Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Extraction, Synthesis of cDNA Library and Sequencing
2.2. Sequence Assembly and Annotation
2.3. Rearing of S. marcescens
2.4. Rearing and Treatment of H. cunea
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Statistical Analysis
3. Results
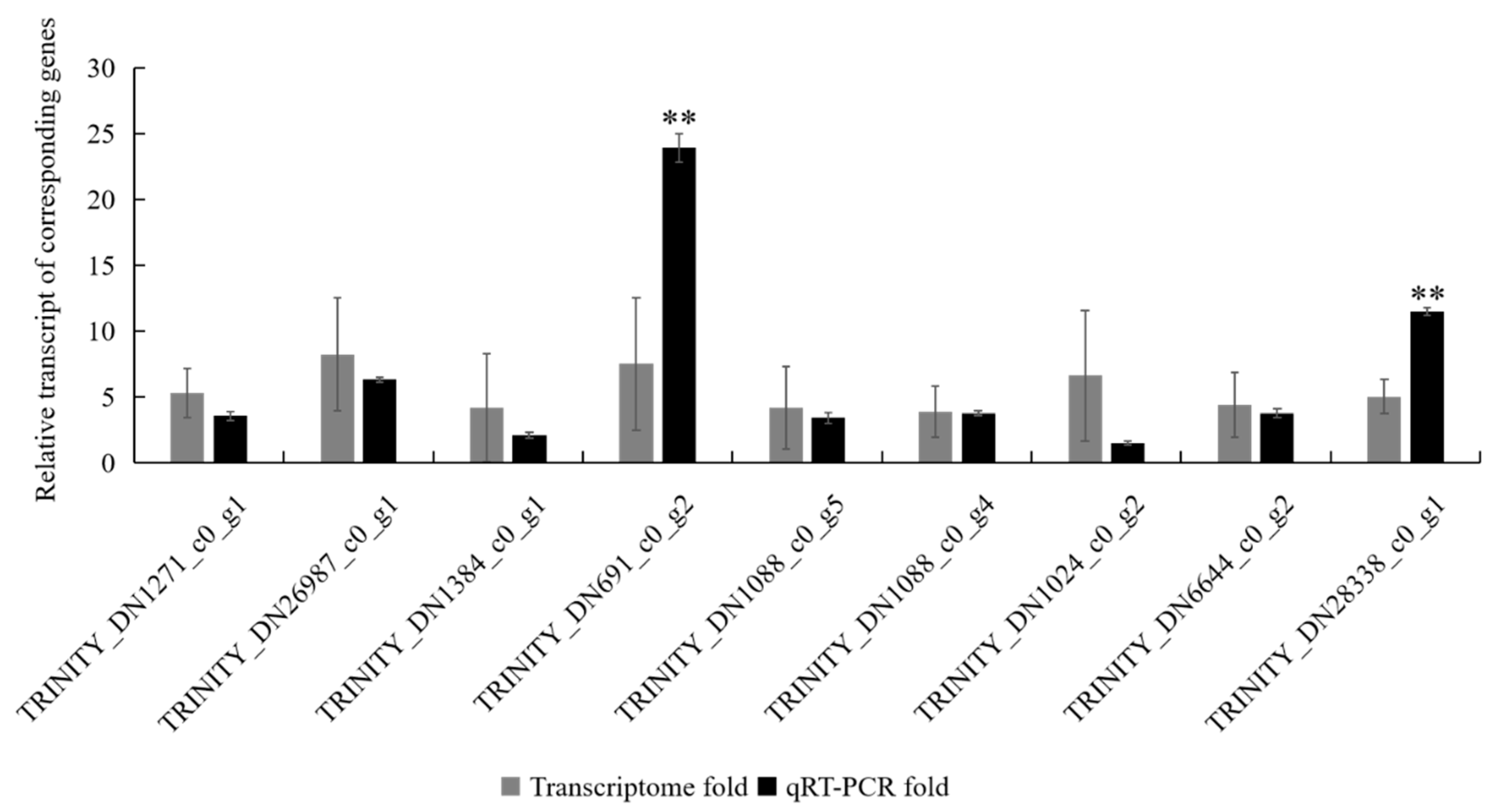
3.1. Verification of Differentially Expressed Genes (DEGs) from the Transcriptomes by qRT-PCR
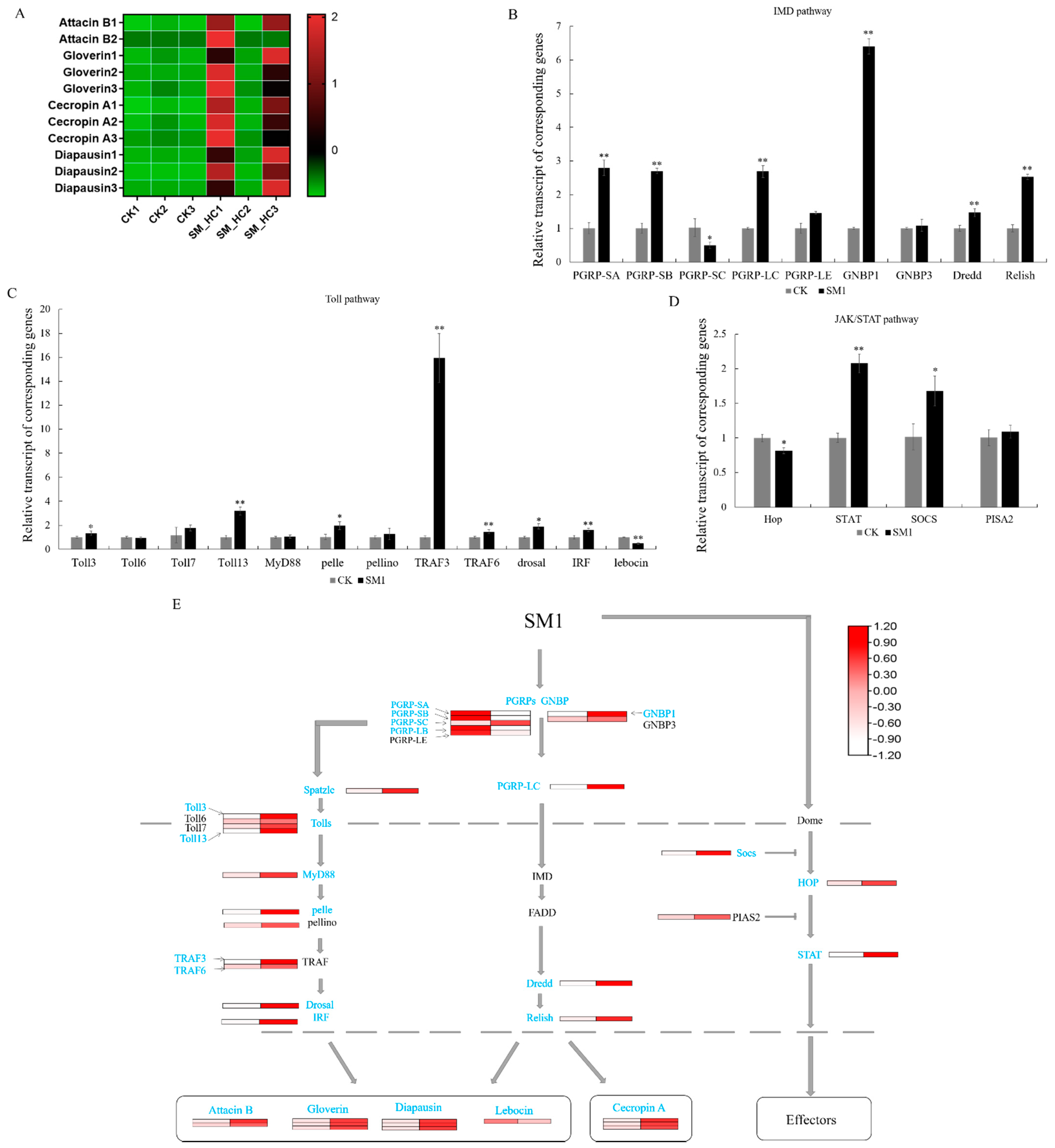
3.2. Identification of the Immune-Related DEGs in H. cunea
3.3. Antimicrobial Peptides Induced by SM1 in H. cunea
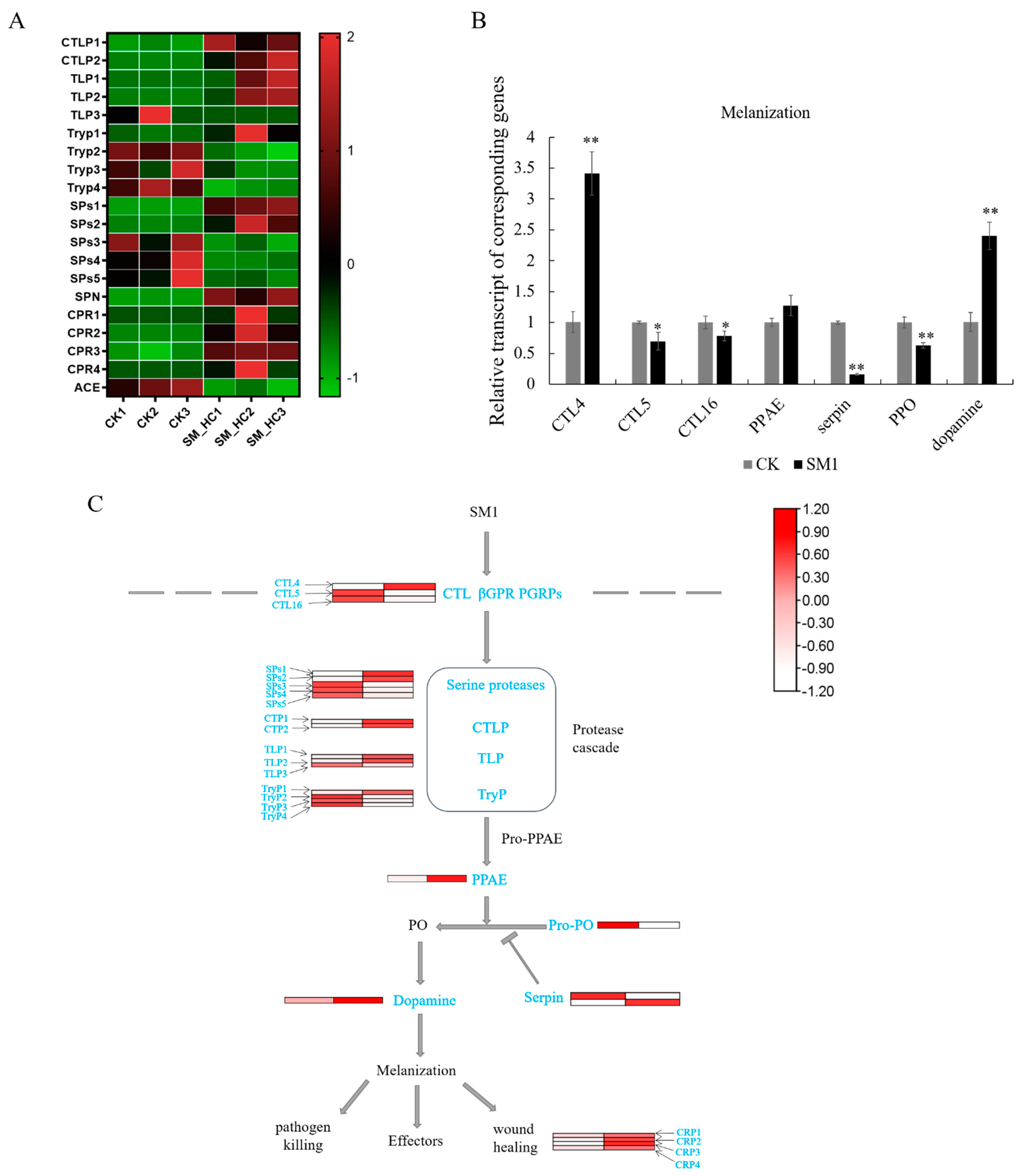
3.4. Genes of the Melanization Pathway Induced by SM1 in H. cunea
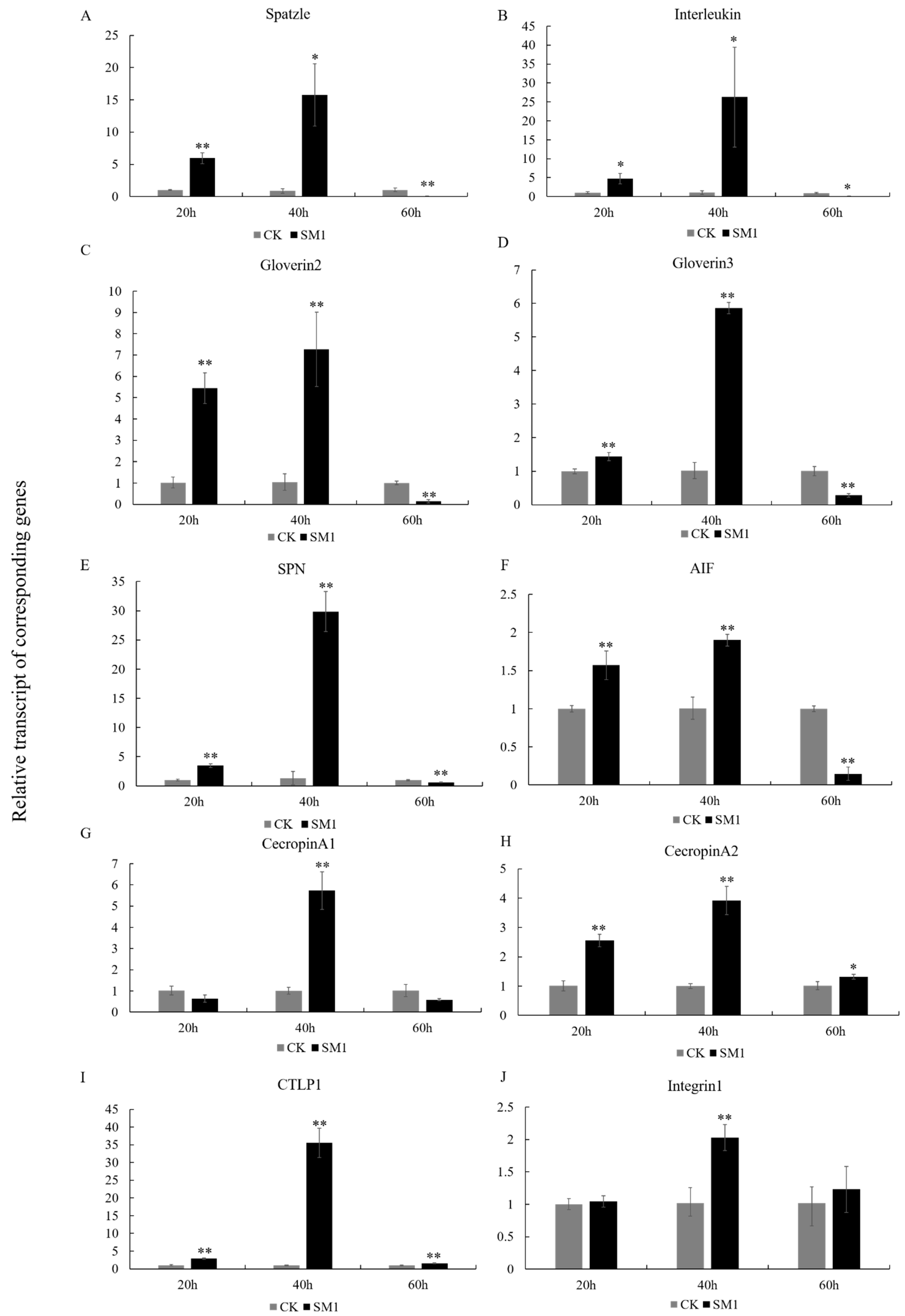
3.5. Induction of the Cellular Immune Response by SM1
3.6. The Response of Immune-Related Genes to SM1 in H. cunea
4. Discussion
4.1. AMPs Synthesis Pathway Response to SM1 in H. cunea
4.2. SM1 Infection Induced the Melanization in H. cunea
4.3. SM1 Infection Affects Cell Responses in H. cunea
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morris, R.F. Synonymy and Color Variation in the Fall Webworm, Hyphantria cunea Drury (Lepidoptera: Arctiidae). Can. Entomol. 1963, 95, 1217–1223. [Google Scholar] [CrossRef]
- Gomi, T. Seasonal adaptations of the fall webworm Hyphantria cunea (Drury) (Lepidoptera: Arctiidae) following its invasion of Japan. Ecol. Res. 2007, 22, 855–861. [Google Scholar] [CrossRef]
- Sullivan, G.; Ozman-Sullivan, S. Tachinid (Diptera) parasitoids of Hyphantria cunea (Lepidoptera: Arctiidae) in its native North America and in Europe and Asia—A literature review. Entomol. Fenn. 2012, 23, 181–192. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.W.; Kang, K.; Jiang, S.C.; Zhang, Y.N.; Wang, T.T.; Zhang, J.; Sun, L.; Yang, Y.Q.; Huang, C.C.; Jiang, L.Y.; et al. Analysis of the Antennal Transcriptome and Insights into Olfactory Genes in Hyphantria cunea (Drury). PLoS ONE 2016, 11, e0164729. [Google Scholar]
