DBSCAN and GIE, Two Density-Based “Grid-Free” Methods for Finding Areas of Endemism: A Case Study of Flea Beetles (Coleoptera, Chrysomelidae) in the Afrotropical Region
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
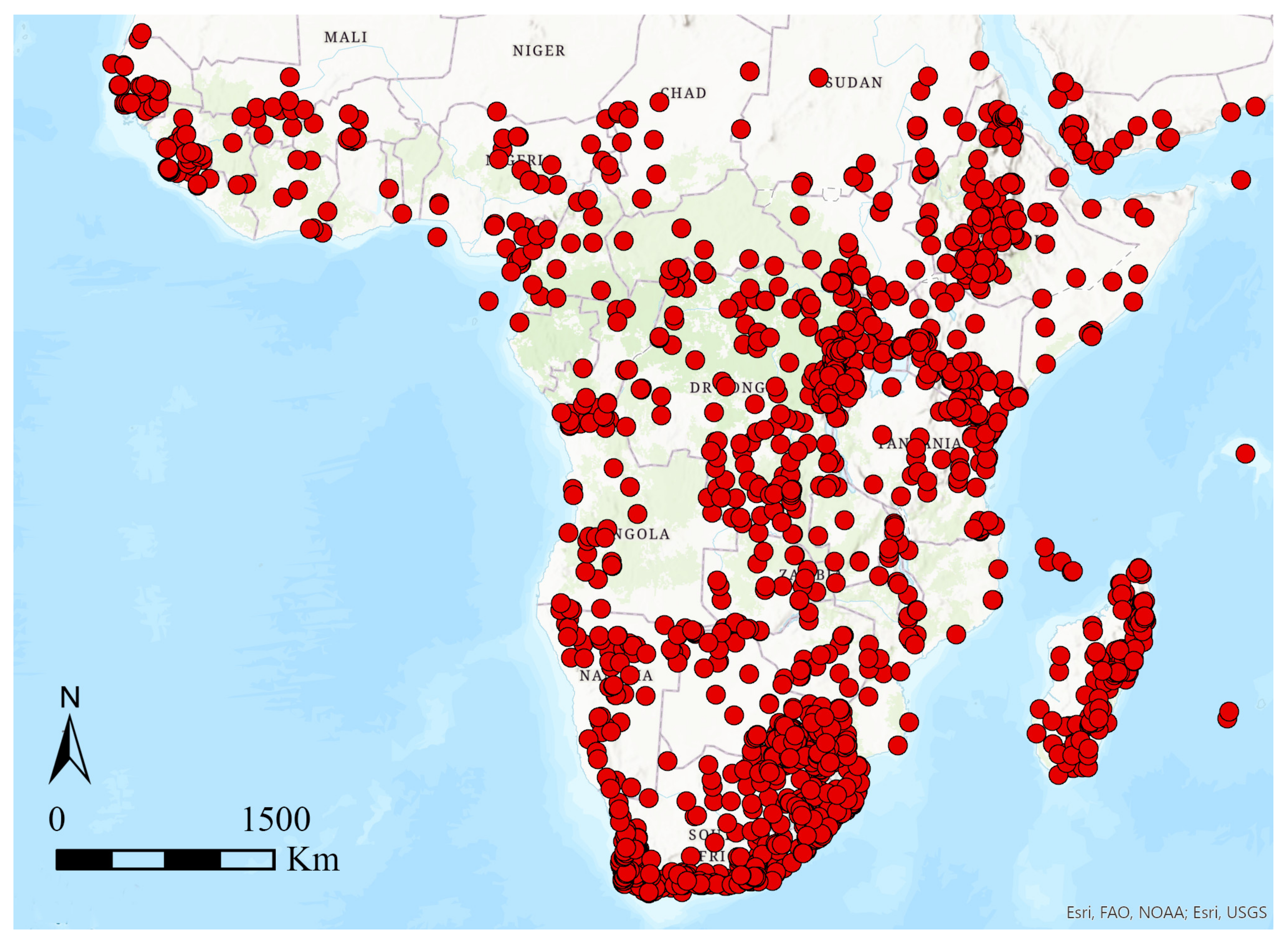
2.1. Study Area and Species Database
2.2. Abbreviations Used
2.3. Geographical Interpolation of Endemism (GIE)
2.4. Density-Based Spatial Clustering of Application with Noise (DBSCAN)
2.5. The CLUENDA Tool
3. Results
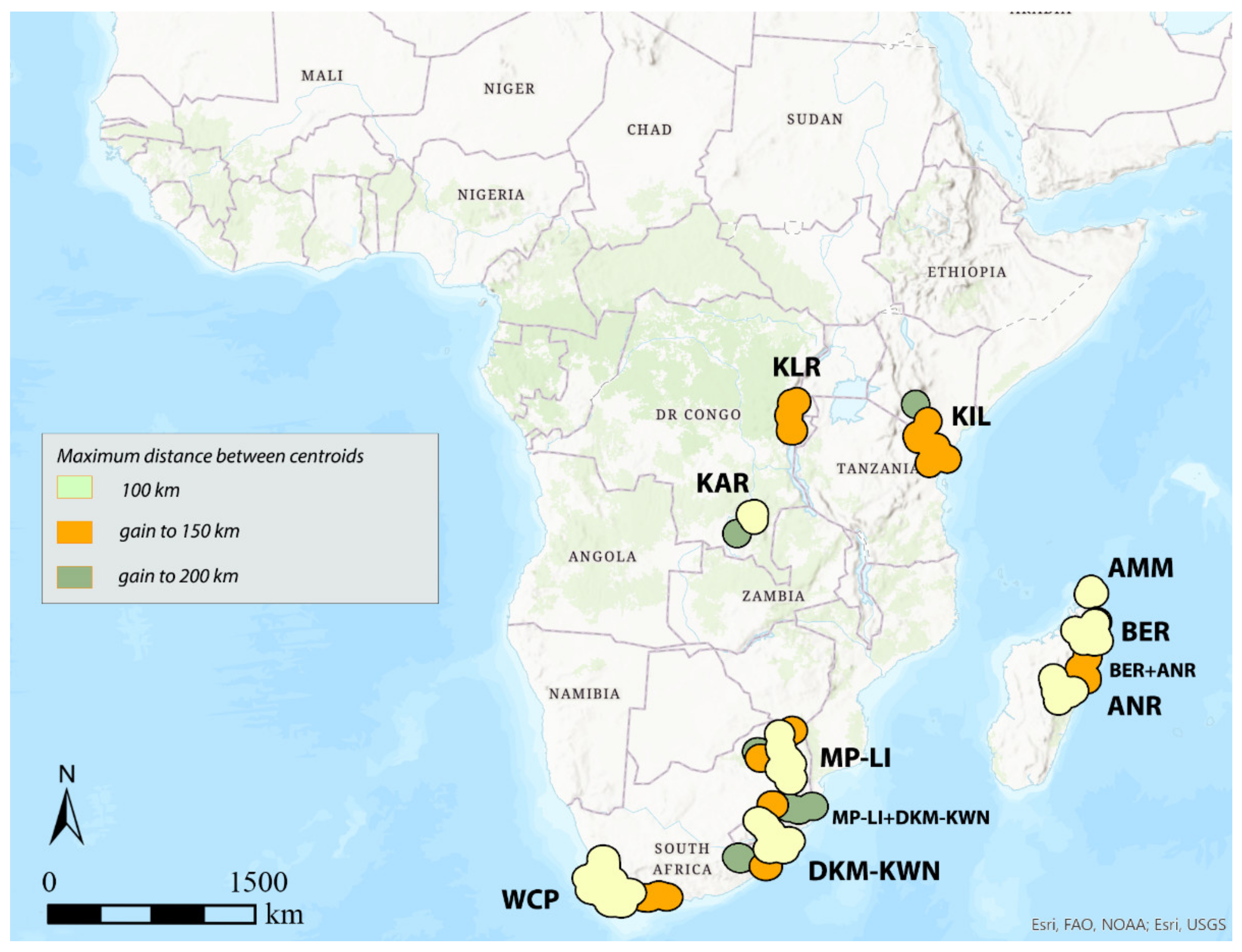
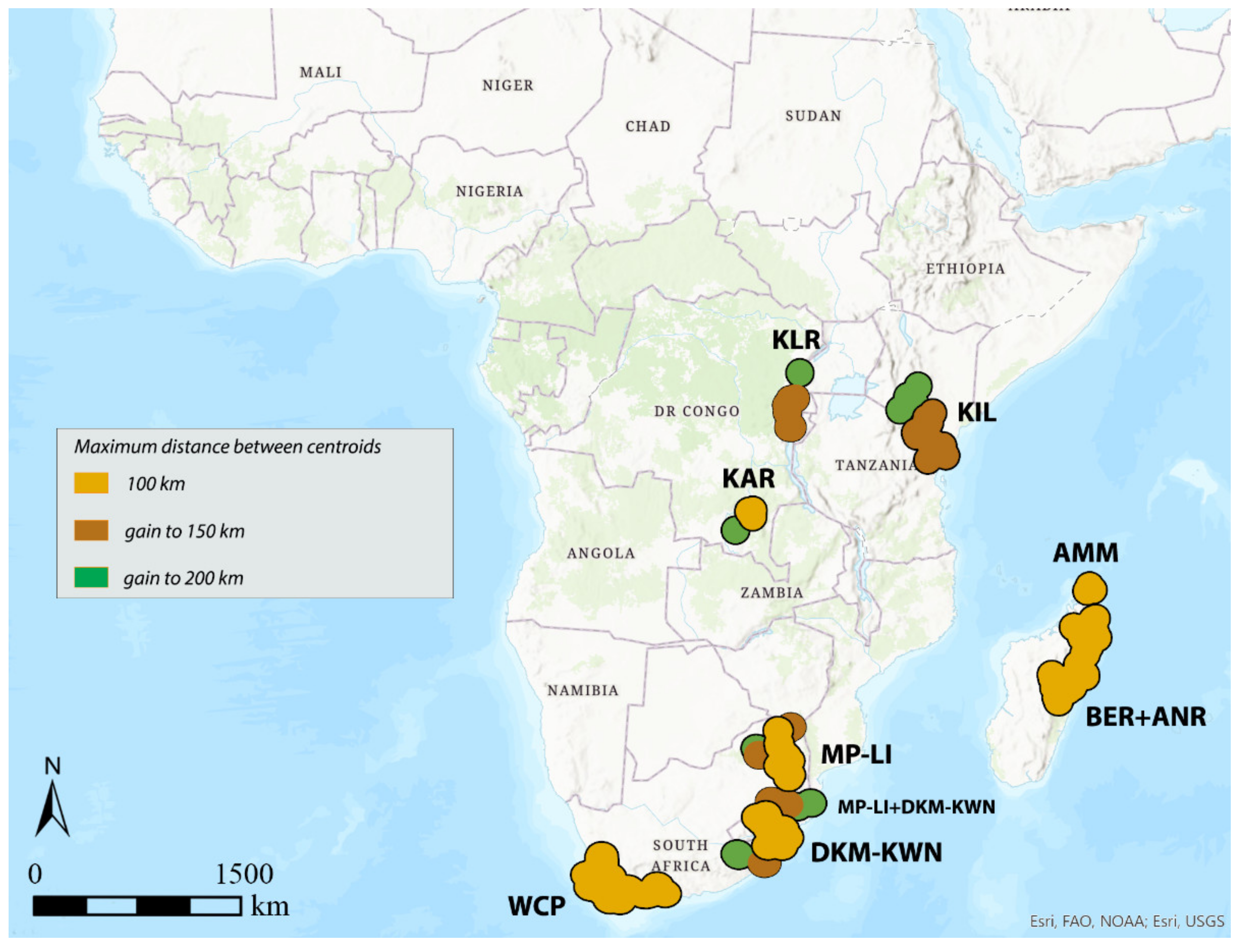
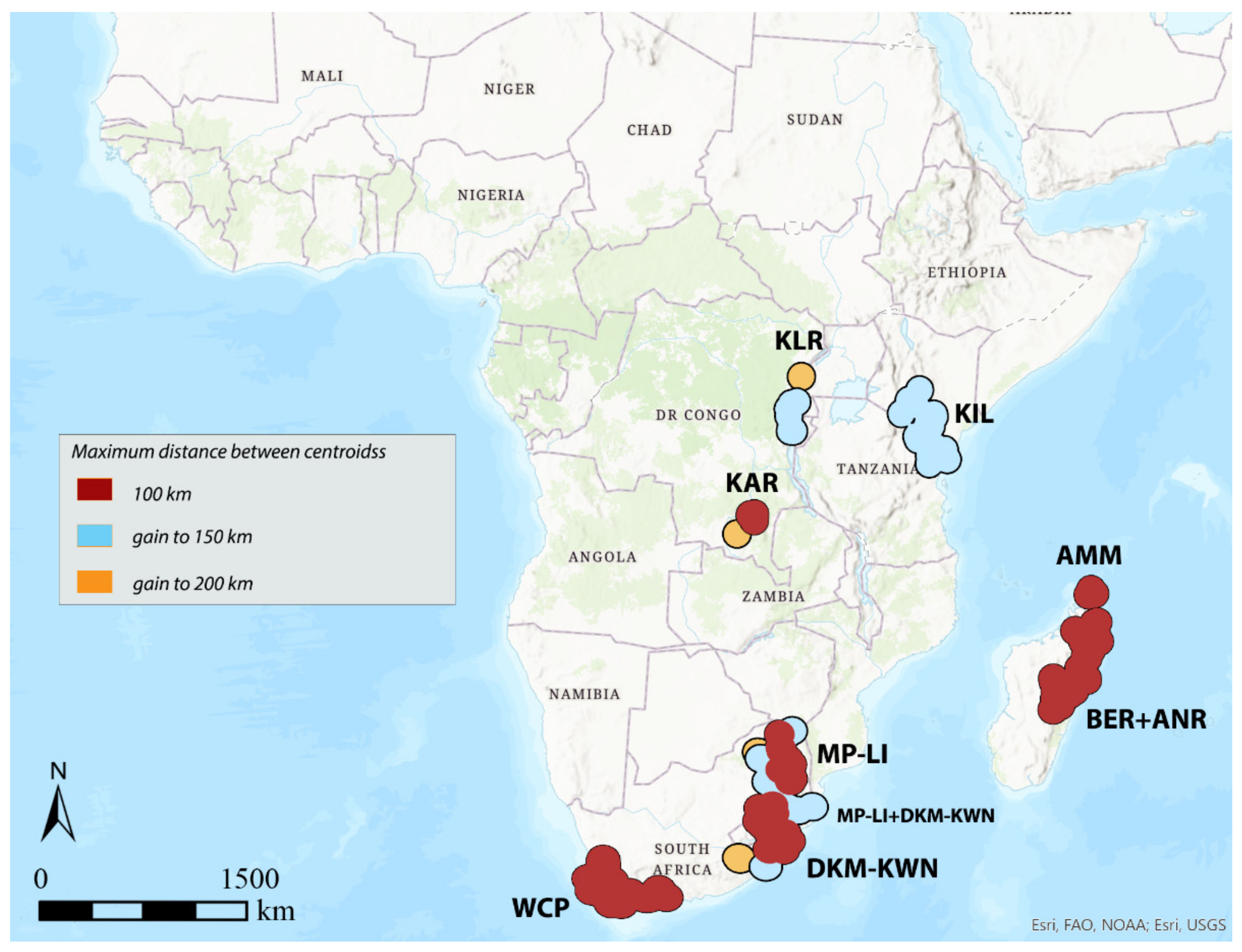
3.1. Areas of Endemism (AoEs)
- -
- AMM (Amber Mountain): this refers to the Amber Mountain, a famous protected area in the Diana region of Northern Madagascar. AMM is well known for its endemic flora and fauna. AMM is a part of the “Madagascar and the Indian Ocean Islands” biodiversity hotspot [50].
- -
- ANR (Antananarivo region): this AoE is located in Central Madagascar and mainly includes Anamalanga, Bongolava, Itasy, and Vakinankaratra regions. ANR is a part of the “Madagascar and the Indian Ocean Islands” biodiversity hotspot.
- -
- BER (Betsiboka region): this AoE is located in Northern Madagascar and includes, in addition to the Betsiboka, the Sofia and Analanjirofo regions. BER is a part of the “Madagascar and the Indian Ocean Islands” biodiversity hotspot.
- -
