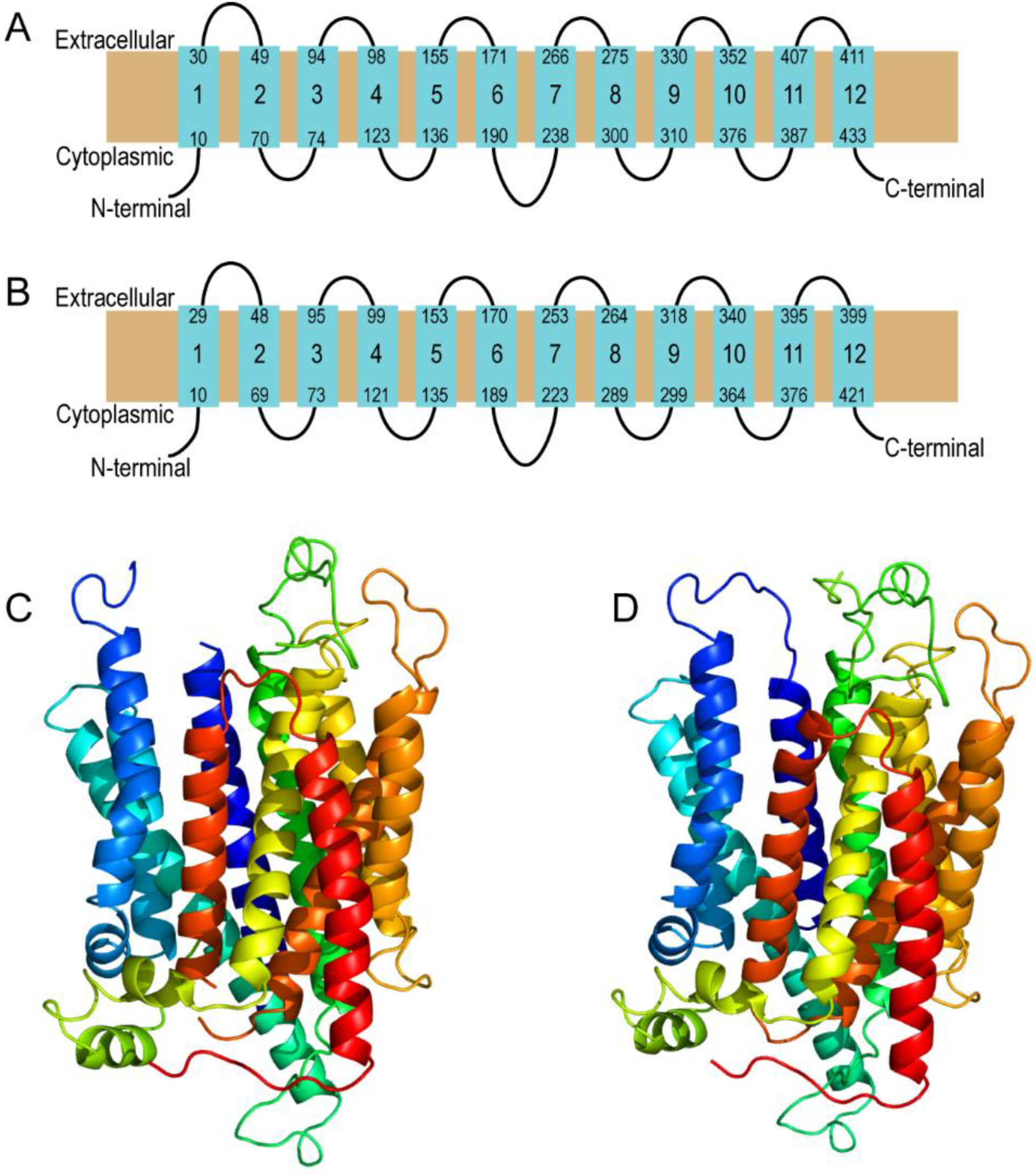
Figure 1.
Prediction of secondary and tertiary structure of human MFSD11 and
D. melanogaster CG18549. Homology modeling and predictions were performed using Phyre2 [
23]. Both proteins were modeled against unc-93 homolog b1 from
Mus musculus (PDB id c7c77B) [
24]. (
A) Secondary structure of human MFSD11. It was modeled with 11% identity, 100% confidence and 94% amino acid coverage. (
B) Secondary structure of
D. melanogaster CG18549. It was modeled with 12% identity, 100% confidence and 97% amino acid coverage. (
C) Tertiary structure of human MFSD11 and (
D)
D. melanogaster CG18549. The gradient of the tertiary structures displays helices from N-terminal (dark blue) to C-terminal (red). Unconnected loops in the predictions are parts of the 3D structure that are modeled with uncertainties and, therefore, they cannot be fully modeled.
Figure 1.
Prediction of secondary and tertiary structure of human MFSD11 and
D. melanogaster CG18549. Homology modeling and predictions were performed using Phyre2 [
23]. Both proteins were modeled against unc-93 homolog b1 from
Mus musculus (PDB id c7c77B) [
24]. (
A) Secondary structure of human MFSD11. It was modeled with 11% identity, 100% confidence and 94% amino acid coverage. (
B) Secondary structure of
D. melanogaster CG18549. It was modeled with 12% identity, 100% confidence and 97% amino acid coverage. (
C) Tertiary structure of human MFSD11 and (
D)
D. melanogaster CG18549. The gradient of the tertiary structures displays helices from N-terminal (dark blue) to C-terminal (red). Unconnected loops in the predictions are parts of the 3D structure that are modeled with uncertainties and, therefore, they cannot be fully modeled.