- Zhang, Y.K.; Zhao, D.; Yan, X.P.; Guo, W.; Bao, Y.J.; Wang, W.; Wang, X.Y. Identification and Characterization of Hyphantria cunea Aminopeptidase N as a Binding Protein of Bacillus thuringiensis Cry1Ab35 Toxin. Int. J. Mol. Sci. 2017, 18, 2575. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Liu, P.; Sun, S.H.; Yan, S.C.; Cao, C.W. Transcriptomic analysis of interactions between Hyphantria cunea larvae and nucleopolyhedrovirus. Pest Manag. Sci. 2019, 75, 1024–1033. [Google Scholar] [CrossRef]
- Nehme, N.T.; Liégeois, S.; Kele, B.; Giammarinaro, P.; Pradel, E.; Hoffmann, J.A.; Ewbank, J.J.; Ferrandon, D. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007, 3, e173. [Google Scholar] [CrossRef]
- Babashpour, S.; Aminzadeh, S.; Farrokhi, N.; Karkhane, A.; Haghbeen, K. Characterization of a chitinase (Chit62) from Serratia marcescens B4A and its efficacy as a bioshield against plant fungal pathogens. Biochem. Genet. 2012, 50, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yan, P.S.; Cao, L.X.; Ding, Q.L.; Shao, C.; Zhao, T.F. Potential of chitinolytic Serratia marcescens strain JPP1 for biological control of Aspergillus parasiticus and aflatoxin. Biomed. Res. Int. 2013, 2013, 397142. [Google Scholar]
- Aggarwal, C.; Paul, S.; Tripathi, V.; Paul, B.; Khan, M.A. Characterization of putative virulence factors of Serratia marcescens strain SEN for pathogenesis in Spodoptera litura. J. Invertebr. Pathol. 2017, 143, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Z.; Ou, X.K.; Zhang, J.F.; Jing, Y.B.; Zhang, Z.Y.; Cao, J.X.; Li, Y.P.; Ma, S.Y.; Li, R.B. Preliminary researches on the aphides bio-control with bacteria. Guangxi Agric. Sci. 2010, 41, 226–230. (In Chinese) [Google Scholar]