- DKM-KWN (Drakensberg Mountains–KwaZulu Natal): this AoE comprehends the Drakensberg Mountains, which are main mountain range in southern Africa, and the coastal areas of KwaZulu Natal. The Drakensberg Mountains rise to more than 3475 m a.s.l., extending roughly from northeast to southwest for over 1100 km parallel to the southeastern coast of South Africa. They are part of the Great Escarpment and separate the extensive highlands of the South African interior from the lower lands along the coast. In the Drakensberg Mountains, alpine grasslands and small pockets of Afromontane Forest are present. The coastal regions of KwaZulu-Natal typically have subtropical thickets and deeper ravines; steep slopes host small spots of Afromontane Forest. The midlands have moist grasslands. The northern area has a primarily moist savannah habitat. DKM-KWN is a part of the “Maputaland–Pondoland–Albany” biodiversity hotspot.
- -
- KAR (Katanga region): this AoE is located in the Democratic Republic of Congo and mainly includes the ridges of the plateaus of Katanga (Shaba) province. They include Kundelungu (1600 m a.s.l.), Mitumba (1500 m), and Hakansson (1100 m) mountains. The Katanga plateaus reach as far north as the Lukuga River and contain the Manika Plateau, the Kibara and the Bia mountains, and the high plains of Marungu. Despite the high plant diversity present [51], KAR is not included in any biodiversity hotspot.
- -
