A Novel Reference for Bt-Resistance Mechanism in Plutella xylostella Based on Analysis of the Midgut Transcriptomes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Sample Collection
2.2. Cry1Ac Protoxin Purification
2.3. RNA Extraction, Library Construction, and Illumina Sequencing
2.4. Bioinformatics Analysis of the Transcriptome
2.5. Gene Function Annotation and Characterization
2.6. Differentially Expressed Gene in the Susceptible and Resistant Strains of P. xylostella
2.7. Real-Time Quantitative PCR Analysis of Gene Expression
3. Results
3.1. Illumina Sequencing Analysis
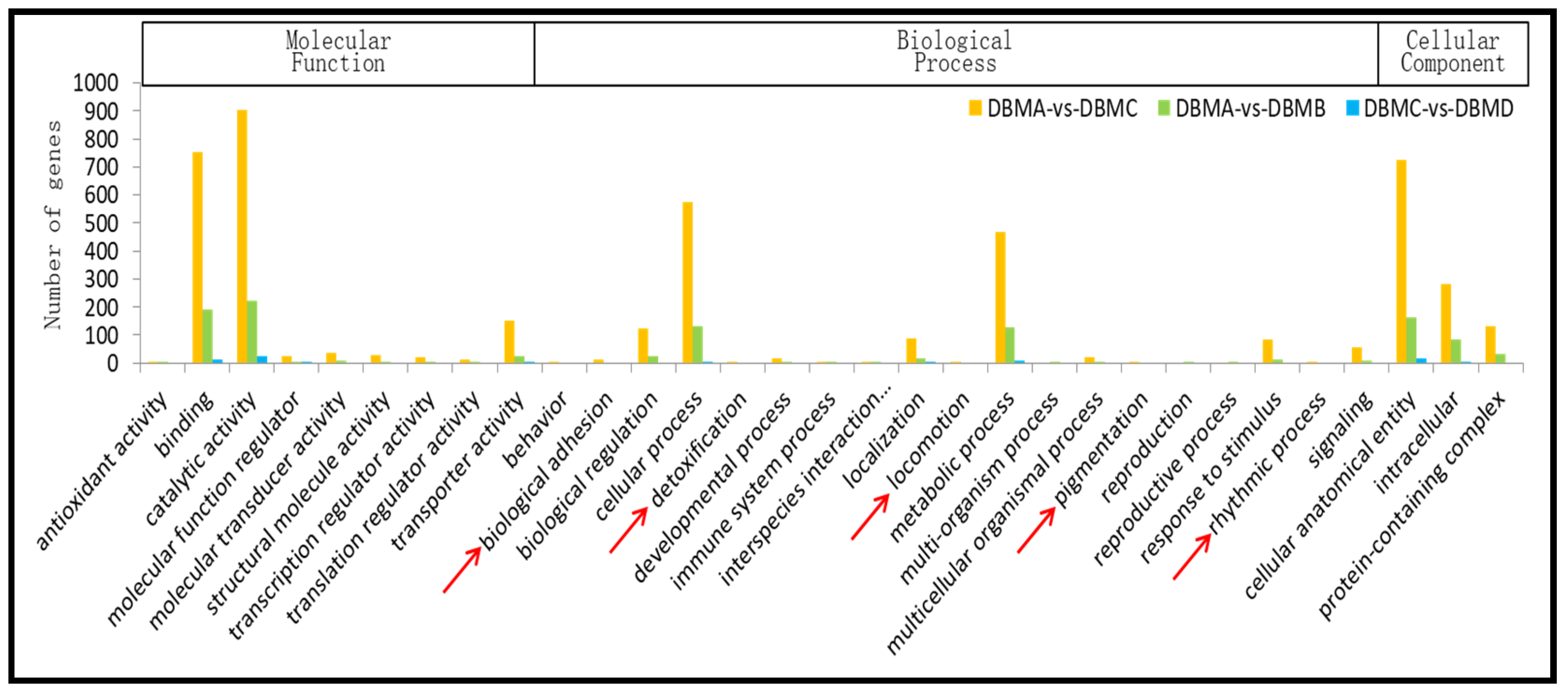
3.2. Gene Ontology (GO) Classification
3.3. Functional Classification by KEGG
3.4. Differentially Expressed Genes (DEGs) in Midgut Transcripts
3.5. Differentially Expressed Genes (DEGs) Involved in Insecticide Resistance
3.6. Validation of Differentially Expressed Genes by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Cushing, N.L.; Finson, N.; Johnson, M.W. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 1990, 83, 1671–1676. [Google Scholar] [CrossRef]
- Bravo, A.; Gill, S.S.; Soberon, M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 2007, 49, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardo-Lopez, L.; Soberon, M.; Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2013, 37, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Insect Midgut Insecticidal Proteins 2014, 47, 39–87. [Google Scholar]
- Zhang, X.; Candas, M.; Griko, N.B.; Taussig, R.; Bulla, L.A. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2006, 103, 9897–9902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vachon, V.; Laprade, R.; Schwartz, J.L. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: A critical review. J. Invertebr. Pathol. 2012, 111, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Zhang, M.; Fabrick, J.A.; Wu, Y.; Gao, M.; Huang, F.; Wei, J.; Zhang, J.; Yelich, A.; Unnithan, G.C.; et al. Dual mode of action of Bt proteins: Protoxin efficacy against resistant insects. Sci. Rep. 2015, 5, 15107. [Google Scholar] [CrossRef] [Green Version]
- Soberón, M.; Monnerat, R.G.; Bravo, A. Mode of action of Cry toxins from Bacillus thuringiensis and resistance mechanisms. Microb. Toxins 2016, 1, 1–13. [Google Scholar]
- Guo, Z.J.; Kang, S.; Chen, D.F.; Wu, Q.J.; Wang, S.; Xie, W.; Zhu, X.G.; Baxter, S.W.; Zhou, X.; Jurat-Fuentes, J.L.; et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 2015, 11, e1005124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Kang, S.; Sun, D.; Gong, L.; Zhou, J.; Qin, J.; Guo, L.; Zhu, L.; Bai, Y.; Ye, F.; et al. MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kang, S.; Wu, Q.; Wang, S.; Crickmore, N.; Zhou, X.; Bravo, A.; Soberon, M.; Zhang, Y. The regulation landscape of MAPK signaling cascade for thwarting Bacillus thuringiensis infection in an insect host. PLoS Pathog. 2021, 17, e1009917. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Yaoi, K.; Nagino, Y.; Hara, H.; Kitami, M.; Atsumi, S.; Miura, N.; Sato, R. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella—Their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002, 519, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Tiewsiri, K.; Wang, P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA 2011, 108, 14037–14042. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Zhang, M.; Liang, G.; Wu, K.; Guo, Y.; Ni, X.; Li, X. APN1 is a functional receptor of Cry1Ac but not Cry2Ab in Helicoverpa zea. Sci. Rep. 2016, 6, 19179. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, P.; Jin, H.; Liu, H.; Zhou, H.; Qiu, L.; Lin, Y.; Ma, W. Knockdown of the aminopeptidase N genes decreases susceptibility of Chilo suppressalis larvae to Cry1Ab/Cry1Ac and Cry1Ca. Pestic. Biochem. Physiol. 2020, 162, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, X.; He, H.; Chen, J.; Liu, Y.; Huang, W.; Ou, L.; Yang, Z.; Guan, X.; Zhang, L.; et al. Knockout of two Cry-binding aminopeptidase N isoforms does not change susceptibility of aedes aegypti larvae to Bacillus thuringiensis subsp. israelensis Cry4Ba and Cry11Aa toxins. Insects 2021, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Atsumi, S.; Yaoi, K.; Nakanishi, K.; Higurashi, S.; Miura, N.; Tabunoki, H.; Sato, R. A cadherin-like protein functions as a receptor for Bacillus thuringiensis Cry1Aa and Cry1Ac toxins on midgut epithelial cells of Bombyx mori larvae. FEBS Lett. 2003, 538, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Morin, S.; Biggs, R.W.; Sisterson, M.S.; Shriver, L.; Ellers-Kirk, C.; Higginson, D.; Holley, D.; Gahan, L.J.; Heckel, D.G.; Carrière, Y.; et al. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 2003, 100, 5004–5009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, D.; Xu, X.; Ye, W.; Yu, Z.; Sun, M. Helicoverpa armigera cadherin fragment enhances Cry1Ac insecticidal activity by facilitating toxin-oligomer formation. Appl. Microbiol. Biotechnol. 2010, 85, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, J.; Xiao, Y.; Yang, Y.; Liu, C.; Peng, R.; Yang, Y.; Bravo, A.; Soberon, M.; Liu, K. The cadherin Cry1Ac binding-region is necessary for the cooperative effect with ABCC2 transporter enhancing insecticidal activity of Bacillus thuringiensis Cry1Ac toxin. Toxins 2019, 11, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Ma, Y.; Guo, X.; Wan, P.; Liu, K.; Cong, S.; Wang, J.; Xu, D.; Xiao, Y.; Li, X.; et al. Pink bollworm resistance to Bt toxin Cry1Ac associated with an insertion in aadherin exon 20. Toxins 2019, 11, 186. [Google Scholar] [CrossRef] [Green Version]
- Jurat-Fuentes, J.L.; Adang, M.J. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 2004, 271, 3127–3135. [Google Scholar] [CrossRef]
- Arenas, I.; Bravo, A.; Soberon, M.; Gomez, I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J. Biol. Chem. 2010, 285, 12497–12503. [Google Scholar] [CrossRef] [Green Version]
- Dechklar, M.; Tiewsiri, K.; Angsuthanasombat, C.; Pootanakit, K. Functional expression in insect cells of glycosylphosphatidylinositol-linked alkaline phosphatase from Aedes aegypti larval midgut: A Bacillus thuringiensis Cry4Ba toxin receptor. Insect Biochem. Mol. Biol. 2011, 41, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, M.; Liang, G.; Li, X. Alkaline phosphatase 2 is a functional receptor of Cry1Ac but not Cry2Ab in Helicoverpa zea. Insect Biochem. Mol. Biol. 2019, 28, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [Green Version]