- Ishii, K.; Adachi, T.; Hara, T.; Hamamoto, H.; Sekimizu, K. Identification of a Serratia marcescens virulence factor that promotes hemolymph bleeding in the silkworm, Bombyx mori. J. Invertebr. Pathol. 2014, 117, 61–67. [Google Scholar] [CrossRef]
- Lee, D.J.; Lee, J.B.; Jang, H.A.; Ferrandon, D.; Lee, B.L. An antimicrobial protein of the Riptortus pedestris salivary gland was cleaved by a virulence factor of Serratia marcescens. Dev. Comp. Immunol. 2017, 67, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.T.; Wang, N.; Liu, B.S.; Xiao, L.J.J.; Wang, L.H.; Guo, H.F. Synergistic and additive interactions of Serratia marcescens S-JS1 to the chemical insecticides for controlling Nilaparvata lugens (Hemiptera: Delphacidae). J. Econ. Entomol. 2018, 111, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.J.; Luo, J.; Feng, K.; Lu, X.Y.; Tang, F. Effects of Serratia marcescens (SM1) and its interaction with common biocontrol agents on the termite, Odontotermes formosanus (Shiraki). J. For. Res. 2021, 32, 1263–1267. [Google Scholar] [CrossRef]
- Feng, K.; Luo, J.; Ding, X.; Tang, F. Transcriptome analysis and response of three important detoxifying enzymes to Serratia marcescens Bizio (SM1) in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 2021, 178, 104922. [Google Scholar] [CrossRef] [PubMed]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Choe, K.M.; Lee, H.; Anderson, K.V. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA 2005, 102, 1122–1126. [Google Scholar] [CrossRef]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef]