- KIL (Kilimanjaro region): this AoE has the Kilimanjaro region as its central area. Towards the southeast, it extends to the Tsavo West National Park in Kenya and Mkomazi National Park in Tanzania, while northwards it extends to the Amboseli National Park. KIL comprehends both montane and savannah areas. The Kilimanjaro area is a part of the “Eastern Afromontane” biodiversity hotspot.
- -
- KLR (Kivu Lake region): this AoE is located in the area of the Kivu Lake in the Albertine Rift Valley and also includes the mountain areas of Birunga, Volcan Mikeno, and Volcan Karisimbi, between Uganda, Rwanda, and the Democratic Republic of Congo (Kivu Sud). KLR is a part of the “Eastern Afromontane” biodiversity hotspot.
- -
- MP-LI (Mpumalanga–Limpopo): this AoE comprehends part of the Mpumalanga and Limpopo provinces. Mpumalanga is divided by the Drakensberg escarpment into a westerly half consisting mainly of high-altitude grassland called the Highveld and an eastern half situated in low-altitude subtropical Lowveld/Bushveld, mostly savannah habitat. The Lowveld is relatively flat with interspersed rocky outcrops. The Lebombo Mountains form a low range in the far east, on the border with Mozambique. Limpopo contains much of the Waterberg Biosphere, a massif of approximately 15,000 km2 which is the first region in the northern part of South Africa to be named a UNESCO Biosphere Reserve (http://waterbergbiospherereserve.org/why-the-waterberg-is-a-biosphere.html, accessed on 12 October 2021). MP-LI is partially included in the “Maputaland–Pondoland–Albany” biodiversity hotspot.