Figure 2.
CG18549 gene expression images of the third instar larvae CNS and the adult brain. (A–D) Dissected brains and ventral nerve cord from third instar larvae, CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 20 µm. (A,B) CG18549-GAL4 > Pin/Pin-UAS-GFP, (C) CG18549-GAL4 and (D) Pin/Pin-UAS-GFP larvae. (E–G) Dissected brains from seven-days-old adult flies, CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 100 µm. (E) CG18549-GAL4 > Pin/Pin-UAS-GFP, (F) CG18549-GAL4 and (G) Pin/Pin-UAS-GFP adult flies. (H–J) 2-photon, high resolution images of CG18549-GAL4 > Pin/Pin-UAS-GFP (n = 3) seven-days-old flies. CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 100 µm. Brains were imaged with Z-stacks and the pictures were merged displaying the average intensity. (H) Picture include 60 (1 µm) slides, (I) picture include 68 (1 µm) slides, (J) Picture include 29 (1 µm) slides.
Figure 2.
CG18549 gene expression images of the third instar larvae CNS and the adult brain. (A–D) Dissected brains and ventral nerve cord from third instar larvae, CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 20 µm. (A,B) CG18549-GAL4 > Pin/Pin-UAS-GFP, (C) CG18549-GAL4 and (D) Pin/Pin-UAS-GFP larvae. (E–G) Dissected brains from seven-days-old adult flies, CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 100 µm. (E) CG18549-GAL4 > Pin/Pin-UAS-GFP, (F) CG18549-GAL4 and (G) Pin/Pin-UAS-GFP adult flies. (H–J) 2-photon, high resolution images of CG18549-GAL4 > Pin/Pin-UAS-GFP (n = 3) seven-days-old flies. CG18549 expression was visualized with green fluorescence and blue (DAPI) marks the nucleus, scale bar 100 µm. Brains were imaged with Z-stacks and the pictures were merged displaying the average intensity. (H) Picture include 60 (1 µm) slides, (I) picture include 68 (1 µm) slides, (J) Picture include 29 (1 µm) slides.
![Insects 12 01024 g002 Insects 12 01024 g002]()
Figure 3.
Validation of RNAi line functionality and counting of F1 progenies. CG18549 knockdown was verified for two CG18549 RNAi lines: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMS01385}attP2 (CG18549 RNAi line 1, Stock no. 39341) and P{KK102196}VIE-260B (CG18549 RNAi line 2, Stock no. v107272) using the ubiquitous driver da-GAL4. Gene expression was measured with qPCR, log2 fold change was calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * or § p < 0.0491, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = against Driver control, § = against RNAi control). (A) The Log2 fold mean differences for 6 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. CG18549 RNAi line 1: Kruskal–Wallis p = 0.0155, Mann–Whitney (MW) CG18549 knockdown * p = 0.0317, §§ p = 0.0079, the unpaired mean difference between Driver ctrl and RNAi ctrl is 0.305 [95.0% CI −0.57, 1.19], p-value of the two-sided permutation t-test is 0.524; Driver ctrl and CG18549 knockdown is −1.02 [95.0% CI −1.71, −0.382], p-value of the two-sided permutation t-test is 0.021; RNAi ctrl and CG18549 knockdown is −1.33 [95.0% CI −2.05, −0.659], p-value of the two-sided permutation t-test is 0.0004. CG18549 RNAi line 2: Kruskal–Wallis < 0.0001, MW CG18549 knockdown p ≤ 0.0001, RNAi ctrl p = 0.0006, the unpaired mean difference between Driver ctrl and RNAi ctrl is −3.53 [95.0% CI −4.93, −1.94], p-value of the two-sided permutation t-test is 0.0012; Driver ctrl and CG18549 knockdown is −3.93 [95.0% CI −5.05, −2.65], p-value of the two-sided permutation t-test is 0.0006; RNAi ctrl and CG18549 knockdown is −0.404 [95.0% CI −1.62, 0.789], p-value of the two-sided permutation t-test is 0.556. (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (B) Summary of p-values in the statistical analysis. (C) F1 progenies were counted from both RNAi lines and controls, the males and females of F1 progenies are shown in the above scatter plot.
Figure 3.