- Brown, G.D.; Gordon, S. Immune recognition of fungal beta-glucans. Cell. Microbiol. 2005, 7, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Kurata, S. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 2014, 42, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Warr, E.; Das, S.; Dong, Y.; Dimopoulos, G. The Gram-negative bacteria-binding protein gene family: Its role in the innate immune system of anopheles gambiae and in anti-Plasmodium defence. Insect Mol. Biol. 2008, 17, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.F.; You, M.S.; Rao, X.J.; Yu, X.Q. Insect C-type lectins in innate immunity. Dev. Comp. Immunol. 2018, 83, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Kim, M.S.; Kim, H.E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.H. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006, 281, 8286–8295. [Google Scholar] [CrossRef]
- Gorman, M.J.; Wang, Y.; Jiang, H.B.; Kanost, M.R. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase-activating proteinase 3 in an insect innate immune response proteinase cascade. J. Biol. Chem. 2007, 282, 11742–11749. [Google Scholar] [CrossRef]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Hillyer, J.F.; Strand, M.R. Mosquito hemocyte-mediated immune responses. Curr. Opin. Insect Sci. 2014, 3, 14–21. [Google Scholar] [CrossRef]
- Lamiable, O.; Imler, J.L. Induced antiviral innate immunity in Drosophila. Curr. Opin. Microbiol. 2014, 20, 62–68. [Google Scholar] [CrossRef]
- Moy, R.H.; Cherry, S. Antimicrobial autophagy: A conserved innate immune response in Drosophila. J. Innate Immun. 2013, 5, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; Saitoh, T.; Nowag, H.; Münz, C.; Yoshimori, T. Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 2015, 16, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Clem, R.J. Baculoviruses and apoptosis: A diversity of genes and responses. Curr. Drug Targets 2007, 8, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gort, T.; Boyle, D.L.; Clem, R.J. Effects of manipulating apoptosis on Sindbis virus infection of Aedes aegypti mosquitoes. J. Virol. 2012, 86, 6546–6554. [Google Scholar] [CrossRef] [PubMed]