- Atsumi, S.; Miyamoto, K.; Yamamoto, K.; Narukawa, J.; Kawai, S.; Sezutsu, H.; Kobayashi, I.; Uchino, K.; Tamura, T.; Mita, K.; et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2012, 109, e1591. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Miyamoto, K.; Noda, H.; Jurat-Fuentes, J.L.; Yoshizawa, Y.; Endo, H.; Sato, R. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J. 2013, 280, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, T.; Liu, C.; Heckel, D.G.; Li, X.; Tabashnik, B.E.; Wu, K. Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci. Rep. 2014, 4, 6184. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.J.; Kang, S.; Zhu, X.; Xia, J.X.; Wu, Q.; Wang, S.L.; Xie, W.; Zhang, Y.J. The novel ABC transporter ABCH1 is a potential target for RNAi-based insect pest control and resistance management. Sci. Rep. 2015, 5, 13728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, T.; Song, S.; Bruning, J.B.; Choo, A.; Baxter, S.W. Expressing a moth abcc2 gene in transgenic Drosophila causes susceptibility to Bt Cry1Ac without requiring a cadherin-like protein receptor. Insect Biochem. Mol. Biol. 2017, 80, 61–70. [Google Scholar] [CrossRef]
- Liu, Z.X.; Fu, S.; Ma, X.L.; Baxter, S.W.; Vasseur, L.; Xiong, L.; Huang, Y.P.; Yang, G.; You, S.J.; You, M.S. Resistance to Bacillus thuringiensis Cry1Ac toxin requires mutations in two Plutella xylostella ATP-binding cassette transporter paralogs. PLoS Pathog. 2020, 16, e1008697. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, W.; Song, X.; Ma, X.; Cotto-Rivera, R.O.; Kain, W.; Chu, H.; Chen, Y.R.; Fei, Z.; Wang, P. Mutation of ABC transporter ABCA2 confers resistance to Bt toxin Cry2Ab in Trichoplusia ni. Insect Biochem. Mol. Biol. 2019, 112, 103209–103220. [Google Scholar] [CrossRef]
- Wang, J.; Ma, H.; Zhao, S.; Huang, J.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Functional redundancy of two ABC transporter proteins in mediating toxicity of Bacillus thuringiensis to cotton bollworm. PLoS Pathog. 2020, 16, e1008427. [Google Scholar] [CrossRef]
- Xu, H.Q.; Ma, M.; Ma, Y.P.; Zhang, S.Y.; Li, W.J.; Wei, D.; Wang, J.J. Identification and expression characterization of ATP-binding cassette (ABC) transporter genes in melon fly. Insects 2021, 12, 270. [Google Scholar] [CrossRef]
- Griffitts, J.S.; Haslam, S.M.; Yang, T.; Garczynski, S.F.; Mulloy, B.; Morris, H.; Cremer, P.S.; Dell, A.; Adang, M.J.; Aroian, R.V. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science 2005, 307, 922–925. [Google Scholar] [CrossRef] [Green Version]
- McNall, R.J.; Adang, M.J. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 2003, 33, 999–1010. [Google Scholar] [CrossRef]
- García-Gómez, B.I.; Cano, S.N.; Zagal, E.E.; Dantán-Gonzalez, E.; Bravo, A.; Soberón, M. Insect hsp90 chaperone assists Bacillus thuringiensis Cry toxicity by enhancing protoxin binding to the receptor and by protecting protoxin from gut protease degradation. mBio 2019, 10, e02775. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.J.; Kang, S.; Zhu, X.; Xia, J.X.; Wu, Q.J.; Wang, S.L.; Xie, W.; Zhang, Y.J. Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2015, 59, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Hickman, R.; Pieterse, C.M.; Van Wees, S.C. RNA-Seq: Revelation of the messengers. Trends Plant Sci. 2013, 18, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Zhu, X.; Xie, W.; Wu, Q.; Wang, S.; Guo, Z.; Xu, B.; Li, X.; Zhou, X.; Zhang, Y. Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 2014, 533, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Y.; Liu, Y.; Chen, F.; Han, L. Differences in midgut transcriptomes between resistant and susceptible strains of Chilo suppressalis to Cry1C toxin. BMC Genom. 2020, 21, 634–653. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yang, S.; Chen, L.; Liu, X.; Du, M.; An, S.; Liang, G. Transcriptomic responses to different Cry1Ac selection stresses in Helicoverpa armigera. Front. Physiol. 2018, 9, 1653–1670. [Google Scholar] [CrossRef]