Validation of RNAi line functionality and counting of F1 progenies. CG18549 knockdown was verified for two CG18549 RNAi lines: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMS01385}attP2 (CG18549 RNAi line 1, Stock no. 39341) and P{KK102196}VIE-260B (CG18549 RNAi line 2, Stock no. v107272) using the ubiquitous driver da-GAL4. Gene expression was measured with qPCR, log2 fold change was calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * or § p < 0.0491, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = against Driver control, § = against RNAi control). (A) The Log2 fold mean differences for 6 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. CG18549 RNAi line 1: Kruskal–Wallis p = 0.0155, Mann–Whitney (MW) CG18549 knockdown * p = 0.0317, §§ p = 0.0079, the unpaired mean difference between Driver ctrl and RNAi ctrl is 0.305 [95.0% CI −0.57, 1.19], p-value of the two-sided permutation t-test is 0.524; Driver ctrl and CG18549 knockdown is −1.02 [95.0% CI −1.71, −0.382], p-value of the two-sided permutation t-test is 0.021; RNAi ctrl and CG18549 knockdown is −1.33 [95.0% CI −2.05, −0.659], p-value of the two-sided permutation t-test is 0.0004. CG18549 RNAi line 2: Kruskal–Wallis < 0.0001, MW CG18549 knockdown p ≤ 0.0001, RNAi ctrl p = 0.0006, the unpaired mean difference between Driver ctrl and RNAi ctrl is −3.53 [95.0% CI −4.93, −1.94], p-value of the two-sided permutation t-test is 0.0012; Driver ctrl and CG18549 knockdown is −3.93 [95.0% CI −5.05, −2.65], p-value of the two-sided permutation t-test is 0.0006; RNAi ctrl and CG18549 knockdown is −0.404 [95.0% CI −1.62, 0.789], p-value of the two-sided permutation t-test is 0.556. (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (B) Summary of p-values in the statistical analysis. (C) F1 progenies were counted from both RNAi lines and controls, the males and females of F1 progenies are shown in the above scatter plot.
![Insects 12 01024 g003 Insects 12 01024 g003]()
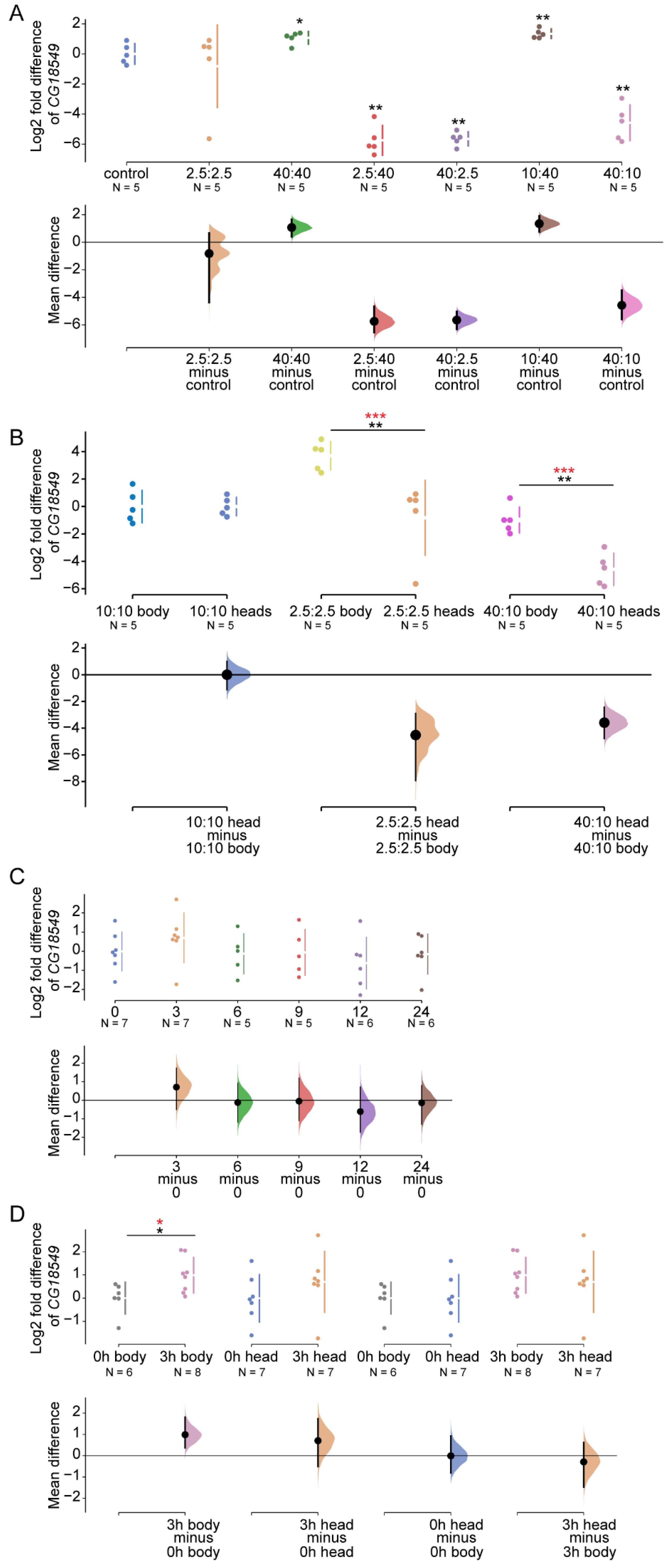
Figure 4.