- Nainu, F.; Tanaka, Y.; Shiratsuchi, A.; Nakanishi, Y. Protection of Insects against Viral Infection by Apoptosis-Dependent Phagocytosis. J. Immunol. 2015, 195, 5696–5706. [Google Scholar] [CrossRef] [PubMed]
- Gesellchen, V.; Kuttenkeuler, D.; Steckel, M.; Pelte, N.; Boutros, M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005, 6, 979–984. [Google Scholar] [CrossRef]
- Giulietti, A.; Overbergh, L.; Valckx, D.; Decallonne, B.; Bouillon, R.; Mathieu, C. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods 2001, 25, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Zheng, Y.J.; Zhong, Y.D.; Wu, Y.F.; Li, Z.T.; Xu, L.A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef]
- Iwanaga, S.; Lee, B.L. Recent advances in the innate immunity of invertebrate animals. J. Biochem. Mol. Biol. 2005, 38, 128–150. [Google Scholar] [CrossRef] [PubMed]
- Pascual, L.; Jakubowska, A.K.; Blanca, J.M.; Cañizares, J.; Ferré, J.; Gloeckner, G.; Vogel, H.; Herrero, S. The transcriptome of Spodoptera exigua larvae exposed to different types of microbes. Insect Biochem. Mol. Biol. 2012, 42, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Je, H.J.; Park, S.Y.; Lee, I.H.; Jin, B.R.; Yun, H.K.; Yun, C.Y.; Han, Y.S.; Kang, Y.J.; Seo, S.J. Immune activation of apolipophorin-III and its distribution in hemocyte from Hyphantria cunea. Insect Biochem. Mol. Biol. 2004, 34, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Zhang, Y.Q.; Zhang, R.; Zhang, J.H. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr. Opin. Insect Sci. 2019, 33, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Goldman, W.E.; Mellroth, P.; Steiner, H.; Fukase, K.; Kusumoto, S.; Harley, W.; Fox, A.; Golenbock, D.; Silverman, N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 2004, 20, 637–649. [Google Scholar] [CrossRef]