- -
- WCP (Western Cape Province): this AoE is mainly restricted to South Africa’s Western Cape Province. Most of the region is covered with fynbos, a sclerophyllous shrubland occurring on acid sands or nutrient-poor soils derived from Table Mountain sandstones. This area covers the Mediterranean climate region of South Africa in the southwestern corner of the country and extends eastward into the Eastern Cape Province. WCP is a part of one of the world’s six floral kingdoms, the “Cape Floristic Region” biodiversity hotspot.
3.2. GIE
3.3. DBSCAN
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Prado, J.R.; Brennand, P.G.G.; Godoy, L.P.; Libardi, G.S.; Abreu-Júnior, E.F.; Roth, P.R.O.; Chiquito, E.A.; Percequillo, A.R. Species richness and areas of endemism of oryzomyine rodents (Cricetidae, Sigmodontinae) in South America: An NDM/VNDM approach. J. Biogeogr. 2015, 42, 540–551. [Google Scholar] [CrossRef]
- DaSilva, M.B.; Pinto-da-Rocha, R.; de Carvalho, C.; Almeida, E.A.B. Biogeografia Da América Do Sul: Padrões E Processos; Editora Roca: São Paulo, Brazil, 2011. [Google Scholar]
- Da Cardoso Silva, J.M.; de Cardoso Sousa, M.; Castelletti, C.H. Areas of endemism for passerine birds in the Atlantic forest, South America. Glob. Ecol. Biogeogr. 2004, 13, 85–92. [Google Scholar] [CrossRef]
- Hurdu, B.-I.; Escalante, T.; Pușcaș, M.; Novikoff, A.; Bartha, L.; Zimmermann, N.E. Exploring the different facets of plant endemism in the South-Eastern Carpathians: A manifold approach for the determination of biotic elements, centres and areas of endemism. Biol. J. Linn. Soc. 2016, 119, 649–672. [Google Scholar] [CrossRef]
- Brown, J.H.; Stevens, G.C.; Kaufman, D.M. The geographic range: Size, shape, boundaries, and internal structure. Annu. Rev. Ecol. Syst. 1996, 27, 597–623. [Google Scholar] [CrossRef] [Green Version]
- Morrone, J.J. Evolutionary Biogeography: An Integrative Approach with Case Studies; Columbia University Press: New York, NY, USA, 2008; ISBN 0-231-51283-X. [Google Scholar]
- Fattorini, S. Endemism in historical biogeography and conservation biology: Concepts and implications. Biogeogr. J. Integr. Biogeogr. 2017, 32, 47–75. [Google Scholar] [CrossRef] [Green Version]
- Morrone, J.J.; Crisci, J.V. Historical biogeography: Introduction to methods. Annu. Rev. Ecol. Syst. 1995, 26, 373–401. [Google Scholar] [CrossRef]
- Platnick, N.I. On areas of endemism. Aust. Syst. Bot. 1991, 4, 11–12. [Google Scholar]
- Risp, M.D.; Linder, H.P.; Weston, P.H. Cladistic biogeography of plants in Australia and New Guinea: Congruent pattern reveals two endemic tropical tracks. Syst. Biol. 1995, 44, 457–473. [Google Scholar] [CrossRef]
- Harold, A.S.; Mooi, R.D. Areas of endemism: Definition and recognition criteria. Syst. Biol. 1994, 43, 261–266. [Google Scholar] [CrossRef]
- Cracraft, J. Species diversity, biogeography, and the evolution of biotas. Am. Zool. 1994, 34, 33–47. [Google Scholar] [CrossRef]
- Anderson, S. Area and endemism. Q. Rev. Biol. 1994, 69, 451–471. [Google Scholar] [CrossRef]
- DaSilva, M.B.; Pinto-da-Rocha, R.; DeSouza, A.M. A protocol for the delimitation of areas of endemism and the historical regionalization of the Brazilian Atlantic Rain Forest using harvestmen distribution data. Cladistics 2015, 31, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Crother, B.I.; Murray, C.M. Ontology of Areas of Endemism; Wiley Online Library: Hoboken, NJ, USA, 2011; ISBN 0305-0270. [Google Scholar]