- Xu, L.N.; Wang, Y.Q.; Wang, Z.Y.; Hu, B.J.; Ling, Y.H.; He, K.L. Transcriptome differences between Cry1Ab resistant and susceptible strains of Asian corn borer. BMC Genom. 2015, 16, 173–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, B.; Sanko, T.J.; Bezuidenhout, C.C.; Berg, J.V.D. Transcriptome and differentially expressed genes of Busseola fusca (Lepidoptera: Noctuidae) larvae challenged with Cry1Ab toxin. Gene 2019, 710, 387–398. [Google Scholar] [CrossRef]
- Lawrie, R.D.; Mitchell Iii, R.D.; Deguenon, J.M.; Ponnusamy, L.; Reisig, D.; Pozo-Valdivia, A.D.; Kurtz, R.W.; Roe, R.M. Multiple known mechanisms and a possible role of an enhanced immune system in Bt-resistance in a field population of the bollworm, Helicoverpa zea: Differences in gene expression with RNAseq. Int. J. Mol. Sci. 2020, 21, 6528. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Lei, Y.; Fu, W.; Yang, Z.; Zhu, X.; Guo, Z.; Wu, Q.; Wang, S.; Xu, B.; Zhou, X.; et al. Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut. Int. J. Biol. Sci. 2012, 8, 1142–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellichirammal, N.N.; Wang, H.; Eyun, S.I.; Moriyama, E.N.; Coates, B.S.; Miller, N.J.; Siegfried, B.D. Transcriptional analysis of susceptible and resistant European corn borer strains and their response to Cry1F protoxin. BMC Genom. 2015, 16, 558. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.L.; Wang, Y.Y.; Ma, Y.J.; Jiang, W.L.; Ma, X.Y.; Hu, H.Y.; Wang, D.; Ma, Y. Midgut de novo transcriptome analysis and gene expression profiling of larvae exposed with sublethal concentrations of Cry1Ca protein. 3 Biotech. 2020, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- He, W.; You, M.; Vasseur, L.; Yang, G.; Xie, M.; Cui, K.; Bai, J.; Liu, C.; Li, X.; Xu, X.; et al. Developmental and insecticide-resistant insights from the de novo assembled transcriptome of the diamondback moth, Plutella xylostella. Genomics 2012, 99, 169–177. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, X.; Liu, Z.; Shao, E.; Rebeca, C.L.; Guo, Y.; Xiong, Y.; Mou, Y.; Xu, R.; Hu, X.; et al. Identification of genes relevant to pesticides and biology from global transcriptome data of monochamus alternatus hope (Coleoptera: Cerambycidae) larvae. PLoS ONE 2016, 11, e0147855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aioub, A.A.A.; Zuo, Y.; Li, Y.; Qie, X.; Zhang, X.; Essmat, N.; Wu, W.; Hu, Z. Transcriptome analysis of Plantago major as a phytoremediator to identify some genes related to cypermethrin detoxification. Environ. Sci. Pollut. Res. Int. 2020, 28, 5101–5115. [Google Scholar] [CrossRef]
- Chen, X.D.; Neupane, S.; Gill, T.A.; Gossett, H.; Pelz-Stelinski, K.S.; Stelinski, L.L. Comparative transcriptome analysis of thiamethoxam susceptible and resistant Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae), using RNA-sequencing. Insect Sci. 2021, 12, 901. [Google Scholar] [CrossRef]