CG18549 expression alterations as a response to sugar and protein imbalanced diets, and starvation. qPCR was used to measure the gene expression of
CG18549 in heads from five- to seven-days-old CSORC flies (n = 5) fed diets with different sugar and yeast ratio: 10:10 g/dL (control), 2.5:2.5 g/dL, 40:40 g/dL, 2.5:40 g/dL, 10:40 g/dL, 40:2.5 g/dL and 40:10 g/dL diets or subjected to 0 (n = 7), 3 (n = 7), 6 (n = 5), 9 (n = 5), 12 (n = 6) and 24 (n = 6) hours of starvation. The log2 fold change was calculated with the delta Ct method against three reference genes (
Actin42a,
Rpl11,
Rp49), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact
p-values were calculated using multiple corrected Mann–Whitney as a post hoc test *
p < 0.0489, **
p < 0.0099, ***
p < 0.0001. (
A) Log2 fold mean differences of
CG18549 for 6 comparisons against the shared control 10:10 g/dL sugar:yeast are shown in the a Cumming estimation plot. The raw data is plotted on the upper axes. On the lower axes, mean differences are plotted as bootstrap sampling distributions. Each mean difference is depicted as a dot. Each 95% confidence interval is indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal–Wallis
p = 0.0001, the unpaired mean difference between control and 2.5:2.5 g/dL is Mann–Whitney (MW)
p = 0.6905, effect size −0.822 [95.0% CI −4.38, 0.664],
p-value of the two-sided permutation t-test is 0.822; control and 40:40 g/dL is MW
p = 0.0317, effect size 1.07 [95.0% CI 0.408, 1.65],
p-value of the two-sided permutation
t-test is 0.019; control and 2.5:40 g/dL is MW
p = 0.0079, effect size −5.75 [95.0% CI −6.55, −4.66],
p-value of the two-sided permutation
t-test is 0.0; control and 40:2.5 g/dL is MW
p = 0.0079, effect size −5.65 [95.0% CI −6.32, −5.04],
p-value of the two-sided permutation
t-test is 0.003; control and 10:40 g/dL is MW
p = 0.0079, effect size 1.35 [95.0% CI 0.758, 1.91],
p-value of the two-sided permutation
t-test is 0.0022; control and 40:10 g/dL is MW
p = 0.0079, effect size −4.58 [95.0% CI −5.59, −3.49],
p-value of the two-sided permutation
t-test is 0.0018. Previous published data [
20] was, with permission of all authors, reanalyzed and plotted against the results obtained from head tissue. (
B)
CG18549 expression in body and head: 10:10 g/dL, 2.5:2.5 g/dL and 40:10 g/dL. The mean differences for 3 comparisons are shown in the above Cumming estimation plot with same settings as in (
B). Kruskal–Wallis
p = 0.0004, the unpaired mean difference between 10:10 g/dL in body and 10:10 g/dL in head is MW
p = 0.8413, effect size 1.4 × 10
−6 [95.0% CI −1.13, 1.0],
p-value of the two-sided permutation
t-test is 0.997; 2.5:2.5 g/dL in body and 2.5:2.5 g/dL in head is MW
p = 0.0079, effect size −4.52 [95.0% CI −7.92, −2.9],
p-value of the two-sided permutation
t-test is 0.0; 40:10 g/dL in body and 40:10 g/dL in head is MW
p = 0.0079, effect size −3.59 [95.0% CI −4.78, −2.44],
p-value of the two-sided permutation
t-test is 0.0. (
C) The mean Log2 fold differences of
CG18549 for 5 comparisons against the shared control 0 are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes. On the lower axes, mean differences are plotted as bootstrap sampling distributions. Each mean difference is depicted as a dot. Each 95% confidence interval is indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal Wallis
p = 0.5194, the unpaired mean difference between 0 and 3 is 0.705 [95.0% CI −0.505, 1.74],
p-value of the two-sided permutation t-test is 0.269; 0 and 6 is −0.124 [95.0% CI −1.19, 0.912],
p-value of the two-sided permutation
t-test is 0.844; 0 and 9 is −0.0533 [95.0% CI −1.1, 1.19],
p-value of the two-sided permutation
t-test is 0.928; 0 and 12 is −0.611 [95.0% CI −1.73, 0.716],
p-value of the two-sided permutation
t-test is 0.366; 0 and 24 is −0.139 [95.0% CI −1.29, 0.793],
p-value of the two-sided permutation
t-test is 0.804. Previous published data [
20] was, with permission of all authors, reanalyzed and plotted against the results obtained from head tissue. (
D)
CG18549 expression in body and head: 0 h and 3 h of starvation, the mean difference for 4 comparisons of
CG18549 expression are shown in the above Cumming estimation plot. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal–Wallis
p = 0.0325, the unpaired mean difference between 0 h body and 3 h body is MW
p = 0.0293, effect size 0.987 [95.0% CI 0.371, 1.81],
p-value of the two-sided permutation
t-test is 0.0248; 0 h head and 3 h head is MW
p = 0.2593, effect size 0.705 [95.0% CI −0.505, 1.74],
p-value of the two-sided permutation
t-test is 0.268; 0 h body and 0 h head is MW
p = 0.7308, effect size −0.0074 [95.0% CI −0.806, 0.931],
p-value of the two-sided permutation
t-test is 0.988; 3 h body and 3 h head is MW
p = 0.8665, effect size −0.29 [95.0% CI −1.48, 0.622],
p-value of the two-sided permutation
t-test is 0.622.
Figure 4.
