-D.; Hornitzky, M.A.Z. Standard methods for European foulbrood research. J. Apic. Res. 2013, 52, 1–14. [Google Scholar] [CrossRef]
- Schafer, M.O.; Genersch, E.; Funfhaus, A.; Poppinga, L.; Formella, N.; Bettin, B.; Karger, A. Rapid identification of differentially virulent genotypes of Paenibacillus larvae, the causative organism of American foulbrood of honey bees, by whole cell Maldi-Tof Mass Spectrometry. Vet. Microbiol. 2014, 170, 291–297. [Google Scholar] [CrossRef]
- Sopko, B.; Zitek, J.; Nesvorna, M.; Markovic, M.; Kamler, M.; Titera, D.; Erban, T.; Hubert, J. Detection and quantification of Melissococcus plutonius in honey bee workers exposed to European foulbrood in czechia through conventional PCR, qPCR, and barcode sequencing. J. Apic. Res. 2019, 59, 503–514. [Google Scholar] [CrossRef]
- Polachova, V.; Pastucha, M.; Mikusova, Z.; Mickert, M.J.; Hlavacek, A.; Gorris, H.H.; Skladal, P.; Farka, Z. Click-conjugated photon-upconversion nanoparticles in an immunoassay for honeybee pathogen Melissococcus Plutonius. Nanoscale 2019, 11, 8343–8351. [Google Scholar] [CrossRef]
- Farka, Z.; Jurik, T.; Kovar, D.; Trnkova, L.; Skladal, P. Nanoparticle-based immunochemical biosensors and assays: Recent advances and challenges. Chem. Rev. 2017, 117, 9973–10042. [Google Scholar] [CrossRef] [PubMed]
- Mikušová, Z.; Farka, Z.; Pastucha, M.; Poláchová, V.; Obořilová, R.; Skládal, P. Amperometric immunosensor for rapid detection of honeybee pathogen Melissococcus Plutonius. Electroanalysis 2019, 31, 1969–1976. [Google Scholar] [CrossRef]
- Tomkies, V.; Flint, J.; Johnson, G.; Waite, R.; Wilkins, S.; Danks, C.; Watkins, M.; Cuthbertson, A.G.S.; Carpana, E.; Marris, G.; et al. Development and validation of a novel field test kit for European foulbrood. Apidologie 2008, 40, 63–72. [Google Scholar] [CrossRef]
- Owen, R. Role of human action in the spread of honey bee (hymenoptera: Apidae) pathogens. J. Econ. Èntomol. 2017, 110, 797–801. [Google Scholar] [CrossRef]
- Jacques, A.; Laurent, M.; Consortium, E.; Ribiere-Chabert, M.; Saussac, M.; Bougeard, S.; Budge, G.E.; Hendrikx, P.; Chauzat, M.P. A pan-european epidemiological study reveals honey bee colony survival depends on beekeeper education and disease control. PLoS ONE 2017, 12, e0172591. [Google Scholar] [CrossRef]
- Budge, G.E.; Barrett, B.; Jones, B.; Pietravalle, S.; Marris, G.; Chantawannakul, P.; Thwaites, R.; Hall, J.; Cuthbertson, A.G.; Brown, M.A. The occurrence of Melissococcus plutonius in healthy colonies of Apis mellifera and the efficacy of European foulbrood control measures. J. Invertebr. Pathol. 2010, 105, 164–170. [Google Scholar] [CrossRef] [PubMed]
- von Buren, R.S.; Oehen, B.; Kuhn, N.J.; Erler, S. High-resolution maps of swiss apiaries and their applicability to study spatial distribution of bacterial honey bee brood diseases. PeerJ 2019, 7, e6393. [Google Scholar] [CrossRef]
- Fries, I.; Camazine, S. Implications of horizontal and vertical pathogen transmission for honey bee epidemiology. Apidologie 2001, 32, 199–214. [Google Scholar] [CrossRef]
- Bzdil, J. Detection of Paenibacillus larvae spores in the debris and wax of honey bee by the tween 80 method. Acta Vet. BRNO 2007, 76, 643–648. [Google Scholar] [CrossRef]
- Ryba, S.; Titera, D.; Haklova, M.; Stopka, P. A PCR method of detecting American foulbrood (Paenibacillus larvae) in winter beehive wax debris. Vet. Microbiol. 2009, 139, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Ryba, S.; Kindlmann, P.; Titera, D.; Haklova, M.; Stopka, P. A new low-cost procedure for detecting nucleic acids in low-incidence samples: A case study of detecting spores of Paenibacillus larvae from bee debris. J. Econ. Entomol. 2012, 105, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Amadoro, C.; Ruberto, A.; Ricchiuti, L. Evaluation of quantitative PCR (qPCR) Paenibacillus larvae targeted assays and definition of optimal conditions for its detection/quantification in honey and hive debris. Insects 2018, 9, 165. [Google Scholar] [CrossRef]
- Ward, L.; Brown, M.; Neumann, P.; Wilkins, S.; Pettis, J.; Boonham, N. A DNA method for screening hive debris for the presence of small hive beetle (Aethina tumida). Apidologie 2007, 38, 272–280. [Google Scholar] [CrossRef]