- Linder, H.P. On areas of endemism, with an example from the African Restionaceae. Syst. Biol. 2001, 50, 892–912. [Google Scholar] [CrossRef]
- Henderson, I.M. Biogeography without area? Aust. Syst. Bot. 1991, 4, 59–71. [Google Scholar] [CrossRef]
- Sigrist, M.S.; de Carvalho, C.J.B. Detection of areas of endemism on two spatial scales using Parsimony Analysis of Endemicity (PAE): The Neotropical region and the Atlantic Forest. Biota Neotrop. 2008, 8, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Morrone, J.J. Parsimony analysis of endemicity (PAE) revisited. J. Biogeogr. 2014, 41, 842–854. [Google Scholar] [CrossRef]
- Morrone, J.J. On the identification of areas of endemism. Syst. Biol. 1994, 43, 438–441. [Google Scholar] [CrossRef]
- Cracraft, J. Patterns of diversification within continental biotas: Hierarchical congruence among the areas of endemism of Australian vertebrates. Aust. Syst. Bot. 1991, 4, 211–227. [Google Scholar] [CrossRef]
- Aagesen, L.; Szumik, C.; Goloboff, P. Consensus in the Search for Areas of Endemism; Wiley Online Library: Hoboken, NJ, USA, 2013; ISBN 0305-0270. [Google Scholar]
- Szumik, C.A.; Goloboff, P.A. Areas of endemism: An improved optimality criterion. Syst. Biol. 2004, 53, 968–977. [Google Scholar] [CrossRef] [Green Version]
- Szumik, C.A.; Cuezzo, F.; Goloboff, P.A.; Chalup, A.E. An optimality criterion to determine areas of endemism. Syst. Biol. 2002, 51, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Torres-Miranda, A.; Luna-Vega, I.; Oyama, K. New approaches to the biogeography and areas of endemism of red oaks (Quercus L., Section Lobatae). Syst. Biol. 2013, 62, 555–573. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, U.; Brescovit, A.D.; Santos, A.J. Delimiting areas of endemism through kernel interpolation. PLoS ONE 2015, 10, e0116673. [Google Scholar]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Knowledge Discovery and Data Mining (KDD); University of Munich: Munich, Germany, 1996; Volume 96, pp. 226–231. [Google Scholar]
- Fleming, C.H.; Calabrese, J.M. A new kernel density estimator for accurate home-range and species-range area estimation. Methods Ecol. Evol. 2017, 8, 571–579. [Google Scholar] [CrossRef]
- Bailey, T.C.; Gatrell, A.C. Interactive Spatial Data Analysis; Longman Scientific & Technical Essex: New York, NY, USA, 1995; Volume 413. [Google Scholar]
- Navarro, F.R.; Cuezzo, F.D.C.; Goloboff, P.A.; Szumik, C.A.; Lizarralde, M.S.; Quintana, M.G. Can insect data be used to infer areas of endemism? An example from the Yungas of Argentina. Soc. Biol. Chile 2009, 82, 507–522. [Google Scholar] [CrossRef]
- Biondi, M.; Urbani, F.; D’Alessandro, P. Relationships between the geographic distribution of phytophagous insects and different types of vegetation: A case study of the flea beetle genus Chaetocnema (Coleoptera: Chrysomelidae) in the Afrotropical region. Eur. J. Entomol. 2015, 112, 311–327. [Google Scholar] [CrossRef] [Green Version]