CG18549 expression alterations as a response to sugar and protein imbalanced diets, and starvation. qPCR was used to measure the gene expression of
CG18549 in heads from five- to seven-days-old CSORC flies (n = 5) fed diets with different sugar and yeast ratio: 10:10 g/dL (control), 2.5:2.5 g/dL, 40:40 g/dL, 2.5:40 g/dL, 10:40 g/dL, 40:2.5 g/dL and 40:10 g/dL diets or subjected to 0 (n = 7), 3 (n = 7), 6 (n = 5), 9 (n = 5), 12 (n = 6) and 24 (n = 6) hours of starvation. The log2 fold change was calculated with the delta Ct method against three reference genes (
Actin42a,
Rpl11,
Rp49), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact
p-values were calculated using multiple corrected Mann–Whitney as a post hoc test *
p < 0.0489, **
p < 0.0099, ***
p < 0.0001. (
A) Log2 fold mean differences of
CG18549 for 6 comparisons against the shared control 10:10 g/dL sugar:yeast are shown in the a Cumming estimation plot. The raw data is plotted on the upper axes. On the lower axes, mean differences are plotted as bootstrap sampling distributions. Each mean difference is depicted as a dot. Each 95% confidence interval is indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal–Wallis
p = 0.0001, the unpaired mean difference between control and 2.5:2.5 g/dL is Mann–Whitney (MW)
p = 0.6905, effect size −0.822 [95.0% CI −4.38, 0.664],
p-value of the two-sided permutation t-test is 0.822; control and 40:40 g/dL is MW
p = 0.0317, effect size 1.07 [95.0% CI 0.408, 1.65],
p-value of the two-sided permutation
t-test is 0.019; control and 2.5:40 g/dL is MW
p = 0.0079, effect size −5.75 [95.0% CI −6.55, −4.66],
p-value of the two-sided permutation
t-test is 0.0; control and 40:2.5 g/dL is MW
p = 0.0079, effect size −5.65 [95.0% CI −6.32, −5.04],
p-value of the two-sided permutation
t-test is 0.003; control and 10:40 g/dL is MW
p = 0.0079, effect size 1.35 [95.0% CI 0.758, 1.91],
p-value of the two-sided permutation
t-test is 0.0022; control and 40:10 g/dL is MW
p = 0.0079, effect size −4.58 [95.0% CI −5.59, −3.49],
p-value of the two-sided permutation
t-test is 0.0018. Previous published data [
20] was, with permission of all authors, reanalyzed and plotted against the results obtained from head tissue. (
B)
CG18549 expression in body and head: 10:10 g/dL, 2.5:2.5 g/dL and 40:10 g/dL. The mean differences for 3 comparisons are shown in the above Cumming estimation plot with same settings as in (
B). Kruskal–Wallis
p = 0.0004, the unpaired mean difference between 10:10 g/dL in body and 10:10 g/dL in head is MW
p = 0.8413, effect size 1.4 × 10
−6 [95.0% CI −1.13, 1.0],
p-value of the two-sided permutation
t-test is 0.997; 2.5:2.5 g/dL in body and 2.5:2.5 g/dL in head is MW
p = 0.0079, effect size −4.52 [95.0% CI −7.92, −2.9],
p-value of the two-sided permutation
t-test is 0.0; 40:10 g/dL in body and 40:10 g/dL in head is MW
p = 0.0079, effect size −3.59 [95.0% CI −4.78, −2.44],
p-value of the two-sided permutation
t-test is 0.0. (
C) The mean Log2 fold differences of
CG18549 for 5 comparisons against the shared control 0 are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes. On the lower axes, mean differences are plotted as bootstrap sampling distributions. Each mean difference is depicted as a dot. Each 95% confidence interval is indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal Wallis
p = 0.5194, the unpaired mean difference between 0 and 3 is 0.705 [95.0% CI −0.505, 1.74],
p-value of the two-sided permutation t-test is 0.269; 0 and 6 is −0.124 [95.0% CI −1.19, 0.912],
p-value of the two-sided permutation
t-test is 0.844; 0 and 9 is −0.0533 [95.0% CI −1.1, 1.19],
p-value of the two-sided permutation
t-test is 0.928; 0 and 12 is −0.611 [95.0% CI −1.73, 0.716],
p-value of the two-sided permutation
t-test is 0.366; 0 and 24 is −0.139 [95.0% CI −1.29, 0.793],
p-value of the two-sided permutation
t-test is 0.804. Previous published data [
20] was, with permission of all authors, reanalyzed and plotted against the results obtained from head tissue. (
D)
CG18549 expression in body and head: 0 h and 3 h of starvation, the mean difference for 4 comparisons of
CG18549 expression are shown in the above Cumming estimation plot. The effect sizes and CIs are reported above as: effect size [CI width lower bound; upper bound]. Kruskal–Wallis
p = 0.0325, the unpaired mean difference between 0 h body and 3 h body is MW
p = 0.0293, effect size 0.987 [95.0% CI 0.371, 1.81],
p-value of the two-sided permutation
t-test is 0.0248; 0 h head and 3 h head is MW
p = 0.2593, effect size 0.705 [95.0% CI −0.505, 1.74],
p-value of the two-sided permutation
t-test is 0.268; 0 h body and 0 h head is MW
p = 0.7308, effect size −0.0074 [95.0% CI −0.806, 0.931],
p-value of the two-sided permutation
t-test is 0.988; 3 h body and 3 h head is MW
p = 0.8665, effect size −0.29 [95.0% CI −1.48, 0.622],
p-value of the two-sided permutation
t-test is 0.622.