- Kaneko, T.; Yano, T.; Aggarwal, K.; Lim, J.H.; Ueda, K.; Oshima, Y.; Peach, C.; Erturk-Hasdemir, D.; Goldman, W.E.; Oh, B.H.; et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006, 7, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Meinander, A.; Runchel, C.; Tenev, T.; Chen, L.; Kim, C.H.; Ribeiro, P.S.; Broemer, M.; Leulier, F.; Zvelebil, M.; Silverman, N.; et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012, 31, 2770–2783. [Google Scholar] [CrossRef]
- Weber, A.N.; Moncrieffe, M.C.; Gangloff, M.; Imler, J.L.; Gay, N.J. Ligand-receptor and receptor-receptor interactions act in concert to activate signaling in the Drosophila toll pathway. J. Biol. Chem. 2005, 280, 22793–22799. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Bristow, B.N.; Qu, G.W.; Wasserman, S.A. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 12871–12876. [Google Scholar] [CrossRef]
- Lee, W.J.; Lee, J.D.; Kravchenko, V.V.; Ulevitch, R.J.; Brey, P.T. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 1996, 93, 7888–7893. [Google Scholar] [CrossRef]
- Hanson, M.A.; Lemaitre, B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.M.; Kim, H.J.; Kim, Y.I.; Kang, Y.J.; Lee, I.H.; Jin, B.R.; Han, Y.S.; Cheon, H.M.; Ha, N.G.; Seo, S.J. Comparative analysis of two attacin genes from Hyphantria cunea. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 151, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Al Souhail, Q.; Hiromasa, Y.; Rahnamaeian, M.; Giraldo, M.C.; Takahashi, D.; Valent, B.; Vilcinskas, A.; Kanost, M.R. Characterization and regulation of expression of an antifungal peptide from hemolymph of an insect, Manduca sexta. Dev. Comp. Immunol. 