- Andrew, N.R.; Hughes, L. Species diversity and structure of phytophagous beetle assemblages along a latitudinal gradient: Predicting the potential impacts of climate change. Ecol. Entomol. 2004, 29, 527–542. [Google Scholar] [CrossRef]
- Basset, Y. Invertebrates in the canopy of tropical rain forests How much do we really know? Plant Ecol. 2001, 153, 87–107. [Google Scholar] [CrossRef]
- Thormann, B. Biodiversity of Leaf Beetles (Coleoptera: Chrysomelidae) in a Tropical Montane Rainforest Ecosystem Assessed with DNA Barcoding. Ph.D. Thesis, University of Bonn, Bonn, Germany, 2016. [Google Scholar]
- Flowers, R.W.; Hanson, P.E. Leaf beetle (Coleoptera: Chrysomelidae) diversity in eight Costa Rican habitats. In Special Topics in Leaf Beetle Biology, Proceedings of the 5th International Symposium on the Chrysomelidae, Foz do Iguaçu, Brazil, 25–27 August 2000; Pensoft Publishers: Sofia, Bulgaria, 2003; pp. 25–51. [Google Scholar]
- Ødegaard, F.; Diserud, O.H.; Engen, S.; Aagaard, K. The magnitude of local host specif icity for phytophagous insects and its implications for estimates of global species richness. Conserv. Biol. 2000, 14, 1182–1186. [Google Scholar] [CrossRef] [Green Version]
- Nadein, K.S.; Beždek, J. Galerucinae Latreille, 1802. Morphol. Syst. Phytophaga Gruyter Berl. 2014, 3, 251–259. [Google Scholar]
- Nadein, K.S. Catalogue of Alticini genera of the World (Coleoptera: Chrysomelidae). Available online: http://www.zin.ru/Animalia/Coleoptera/eng/alticinw.htm (accessed on 20 October 2021).
- Ruan, Y.; Konstantinov, A.S.; Shi, G.; Tao, Y.; Li, Y.; Johnson, A.J.; Luo, X.; Zhang, X.; Zhang, M.; Wu, J.; et al. The jumping mechanism of flea beetles (Coleoptera, Chrysomelidae, Alticini), its application to bionics and preliminary design for a robotic jumping leg. ZooKeys 2020, 915, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Nadein, K.; Betz, O. Jumping mechanisms and performance in beetles. I. Flea beetles (Coleoptera: Chrysomelidae: Alticini). J. Exp. Biol. 2016, 219, 2015–2027. [Google Scholar] [CrossRef] [Green Version]
- Iannella, M.; D’Alessandro, P.; de Simone, W.; Biondi, M. Habitat specificity, host plants and areas of endemism for the genera-group Blepharida sl in the afrotropical region (Coleoptera, Chrysomelidae, Galerucinae, Alticini). Insects 2021, 12, 299. [Google Scholar] [CrossRef]
- D’Alessandro, P.; Iannella, M.; Frasca, R.; Biondi, M. Distribution patterns and habitat preference for the genera-group Blepharida sl in Sub-Saharan Africa (Coleoptera: Chrysomelidae: Galerucinae: Alticini). Zool. Anz. 2018, 277, 23–32. [Google Scholar] [CrossRef]
- Jolivet, P.; Verma, K.K. Biology of Leaf Beetles; Intercept Limited: Andover, UK, 2002; ISBN 1-898298-86-6. [Google Scholar]
- Biondi, M.; Iannella, M.; D’Alessandro, P. Adamastoraltica humicola, new genus and new species: The first example of possible moss-inhabiting flea beetle genus from sub-Saharan Africa (Coleoptera, Chrysomelidae, Galerucinae). Zootaxa 2020, 4763, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Biondi, M.; D’Alessandro, P. Ntaolaltica and Pseudophygasia, two new flea beetle genera from Madagascar (Coleoptera: Chrysomelidae: Galerucinae: Alticini). Insect Syst. Evol. 2013, 44, 93–106. [Google Scholar] [CrossRef]