![Insects 12 01024 g004 Insects 12 01024 g004]()
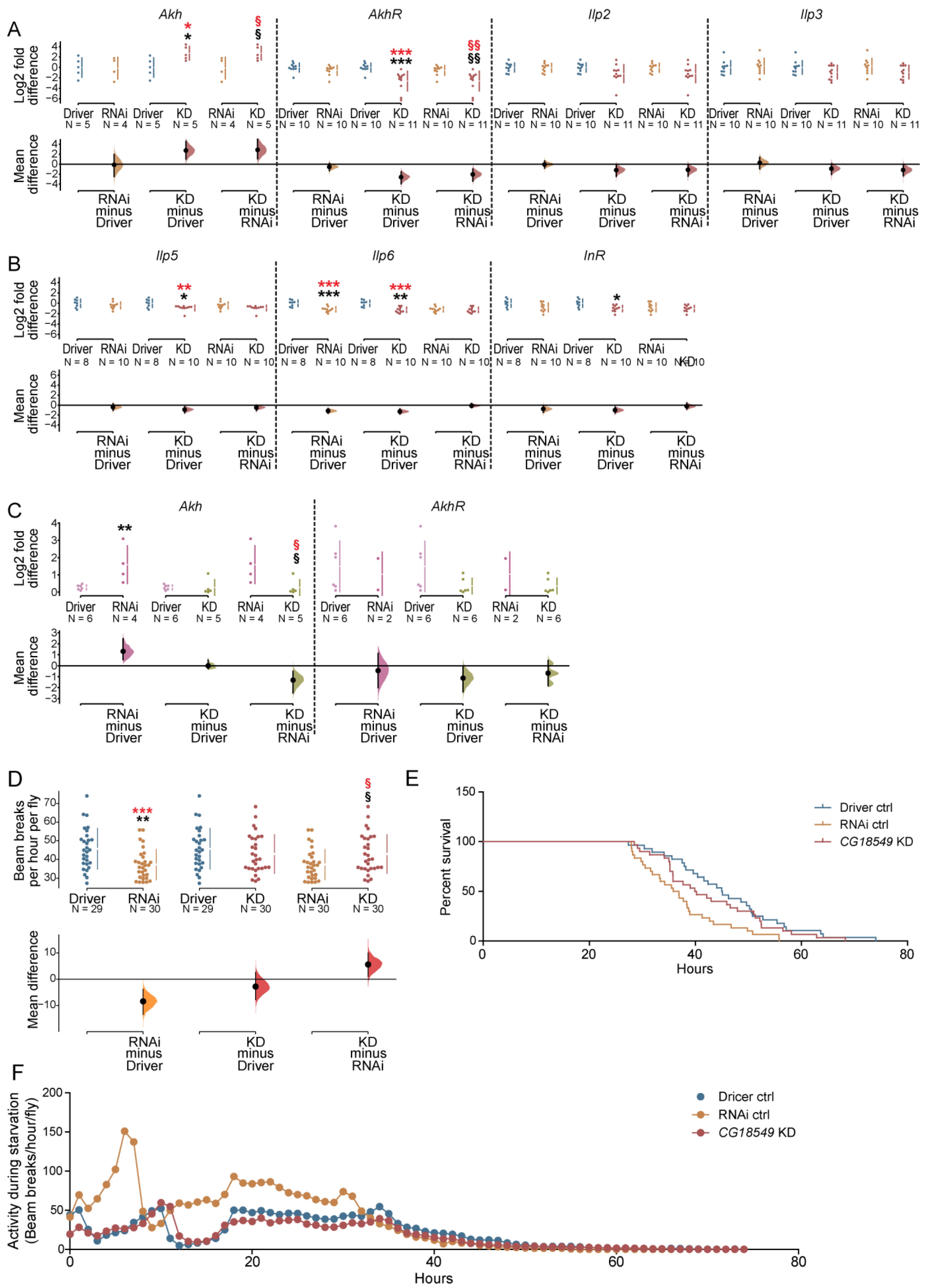
Figure 5.
mRNA expression of glucagon and insulin like peptides and receptors, and starvation resistance in CG18549 knockdown flies. The ubiquitous da-GAL4 driver, w1118 and CG18549 RNAi line 1 and 2 were used. Gene expression of Akh, AkhR, Ilp2, Ilp3, Ilp5, Ilp6 and InR were measured with qPCR, log2 fold changes were calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49) for CG18549 RNAi line 1 and controls, while one reference gene (Actin42a) was used for CG18549 RNAi line 2 and controls. The Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * or § p < 0.0491, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A,B) Data of CG18549 RNAi line 1 (* = significant difference against the Driver control, § = significant difference against the RNAi control. The mean differences for 21 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars: Akh (RNAi ctrl minus Driver ctrl −0.127 [95.0% CI −2.48, 1.95] p = 0.886; CG18549 knockdown minus Driver ctrl 2.75 [95.0% CI 1.03, 4.6] p = 0.0254; CG18549 knockdown minus RNAi ctrl 2.88 [95.0% CI 1.07, 5.16] p = 0.0268); AkhR (RNAi ctrl minus Driver ctrl −0.55 [95.0% CI −1.34 p = 0.197; CG18549 knockdown minus Driver ctrl −2.6 [95.0% CI −3.96, −1.56] p = 0.0006; CG18549 knockdown minus RNAi ctrl is −2.05 [95.0% CI −3.42, −0.971] p = 0.0036); Ilp2 (RNAi ctrl minus Driver ctrl −0.058 [95.0% CI −0.799, 0.665] p = 0.888; CG18549 knockdown minus Driver ctrl −1.19 [95.0% CI −2.46, −0.223] p = 0.0542; CG18549 knockdown minus RNAi ctrl −1.13 [95.0% CI −2.4, −0.166] p = 0.0656); Ilp3 (RNAi ctrl minus Driver ctrl 0.263 [95.0% CI −0.946, 1.41] p = 0.675; CG18549 knockdown minus Driver ctrl −0.904 [95.0% CI −2.05, 0.0776] p = 0.126; CG18549 knockdown minus RNAi ctrl −1.17 [95.0% CI −2.33, −0.051]. p = 0.0712); Ilp5 (RNAi ctrl minus Driver ctrl −0.406 [95.0% CI −1.05, 0.281] p = 0.266; CG18549 knockdown minus Driver ctrl −0.907 [95.0% CI −1.55, −0.339] p = 0.0098; CG18549 knockdown minus RNAi ctrl −0.501 [95.0% CI −1.12, −0.068] p = 0.0946); Ilp6 (RNAi ctrl minus Driver ctrl is −1.16 [95.0% CI −1.64, −0.667] p = 0.001; CG18549 knockdown minus Driver ctrl −1.28 [95.0% CI −1.75, −0.749] p = 0.0004; CG18549 knockdown minus RNAi ctrl −0.117 [95.0% CI −0.519, 0.318] p = 0.615); InR (RNAi ctrl minus Driver ctrl 0.777 [95.0% CI −1.47, −0.0428] p = 0.0628; CG18549 knockdown minus Driver ctrl −1.0 [95.0% CI −1.61, −0.375] p = 0.0084; CG18549 knockdown minus RNAi ctrl −0.223 [95.0% CI −0.8, 0.415] p = 0.503); Akh (up, Kruskal–Wallis p = 0.0340, Mann–Whitney (MW) * p = 0.0317, § p = 0.0317), AkhR (down, Kruskal–Wallis p = 0.0005, MW * p = 0.0006, § p = 0.0035), Ilp2 (Kruskal–Wallis p = 0.1398), Ilp3 (Kruskal–Wallis p = 0.2033), Ilp5 (CG18549 knockdown, down, Kruskal–Wallis p = 0.0566, MW * p = 0.0367), Ilp6 (CG18549 knockdown, down, Kruskal–Wallis p = 0.0015, MW * p = 0.0019; RNAi control, down * p = 0.0009) and InR (CG18549 knockdown, down, Kruskal–Wallis p = 0.0395, MW * p = 0.0117). (C) Data of CG18549 RNAi line 2; Log2 fold change of CG18549 in Driver control, RNAi control and CG18549 knockdown flies (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, §= significant difference against the RNAi control), Cumming estimation plot illustrating the effects size with the same setup as in A and B. Akh: Kruskal–Wallis p = 0.0064, effect size RNAi ctrl minus Driver ctrl 1.31 [95.0% CI 0.552, 2.47] p = 0.0914; CG18549 knockdown minus Driver ctrl −0.00567 [95.0% CI −0.264, 0.535] p = 0.984; CG18549 knockdown minus RNAi ctrl −1.31 [95.0% CI −2.49, −0.497] p = 0.0264; AkhR: Kruskal–Wallis p = 0.1819, effect size RNAi ctrl minus Driver ctrl −0.45 [95.0% CI −2.02, 1.13] p = 0.6, CG18549 knockdown minus Driver ctrl −1.13 [95.0% CI −2.4, −0.118] p = 0.103, CG18549 knockdown minus RNAi ctrl −0.678 [95.0% CI −1.84, 0.47], p = 0.0734): Akh (* p = 0.0095, § p = 0.05), AhkR (no change). (D–F) Five- to seven-days-old male flies from Driver control, RNAi control and CG18549 knockdown (da-GAL4 driver, n = 30) were transferred to glass tubes, prepared with 1% agarose, and contained in the DAMS until the last beam break. Last beam crossing per fly was defined as the timepoint of death. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * p < 0.0492, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (D) The mean difference of starvation resistance is presented in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The unpaired mean difference between Driver ctrl and RNAi ctrl is −8.41 [95.0% CI −13.4, −3.82], p-value of the two-sided permutation t-test is 0.0006; Driver ctrl and CG18549 knockdown is −2.78 [95.0% CI −7.8, 2.64], p-value of the two-sided permutation t-test is 0.321; RNAi ctrl and CG18549 knockdown is 5.63 [95.0% CI 1.01, 10.1], p-value of the two-sided permutation t-test is 0.0206. (E) A survival proportion plot, Log-rank (Mantel-Cox) test (conservative) p = 0.0040, and (F)) a point-connection line graph to illustrate the survival over time and locomotion.
Figure 5.