- Ponting, S.; Tomkies, V.; Stainton, K. Rapid identification of the invasive small hive beetle (Aethina tumida) using lamp. Pest Manag. Sci. 2021, 77, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Govan, V.A.; Brozel, V.; Allsopp, M.H.; Davison, S. A pcr detection method for rapid identification of Melissococcus pluton in honeybee larvae. Appl. Environ. Microbiol. 1998, 64, 1983–1985. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.P.; Budge, G.; Cornman, R.S.; De la Rua, P.; de Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard methods for molecular research in Apis Mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Roetschi, A.; Berthoud, H.; Kuhn, R.; Imdorf, A. Infection rate based on quantitative real-time PCR of Melissococcus plutonius, the causal agent of European foulbrood, in honeybee colonies before and after apiary sanitation. Apidologie 2008, 39, 362–371. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Dainat, B.; Grossar, D.; Ecoffey, B.; Haldemann, C. Triplex real-time PCR method for the qualitative detection of European and american foulbrood in honeybee. J. Microbiol. Methods 2018, 146, 61–63. [Google Scholar] [CrossRef]
- Metz, C.E. Basic principles of roc analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Pagano, M.; Gauvreau, K. Principles of Biostatistics, 2nd ed.; Chapman and Hall/CRC: New York, NY, USA, 2018. [Google Scholar]
- vanEngelsdorp, D.; Lengerich, E.; Spleen, A.; Dainat, B.; Cresswell, J.; Baylis, K.; Nguyen, B.K.; Soroker, V.; Underwood, R.; Human, H.; et al. Standard epidemiological methods to understand and improve Apis mellifera health. J. Apic. Res. 2013, 52, 1–16. [Google Scholar] [CrossRef]
- Erban, T.; Ledvinka, O.; Kamler, M.; Hortova, B.; Nesvorna, M.; Tyl, J.; Titera, D.; Markovic, M.; Hubert, J. Bacterial community associated with worker honey bees (Apis mellifera) affected by European foulbrood. PeerJ 2017, 5, e3816. [Google Scholar] [CrossRef] [PubMed]
- Alippi, A.M.; Lopez, A.C.; Aguilar, O.M. A PCR-based method that permits specific detection of Paenibacillus larvae subsp. Larvae, the cause of American foulbrood of honey bees, at the subspecies level. Lett. Appl. Microbiol. 2004, 39, 25–33. [Google Scholar] [CrossRef]
- Forsgren, E.; Lundhagen, A.C.; Imdorf, A.; Fries, I. Distribution of Melissococcus plutonius in honeybee colonies with and without symptoms of European foulbrood. Microb. Ecol. 2005, 50, 369–374. [Google Scholar] [CrossRef]
- Forsgren, E.; Laugen, A.T. Prognostic value of using bee and hive debris samples for the detection of American foulbrood disease in honey bee colonies. Apidologie 2013, 45, 10–20. [Google Scholar] [CrossRef]
- Bloch, D.A. Comparing two diagnostic tests against the same “gold standard” in the same sample. Biometrics 1997, 53, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Umemneku Chikere, C.M.; Wilson, K.; Graziadio, S.; Vale, L.; Allen, A.J. Diagnostic test evaluation methodology: A systematic review of methods employed to evaluate diagnostic tests in the absence of gold standard—An update. PLoS ONE 2019, 14, e0223832. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.R.; Rocha-Silva, F.; Graciele-Melo, C.; Lafuente, C.R.; Magalhaes, T.; Caligiorne, R.B. Comparison between conventional and real-time PCR assays for diagnosis of visceral leishmaniasis. Biomed Res. Int. 2014, 2014, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Johansson, M.L.; Gao, Y.; Zhang, L.; Haffner, G.D.; MacIsaac, H.J.; Zhan, A. Conventional versus real-time quantitative pcr for rare species detection. Ecol. Evol. 2018, 8, 11799–11807. [Google Scholar] [CrossRef]
- Belloy, L.; Imdorf, A.; Fries, I.; Forsgren, E.; Berthoud, H.; Kuhn, R.; Charrière, J.-D. Spatial distribution of Melissococcus plutonius in adult honey bees collected from apiaries and colonies with and without symptoms of European foulbrood. Apidologie 2007, 38, 136–140. [Google Scholar] [CrossRef]
- Cremer, S.; Armitage, S.A.; Schmid-Hempel, P. Social immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef]
- Simone-Finstrom, M. Social immunity and the superorganism: Behavioral defenses protecting honey bee colonies from pathogens and parasites. Bee World 2017, 94, 21–29. [Google Scholar] [CrossRef]
- Spivak, M.; Reuter, G.S. Resistance to American foulbrood disease by honey bee colonies Apis mellifera bred for hygienic behavior. Apidologie 2001, 32, 555–565. [Google Scholar] [CrossRef]
- Swanson, J.A.; Torto, B.; Kells, S.A.; Mesce, K.A.; Tumlinson, J.H.; Spivak, M. Odorants that induce hygienic behavior in honeybees: Identification of volatile compounds in chalkbrood-infected honeybee larvae. J. Chem. Ecol. 2009, 35, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Palacio, M.A.; Rodriguez, E.; Goncalves, L.; Bedascarrasbure, E.; Spivak, M. Hygienic behaviors of honey bees in response to brood experimentally pin-killed or infected with Ascosphaera apis. Apidologie 2010, 41, 602–612. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).