- Biondi, M.; D’Alessandro, P. Afrotropical flea beetle genera: A key to their identification, updated catalogue and biogeographical analysis (Coleoptera, Chrysomelidae, Galerucinae, Alticini). ZooKeys 2012, 253, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udvardy, M.D.; Udvardy, M.D.F. A Classification of the Biogeographical Provinces of the World; International Union for Conservation of Nature and Natural Resources: Morges, Switzerland, 1975; Volume 8. [Google Scholar]
- Oliveira, U.; Soares-Filho, B.; Leitão, R.F.M.; Rodrigues, H.O. BioDinamica: A toolkit for analyses of biodiversity and biogeography on the Dinamica-EGO modelling platform. PeerJ 2019, 7, e7213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campello, R.J.G.B.; Moulavi, D.; Sander, J. Density-Based Clustering Based on Hierarchical Density Estimates. In Advances in Knowledge Discovery and Data Mining; Pei, J., Tseng, V.S., Cao, L., Motoda, H., Xu, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 160–172. [Google Scholar]
- Habel, J.C.; Rasche, L.; Schneider, U.A.; Engler, J.O.; Schmid, E.; Rödder, D.; Meyer, S.T.; Trapp, N.; del Sos Diego, R.; Eggermont, H. Final countdown for biodiversity hotspots. Conserv. Lett. 2019, 12, e12668. [Google Scholar] [CrossRef] [Green Version]
- Küper, W.; Sommer, J.H.; Lovett, J.C.; Mutke, J.; Linder, H.P.; Beentje, H.J.; van Rompaey, R.S.A.R.; Chatelain, C.; Sosef, M.; Barthlott, W. Africa’s hotspots of biodiversity redefined. Ann. Mo. Bot. Gard. 2004, 1, 525–535. [Google Scholar]
- Morrone, J.J.; Escalante, T. Parsimony analysis of endemicity (PAE) of Mexican terrestrial mammals at different area units: When size matters. J. Biogeogr. 2002, 29, 1095–1104. [Google Scholar] [CrossRef]


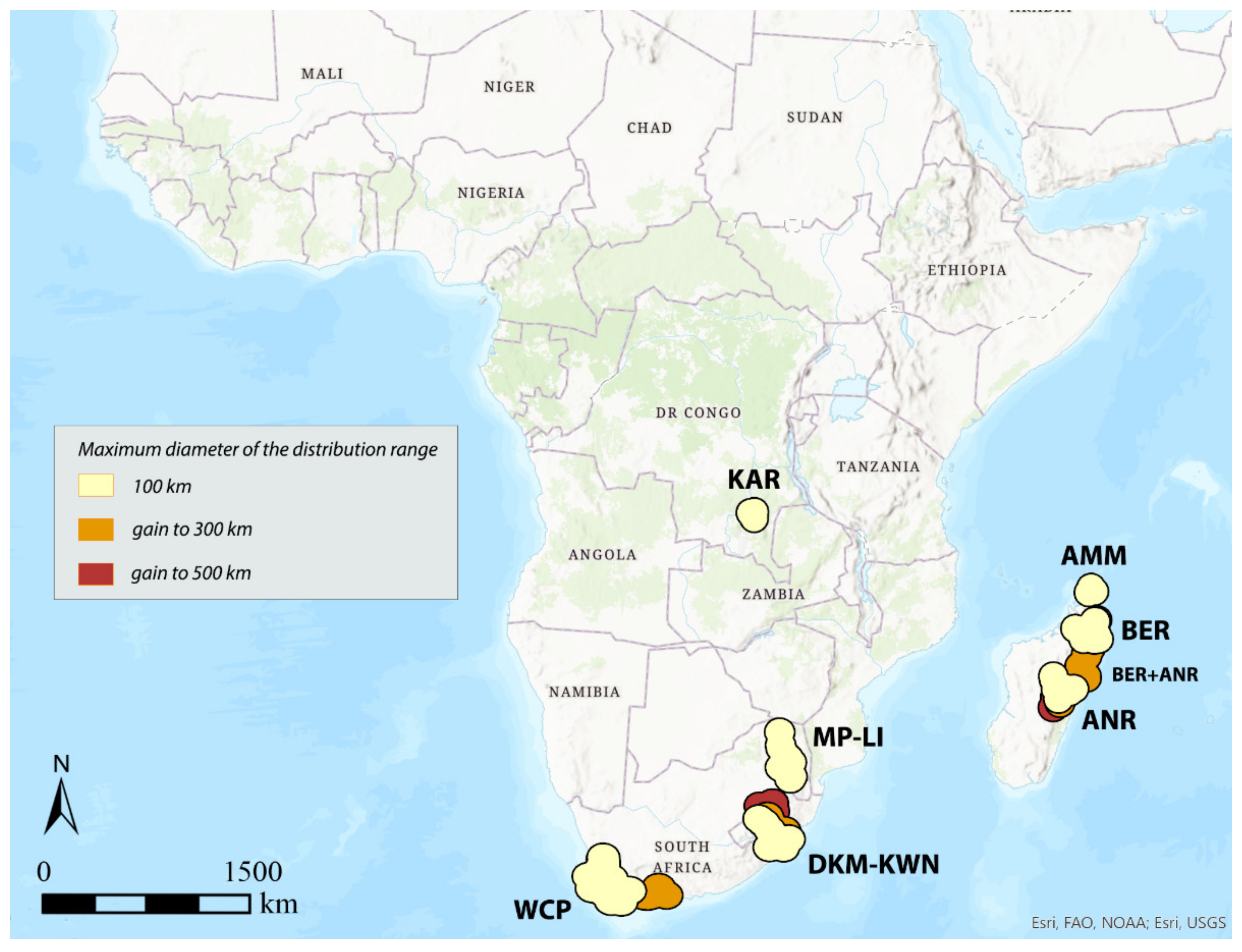
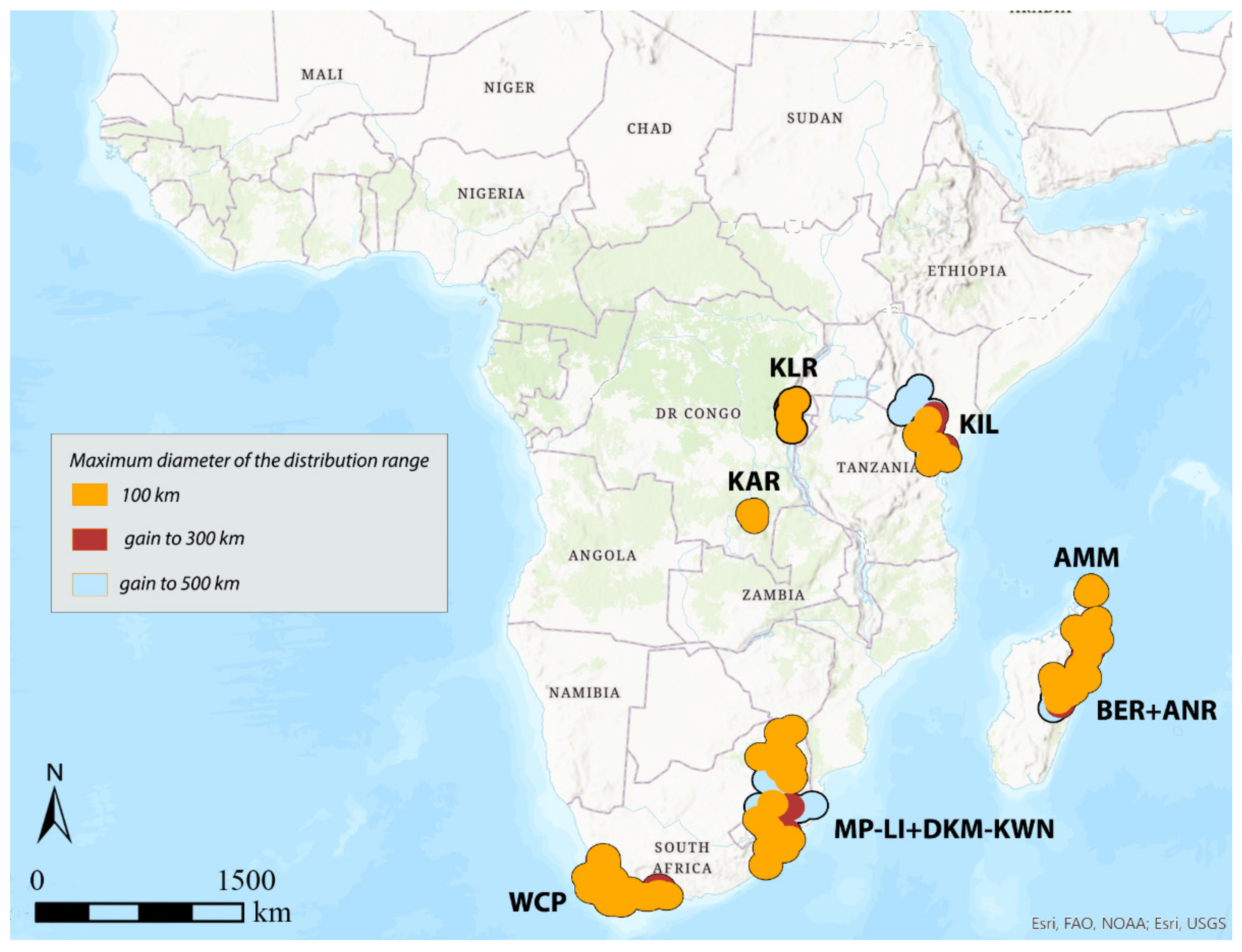
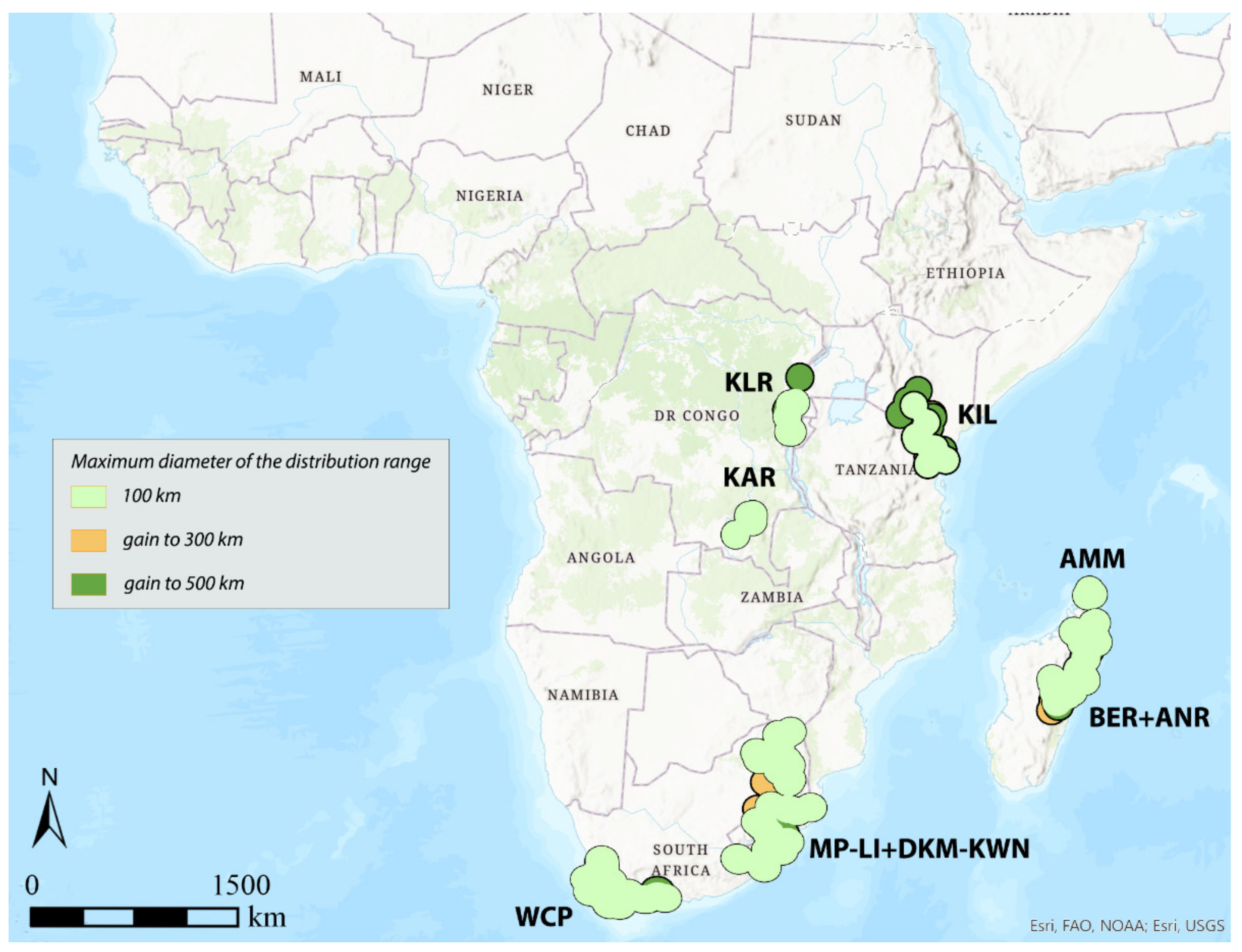

| AoE → | KLR | Area | KIL | Area | KAR | Area | DKM-KWN | Area | MP-LI | Area | DKM-KWN + MP-LI | Area | WCP | Area | AMM | Area | BER | Area | ANR | Area | BER + ANR | Area |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dd-dc ↓ | syn | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 | syn (tot) | km2 |
| 100–100 | <5 | / | <5 | / | 6 (26) | 44,295 | 10 (53) | 102,354 | 11 (55) | 108,354 | ꟷ | / | 21 (57) | 146,352 | 6 (12) | 43,465 | 10 (27) | 91,580 | 10 (34) | 85,126 | ꟷ | / |
| 300–100 | <5 | / | <5 | / | 6 (26) | 44,295 | 13 (53) | 114,482 | 11 (55) | 108,354 | ꟷ | / | 32 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 35 (60) | 231,762 | ||
| 500–100 | <5 | / | <5 | / | 6 (26) | 44,295 | 16 (54) | 143,527 | 11 (55) | 108,354 | ꟷ | / | 35 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 40 (61) | 242,557 | ||
| 100–150 | 5 (31) | 80,810 | 8 (35) | 127,636 | 6 (26) | 44,295 | 13 (58) | 151,118 | 14 (59) | 148,547 | ꟷ | / | 25 (67) | 190,544 | 6 (12) | 43,465 | ► | ► | 25 (60) | 224,984 | ||
| 300–150 | 5 (31) | 80,810 | 11 (36) | 144,800 | 6 (26) | 44,295 | 17 (58) | 175,819 | 14 (59) | 148,547 | ꟷ | / | 32 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 35 (60) | 231,762 | ||
| 500–150 | 6 (31) | 82,179 | 17 (39) | 221,626 | 6 (26) | 44,295 | ► | ► | 38 (94) | 379,608 | 35 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 40 (61) | 242,557 | ||||
| 100–200 | 5 (31) | 80,810 | 9 (38) | 154,709 | 7 (28) | 71,240 | ► | ► | 34 (97) | 389,194 | 25 (67) | 190,544 | 6 (12) | 43,465 | ► | ► | 25 (60) | 224,984 | ||||