- Guo, Z.; Sun, D.; Kang, S.; Zhou, J.; Gong, L.; Qin, J.; Guo, L.; Zhu, L.; Bai, Y.; Luo, L.; et al. CRISPR/Cas9-mediated knockout of both the PxABCC2 and PxABCC3 genes confers high-level resistance to Bacillus thuringiensis Cry1Ac toxin in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2019, 107, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrode, S.D.; Wilsey, W.T.; Kroening, M.K.; Cooley, R.J.; Shelton, A.M. Comparative analysis of two rearing procedures for diamondback moth (Lepidoptera: Plutellidae). J. Entomol. Sci. 1991, 26, 17–26. [Google Scholar]
- Aronson, A.I.; Angelo, N.; Holt, S.C. Regulation of extracellular protease production in Bacillus cereus T: Characterization of mutants producing altered amounts of protease. J. Bacteriol. 1971, 106, 1016–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, L.; Banks, D.; Adang, M.J. Toxicity, binding, and permeability analyses of four Bacillus thuringiensis Cry1 delta-endotoxins using brush border membrane vesicles of Spodoptera exigua and Spodoptera frugiperda. Appl. Environ. Microbiol. 1999, 65, 457–464. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefan, G.; Miguel, G.; Javier, T.; Williams, T.D.; Nagaraj, S.H.; José, N.M.; Montserrat, R.; Manuel, T.; Joaquín, D.; Ana, C. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gafney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Ander, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- You, M.S.; Yue, Z.; He, W.Y.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.J.; Kang, S.; Zhu, X.; Wu, Q.J.; Wang, S.l.; Xie, W.; Zhang, Y.J. The midgut cadherin-like gene is not associated with resistance to Bacillus thuringiensis toxin Cry1Ac in Plutella xylostella (L.). J. Invertebr. Pathol. 2015, 126, 21–30. [Google Scholar] [CrossRef]
- Baxter, S.W.; Zhao, J.Z.; Gahan, L.J.; Shelton, A.M.; Tabashnik, B.E.; Heckel, D.G. Novel genetic basis of field-evolved resistance to Bt toxins in Plutella xylostella. Insect Mol. Biol. 2005, 14, 327–334. [Google Scholar] [CrossRef]
- Baxter, S.W.; Zhao, J.Z.; Shelton, A.M.; Vogel, H.; Heckel, D.G. Genetic mapping of Bt-toxin binding proteins in a Cry1A-toxin resistant strain of diamondback moth Plutella xylostella. Insect Biochem. Mol. Biol. 2008, 38, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Sieku, T.; Hooper, N.M. Inhibition of aminopeptidases N, A and W. Biochem. Pharmacol. 1992, 44, 1725–1730. [Google Scholar]
- Knight, P.J.; Crickmore, N.; Ellar, D.J. The receptor for Bacillus thuringiensis CrylA(c) delta-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 1994, 11, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Cowles, E.A.; Francis, V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J. Biol. Chem. 1995, 270, 27277–27282. [Google Scholar] [CrossRef] [Green Version]
- Luo, K.; Tabashnik, B.E.; Adang, M.J. Binding of Bacillus thuringiensis Cry1Ac toxin to aminopeptidase in susceptible and resistant diamondback moths (Plutella xylostella). Appl. Environ. Microbiol. 1997, 63, 1024–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.K.; You, T.H.; Young, B.A.; Cotrill, J.A.; Valaitis, A.P.; Dean, D.H. Aminopeptidase N purified from gypsy moth brush border membrane vesicles is a specific receptor for Bacillus thuringiensis CryIAc toxin. Appl. Environ. Microbiol. 1996, 62, 2845–2849. [Google Scholar] [CrossRef] [Green Version]