2016, 61, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.L.; Cheng, T.C.; Xu, P.Z.; Cheng, D.J.; Fang, T.; Xia, Q.Y. A genome-wide survey for host response of silkworm, Bombyx mori during pathogen Bacillus bombyseptieus infection. PLoS ONE 2009, 4, e8098. [Google Scholar] [CrossRef]
- Christensen, B.M.; Li, J.; Chen, C.C.; Nappi, A.J. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005, 21, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Zhou, F.; Liu, Y.; Hong, F.; Wang, G.R.; An, C.J. Ostrinia furnacalis serpin-3 regulates melanization cascade by inhibiting a prophenoloxidase-activating protease. Insect Biochem. Mol. Biol. 2015, 61, 53–61. [Google Scholar] [CrossRef]
- Tong, Y.R.; Jiang, H.B.; Kanost, M.R. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J. Biol. Chem. 2005, 280, 14932–14942. [Google Scholar] [CrossRef]
- Huybrechts, R.; Coltura, L. Immune-induced angiotensin-converting enzyme assures the appearance of complementary peptides in Locusta migratoria for fine-tuning the innate immune response by inhibiting immune-activated phenoloxidase. Trends Entomol. 2018. [Google Scholar] [CrossRef]
- Kim, G.S.; Nalini, M.; Kim, Y.; Lee, D.W. Octopamine and 5-hydroxytryptamine mediate hemocytic phagocytosis and nodule formation via eicosanoids in the beet armyworm, Spodoptera exigua. Arch. Insect Biochem. Physiol. 2009, 70, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Nagaosa, K.; Okada, R.; Nonaka, S.; Takeuchi, K.; Fujita, Y.; Miyasaka, T.; Manaka, J.; Ando, I.; Nakanishi, Y. Integrin βν-mediated phagocytosis of apoptotic cells in Drosophila embryos. J. Biol. Chem. 2011, 286, 25770–25777. [Google Scholar] [CrossRef]