| 300–200 | 7 (42) | 112,281 | 15 (39) | 218,954 | 7 (28) | 71,240 | ► | ► | 37 (97) | 393,318 | 32 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 35 (60) | 231,762 | ||||
| 500–200 | 7 (42) | 112,281 | 17 (39) | 221,626 | 7 (28) | 71,240 | ► | ► | 41 (97) | 419,098 | 35 (67) | 197,269 | 6 (12) | 43,465 | ► | ► | 40 (61) | 242,557 |
| KLR | KIL | KAR | DKM-KWN | MP-LI | DKM-KWN + MP-LI | WCP | AMM | BER + ANR | AMM + BER + ANR | |
|---|---|---|---|---|---|---|---|---|---|---|
| GIE | syn | syn | syn | syn | syn | syn | syn | syn | syn | syn |
| 100 km | / | / | 6 | / | 13 | / | 20 | 6 | 28 | / |
| 150 km | 5 | / | 6 | 17 | 13 | / | 31 | 6 | 35 | / |
| 200 km | 5 | 12 | 7 | 19 | 14 | / | 32 | / | / | 51 |
| DBSCAN | ||||||||||
| 100 km | / | / | 6 | 16 | 11 | / | 35 | 6 | 40 | / |
| 150 km | 6 | 17 | 6 | / | / | 38 | 35 | 6 | 40 | / |
| 200 km | 7 | 17 | 7 | / | / | 41 | 35 | 6 | 40 | / |
| DBSCAN | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| dd-dc | 100 km/100 km | 300 km/100 km | 500 km/100 km | 100 km/150 km | 300 km/150 km | 500 km/150 km | 100 km/200 km | 300 km/200 km | 500 km/200 km |
| 100 km/100 km | 1 | 0.522 | 0.459 | 1.000 | 0.522 | 0.459 | 1.000 | 0.522 | 0.459 |
| 300 km/100 km | 0.522 | 1 | 0.920 | 0.522 | 1.000 | 0.920 | 0.522 | 1.000 | 0.920 |
| 500 km/100 km | 0.459 | 0.920 | 1 | 0.459 | 0.920 | 1.000 | 0.459 | 0.920 | 1.000 |
| 100 km/150 km | 1.000 | 0.522 | 0.459 | 1 | 0.522 | 0.459 | 1.000 | 0.522 | 0.459 |
| 300 km/150 km | 0.522 | 1.000 | 0.920 | 0.522 | 1 | 0.920 | 0.522 | 1.000 | 0.920 |
| 500 km/150 km | 0.459 | 0.920 | 1.000 | 0.459 | 0.920 | 1 | 0.459 | 0.920 | 1.000 |
| 100 km/200 km | 1.000 | 0.522 | 0.459 | 1.000 | 0.522 | 0.459 | 1 | 0.522 | 0.459 |
| 300 km/200 km | 0.522 | 1.000 | 0.920 | 0.522 | 1.000 | 0.920 | 0.522 | 1 | 0.920 |
| 500 km/200 km | 0.459 | 0.920 | 1.000 | 0.459 | 0.920 | 1.000 | 0.459 | 0.920 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biondi, M.; D’Alessandro, P.; De Simone, W.; Iannella, M. DBSCAN and GIE, Two Density-Based “Grid-Free” Methods for Finding Areas of Endemism: A Case Study of Flea Beetles (Coleoptera, Chrysomelidae) in the Afrotropical Region. Insects 2021, 12, 1115. https://doi.org/10.3390/insects12121115
Biondi M, D’Alessandro P, De Simone W, Iannella M. DBSCAN and GIE, Two Density-Based “Grid-Free” Methods for Finding Areas of Endemism: A Case Study of Flea Beetles (Coleoptera, Chrysomelidae) in the Afrotropical Region. Insects. 2021; 12(12):1115. https://doi.org/10.3390/insects12121115
Chicago/Turabian StyleBiondi, Maurizio, Paola D’Alessandro, Walter De Simone, and Mattia Iannella. 2021. "DBSCAN and GIE, Two Density-Based “Grid-Free” Methods for Finding Areas of Endemism: A Case Study of Flea Beetles (Coleoptera, Chrysomelidae) in the Afrotropical Region" Insects 12, no. 12: 1115. https://doi.org/10.3390/insects12121115
APA StyleBiondi, M., D’Alessandro, P., De Simone, W., & Iannella, M. (2021). DBSCAN and GIE, Two Density-Based “Grid-Free” Methods for Finding Areas of Endemism: A Case Study of Flea Beetles (Coleoptera, Chrysomelidae) in the Afrotropical Region. Insects, 12(12), 1115. https://doi.org/10.3390/insects12121115