- Valaitis, A.P.; Lee, M.K.; Rajamohan, F.; Dean, D.H. Brush border membrane aminopeptidase-N in the midgut of the gypsy moth serves as the receptor for the CryIA(c) delta-endotoxin of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 1995, 25, 1143–1451. [Google Scholar] [CrossRef]
- Yaoi, K.; Kadotani, T.; Kuwana, H.; Shinkawa, A.; Takahashi, T.; Iwahana, H.; Sato, R. Aminopeptidase N from Bombyx Mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur. J. Biochem. 1997, 246, 652–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorence, A.; Darszon, A.; Bravo, A. Aminopeptidase dependent pore formation of Bacillus thuringiensis Cry1Ac toxin on Trichoplusia ni membranes. FEBS Lett. 1997, 414, 303–307. [Google Scholar] [PubMed] [Green Version]
- Qiu, L.; Fan, J.; Zhang, B.; Liu, L.; Wang, X.; Lei, C.; Lin, Y.; Ma, W. RNA interference knockdown of aminopeptidase N genes decrease the susceptibility of Chilo suppressalis larvae to Cry1Ab/Cry1Ac and Cry1Ca-expressing transgenic rice. J. Invertebr. Pathol. 2017, 145, 9–12. [Google Scholar] [CrossRef]
- Niu, X.; Kassa, A.; Hasler, J.; Griffin, S.; Perez-Ortega, C.; Procyk, L.; Zhang, J.; Kapka-Kitzman, D.M.; Nelson, M.E.; Lu, A. Functional validation of DvABCB1 as a receptor of Cry3 toxins in western corn rootworm, Diabrotica virgifera virgifera. Sci. Rep. 2020, 10, 15830. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Yang, J.; Hou, W.; Xu, B.; Xie, W.; Wang, S.; Zhang, Y.; Zhou, X.; Wu, Q. Molecular cloning and characterization of a P-glycoprotein from the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Mol. Sci. 2013, 14, 22891–22905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.L.; Guo, Z.J.; Kang, S.; Qin, J.Y.; Gong, L.J.; Sun, D.; Guo, L.; Zhu, L.H.; Bai, Y.; Zhang, Z.Z.; et al. Reduced expression of the P-glycoprotein gene PxABCB1 is linked to resistance to Bacillus thuringiensis Cry1Ac toxin in Plutella xylostella (L.). Pest Manag. Sci. 2020, 76, 712–720. [Google Scholar] [CrossRef]
- Wu, C.P.; Hung, C.Y.; Lusvarghi, S.; Huang, Y.H.; Tseng, P.J.; Hung, T.H.; Yu, J.S.; Ambudkar, S.V. Overexpression of ABCB1 and ABCG2 contributes to reduced efficacy of the PI3K/mTOR inhibitor samotolisib (LY3023414) in cancer cell lines. Biochem. Pharmacol. 2020, 180, 114137. [Google Scholar] [CrossRef] [PubMed]
- Ocelotl, J.; Sanchez, J.; Gomez, I.; Tabashnik, B.E.; Bravo, A.; Soberon, M. ABCC2 is associated with Bacillus thuringiensis Cry1Ac toxin oligomerization and membrane insertion in diamondback moth. Sci. Rep. 2017, 7, 2386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.; Wei, D.D.; Xu, H.Q.; Yang, Y.; Miao, Z.Q.; Wang, L.; Wang, J.J. Molecular characterization and transcriptional expression analysis of ABC transporter H subfamily genes in the Oriental Fruit Fly. J. Econ. Entomol. 2021, 114, 1298–1309. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, Z.; Wang, Y.; Ma, H.; Zhu, H.; Liu, J.; Zhou, Y.; Deng, X.; Zhou, X. ABCC2 participates in the resistance of Plutella xylostella to chemical insecticides. Pestic. Biochem. Physiol. 