- Wu, G.Q.; Zhao, Z.Y.; Liu, C.L.; Qiu, L.H. Priming Galleria mellonella (Lepidoptera: Pyralidae) larvae with heat-killed bacterial cells induced an enhanced immune protection against Photorhabdus luminescens TT01 and the role of innate immunity in the process. J. Econ. Entomol. 2014, 107, 559–569. [Google Scholar] [CrossRef]
- Wu, G.Q.; Li, M.; Liu, Y.; Ding, Y.; Yi, Y.H. The specificity of immune priming in silkworm, Bombyx mori, is mediated by the phagocytic ability of granular cells. J. Insect Physiol. 2015, 81, 60–68. [Google Scholar] [CrossRef]
- Best, S.M. Viral subversion of apoptotic enzymes: Escape from death row. Annu. Rev. Microbiol. 2008, 62, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Cashio, P.; Lee, T.V.; Bergmann, A. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 2005, 16, 225–235. [Google Scholar] [CrossRef]
- Steller, H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008, 15, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Mauvezin, C.; Neisch, A.L.; Ayala, C.I.; Kim, J.; Beltrame, A.; Braden, C.R.; Gardner, M.K.; Hays, T.S.; Neufeld, T.P. Coordination of autophagosome-lysosome fusion and transport by a Klp98A-Rab14 complex in Drosophila. J. Cell Sci. 2016, 129, 971–982. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Feng, K.; Tang, F.; Xu, M. Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio. Insects 2021, 12, 983. https://doi.org/10.3390/insects12110983
Wang Z, Feng K, Tang F, Xu M. Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio. Insects. 2021; 12(11):983. https://doi.org/10.3390/insects12110983
Chicago/Turabian StyleWang, Zhiqiang, Kai Feng, Fang Tang, and Meng Xu. 2021. "Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio" Insects 12, no. 11: 983. https://doi.org/10.3390/insects12110983
APA StyleWang, Z., Feng, K., Tang, F., & Xu, M. (2021). Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio. Insects, 12(11), 983. https://doi.org/10.3390/insects12110983