2020, 162, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Kang, S.; Zhou, J.; Sun, D.; Guo, L.; Qin, J.; Zhu, L.; Bai, Y.; Ye, F.; Akami, M.; et al. Reduced expression of a novel midgut trypsin gene involved in protoxin activation correlates with Cry1Ac resistance in a laboratory-selected strain of Plutella xylostella (L.). Toxins 2020, 12, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Liang, G.; Wang, B.; Zhong, F.; Chen, L.; Khaing, M.M.; Zhang, J.; Guo, Y.; Wu, K.; Tabashnik, B.E. Activation of Bt protoxin Cry1Ac in resistant and susceptible Cotton Bollworm. PLoS ONE 2016, 11, e0156560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bautista, M.A.; Miyata, T.; Miura, K.; Tanaka, T. RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem. Mol. Biol. 2009, 39, 38–46. [Google Scholar] [CrossRef]
- Chen, A.; Zhang, H.; Shan, T.; Shi, X.; Gao, X. The overexpression of three cytochrome P450 genes CYP6CY14, CYP6CY22 and CYP6UN1 contributed to metabolic resistance to dinotefuran in melon/cotton aphid, Aphis gossypii Glover. Pestic. Biochem. Physiol. 2020, 167, 104601. [Google Scholar] [CrossRef] [PubMed]
- Dulbecco, A.B.; Moriconi, D.E.; Pedrini, N. Knockdown of CYP4PR1, a cytochrome P450 gene highly expressed in the integument tissue of Triatoma infestans, increases susceptibility to deltamethrin in pyrethroid-resistant insects. Pestic. Biochem. Physiol. 2021, 173, 104781. [Google Scholar] [CrossRef]
- Qin, J.; Ye, F.; Xu, L.; Zhou, X.; Crickmore, N.; Zhou, X.; Zhang, Y.; Guo, Z. A cis-Acting mutation in the PxABCG1 promoter is associated with Cry1Ac resistance in Plutella xylostella (L.). Int. J. Mol. Sci. 2021, 22, 6106. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Guo, L.; Ye, F.; Kang, S.; Sun, D.; Zhu, L.; Bai, Y.; Cheng, Z.; Xu, L.; Ouyang, C.; et al. MAPK-Activated transcription factor PxJun suppresses PxABCB1 expression and confers resistance to Bacillus thuringiensis Cry1Ac toxin in Plutella xylostella (L.). Appl. Environ. Microbiol. 2021, 87, e0046621. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Wang, C.; Yang, Y.; Wu, S.; Wu, Y. Characterization of resistance to Bacillus thuringiensis toxin Cry1Ac in Plutella xylostella from China. J. Invertebr. Pathol. 2010, 104, 90–96. [Google Scholar] [CrossRef]






| Samples | Raw Reads (M) | Clean Reads (M) | Q20 (%) a | Q30 (%) b | Clean Reads Ratio (%) |
|---|---|---|---|---|---|
| G88_1 (DBMA1) | 65.18 | 61.75 | 97.49 | 90.02 | 94.74 |
| G88_2 (DBMA2) | 67.68 | 62.43 | 97.00 | 88.62 | 92.24 |
| G88_3 (DBMA3) | 67.68 | 61.70 | 96.58 | 87.38 | 91.16 |
| G88 + toxin_1 (DBMB1) | 65.18 | 60.99 | 96.77 | 87.91 | 93.58 |
| G88 + toxin_2 (DBMB2) | 67.68 | 64.15 | 97.30 | 89.57 | 94.78 |
| G88 + toxin_3 (DBMB3) | 67.68 | 63.14 | 96.70 | 87.78 | 93.29 |
| Cry1S1000_1 (DBMC1) | 70.19 | 66.30 | 97.00 | 88.61 | 94.47 |
| Cry1S1000_2 (DBMC2) | 67.68 | 63.07 | 96.68 | 87.87 | 93.19 |
| Cry1S1000_3 (DBMC3) | 67.68 | 64.06 | 97.38 | 89.74 | 94.65 |
| Cry1S1000 + toxin_1 (DBMD1) | 67.68 | 62.53 | 96.60 | 87.61 | 92.39 |
| Cry1S1000 + toxin_2 (DBMD2) | 70.12 | 66.35 | 97.36 | 89.71 | 94.62 |
| Cry1S1000 + toxin_3 (DBMD3) | 70.18 | 65.73 | 97.25 | 89.19 | 93.65 |
| Total Novel Transcripts | Coding Transcripts | Novel Transcripts | Novel Isoforms | Novel Genes |
|---|---|---|---|---|
| 19,415 | 14,166 | 5249 | 10,525 | 3641 |
| Genes | DBMA/DBMC | DBMA/DBMB | DBMC/DBMD |
|---|---|---|---|
| Bt resistance | |||
| Cadherin | 5 | 0 | 0 |
| Aminopeptidase N/P | 14 | 4 | 0 |
| Alkaline phosphatase | 0 | 0 | 0 |
| ABC transporter | 14 | 3 | 0 |
| Trypsin | 55 | 22 | 1 |
| Glycolipid | 0 | 0 | 0 |
| Heat-shock proteins | 5 | 6 | 0 |
| Insecticide targets and metabolic insecticide resistance | |||
| Cytochrome P450 (P450s) | 24 | 7 | 3 |
| Carboxylesterase (CarEs) | 4 | 0 | 0 |
| Glutathione S-transferase (GSTs) | 4 | 3 | 1 |
| Acetylcholinesterase | 2 | 1 | 0 |
| Nicotinic acetylcholine receptor | 0 | 0 | 0 |
| GABA receptor | 0 | 0 | 0 |
| Glutamate receptor | 10 | 1 | 0 |
| G-protein coupled receptor | 3 | 0 | 0 |
| Ryanodine receptor | 1 | 0 | 0 |
| Sodium channel | 1 | 1 | 0 |
| Chloride channel | 1 | 0 | 0 |
| Immune-related genes | |||
| Serpin protease | 0 | 0 | 0 |
| Serpin protease inhibitor | 1 | 0 | 0 |
| Gene ID | DBMA-TPM | DBMC-TPM | Log2 (DBMC/DBMA) | Annotation | Up/Down | |
|---|---|---|---|---|---|---|
| 1 | Px002415 | 92.57 | 15.99 | −2.53 | Probable multidrug resistance-associated protein lethal (2) 03659 | Down |
| 2 | Px012806 | 217.58 | 39.78 | −2.45 | Luciferin 4-monooxygenase | Down |
| 3 | Px000515 | 127.45 | 23.13 | −2.46 | Esterase FE4 | Down |
| 4 | Px005972 | 82.80 | 16.80 | −2.30 | N-acetylneuraminate lyase | Down |
| 5 | Px007616 | 9973.06 | 2139.83 | −2.22 | Trypsin CFT-1 | Down |
| 6 | Px013169 | 314.29 | 77.30 | −2.02 | Lactase-phlorizin hydrolase | Down |
| 7 | Px005361 | 12.89 | 58.54 | 2.18 | Probable glutamine-dependent NAD(+) synthetase | Up |
| 8 | Px016564 | 29.39 | 165.13 | 2.49 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | Up |
| 9 | Px015078 | 53.31 | 268.45 | 2.33 | Ecdysteroid UDP-glucosyltransferase | Up |
| 10 | Px011160 | 96.75 | 630.73 | 2.70 | Lactase-phlorizin hydrolase | Up |
| 11 | Px015831 | 71.64 | 488.08 | 2.77 | Zinc carboxypeptidase A 1 | Up |
| 12 | Px004235 | 15.75 | 204.14 | 3.70 | Putative uncharacterized protein | Up |
| 13 | Px007138 | 17.01 | 125.17 | 2.88 | Leucine-rich repeat- containing protein C10orf11 homolog | Up |
| 14 | Px009634 | 734.96 | 6072.207 | 3.04 | Ecdysteroid-regulated protein | Up |
| 15 | Px005853 | 24.69 | 200.48 | 3.02 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mitochondrial | Up |
| 16 | Px007598 | 36.14 | 750.73 | 4.38 | Chymotrypsin−1 | Up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, L.; Liu, Z.; Shen, L.; Xie, C.; Ye, M.; Li, Z.; Zhang, Z.; Li, J.; Dong, Y.; You, M.; et al. A Novel Reference for Bt-Resistance Mechanism in Plutella xylostella Based on Analysis of the Midgut Transcriptomes. Insects 2021, 12, 1091. https://doi.org/10.3390/insects12121091
Xiong L, Liu Z, Shen L, Xie C, Ye M, Li Z, Zhang Z, Li J, Dong Y, You M, et al. A Novel Reference for Bt-Resistance Mechanism in Plutella xylostella Based on Analysis of the Midgut Transcriptomes. Insects. 2021; 12(12):1091. https://doi.org/10.3390/insects12121091
Chicago/Turabian StyleXiong, Lei, Zhaoxia Liu, Lingling Shen, Chao Xie, Min Ye, Zeyun Li, Zhen Zhang, Jingge Li, Yi Dong, Minsheng You, and et al. 2021. "A Novel Reference for Bt-Resistance Mechanism in Plutella xylostella Based on Analysis of the Midgut Transcriptomes" Insects 12, no. 12: 1091. https://doi.org/10.3390/insects12121091
APA StyleXiong, L., Liu, Z., Shen, L., Xie, C., Ye, M., Li, Z., Zhang, Z., Li, J., Dong, Y., You, M., & You, S. (2021). A Novel Reference for Bt-Resistance Mechanism in Plutella xylostella Based on Analysis of the Midgut Transcriptomes. Insects, 12(12), 1091. https://doi.org/10.3390/insects12121091