mRNA expression of glucagon and insulin like peptides and receptors, and starvation resistance in CG18549 knockdown flies. The ubiquitous da-GAL4 driver, w1118 and CG18549 RNAi line 1 and 2 were used. Gene expression of Akh, AkhR, Ilp2, Ilp3, Ilp5, Ilp6 and InR were measured with qPCR, log2 fold changes were calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49) for CG18549 RNAi line 1 and controls, while one reference gene (Actin42a) was used for CG18549 RNAi line 2 and controls. The Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * or § p < 0.0491, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A,B) Data of CG18549 RNAi line 1 (* = significant difference against the Driver control, § = significant difference against the RNAi control. The mean differences for 21 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars: Akh (RNAi ctrl minus Driver ctrl −0.127 [95.0% CI −2.48, 1.95] p = 0.886; CG18549 knockdown minus Driver ctrl 2.75 [95.0% CI 1.03, 4.6] p = 0.0254; CG18549 knockdown minus RNAi ctrl 2.88 [95.0% CI 1.07, 5.16] p = 0.0268); AkhR (RNAi ctrl minus Driver ctrl −0.55 [95.0% CI −1.34 p = 0.197; CG18549 knockdown minus Driver ctrl −2.6 [95.0% CI −3.96, −1.56] p = 0.0006; CG18549 knockdown minus RNAi ctrl is −2.05 [95.0% CI −3.42, −0.971] p = 0.0036); Ilp2 (RNAi ctrl minus Driver ctrl −0.058 [95.0% CI −0.799, 0.665] p = 0.888; CG18549 knockdown minus Driver ctrl −1.19 [95.0% CI −2.46, −0.223] p = 0.0542; CG18549 knockdown minus RNAi ctrl −1.13 [95.0% CI −2.4, −0.166] p = 0.0656); Ilp3 (RNAi ctrl minus Driver ctrl 0.263 [95.0% CI −0.946, 1.41] p = 0.675; CG18549 knockdown minus Driver ctrl −0.904 [95.0% CI −2.05, 0.0776] p = 0.126; CG18549 knockdown minus RNAi ctrl −1.17 [95.0% CI −2.33, −0.051]. p = 0.0712); Ilp5 (RNAi ctrl minus Driver ctrl −0.406 [95.0% CI −1.05, 0.281] p = 0.266; CG18549 knockdown minus Driver ctrl −0.907 [95.0% CI −1.55, −0.339] p = 0.0098; CG18549 knockdown minus RNAi ctrl −0.501 [95.0% CI −1.12, −0.068] p = 0.0946); Ilp6 (RNAi ctrl minus Driver ctrl is −1.16 [95.0% CI −1.64, −0.667] p = 0.001; CG18549 knockdown minus Driver ctrl −1.28 [95.0% CI −1.75, −0.749] p = 0.0004; CG18549 knockdown minus RNAi ctrl −0.117 [95.0% CI −0.519, 0.318] p = 0.615); InR (RNAi ctrl minus Driver ctrl 0.777 [95.0% CI −1.47, −0.0428] p = 0.0628; CG18549 knockdown minus Driver ctrl −1.0 [95.0% CI −1.61, −0.375] p = 0.0084; CG18549 knockdown minus RNAi ctrl −0.223 [95.0% CI −0.8, 0.415] p = 0.503); Akh (up, Kruskal–Wallis p = 0.0340, Mann–Whitney (MW) * p = 0.0317, § p = 0.0317), AkhR (down, Kruskal–Wallis p = 0.0005, MW * p = 0.0006, § p = 0.0035), Ilp2 (Kruskal–Wallis p = 0.1398), Ilp3 (Kruskal–Wallis p = 0.2033), Ilp5 (CG18549 knockdown, down, Kruskal–Wallis p = 0.0566, MW * p = 0.0367), Ilp6 (CG18549 knockdown, down, Kruskal–Wallis p = 0.0015, MW * p = 0.0019; RNAi control, down * p = 0.0009) and InR (CG18549 knockdown, down, Kruskal–Wallis p = 0.0395, MW * p = 0.0117). (C) Data of CG18549 RNAi line 2; Log2 fold change of CG18549 in Driver control, RNAi control and CG18549 knockdown flies (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, §= significant difference against the RNAi control), Cumming estimation plot illustrating the effects size with the same setup as in A and B. Akh: Kruskal–Wallis p = 0.0064, effect size RNAi ctrl minus Driver ctrl 1.31 [95.0% CI 0.552, 2.47] p = 0.0914; CG18549 knockdown minus Driver ctrl −0.00567 [95.0% CI −0.264, 0.535] p = 0.984; CG18549 knockdown minus RNAi ctrl −1.31 [95.0% CI −2.49, −0.497] p = 0.0264; AkhR: Kruskal–Wallis p = 0.1819, effect size RNAi ctrl minus Driver ctrl −0.45 [95.0% CI −2.02, 1.13] p = 0.6, CG18549 knockdown minus Driver ctrl −1.13 [95.0% CI −2.4, −0.118] p = 0.103, CG18549 knockdown minus RNAi ctrl −0.678 [95.0% CI −1.84, 0.47], p = 0.0734): Akh (* p = 0.0095, § p = 0.05), AhkR (no change). (D–F) Five- to seven-days-old male flies from Driver control, RNAi control and CG18549 knockdown (da-GAL4 driver, n = 30) were transferred to glass tubes, prepared with 1% agarose, and contained in the DAMS until the last beam break. Last beam crossing per fly was defined as the timepoint of death. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * p < 0.0492, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (D) The mean difference of starvation resistance is presented in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The unpaired mean difference between Driver ctrl and RNAi ctrl is −8.41 [95.0% CI −13.4, −3.82], p-value of the two-sided permutation t-test is 0.0006; Driver ctrl and CG18549 knockdown is −2.78 [95.0% CI −7.8, 2.64], p-value of the two-sided permutation t-test is 0.321; RNAi ctrl and CG18549 knockdown is 5.63 [95.0% CI 1.01, 10.1], p-value of the two-sided permutation t-test is 0.0206. (E) A survival proportion plot, Log-rank (Mantel-Cox) test (conservative) p = 0.0040, and (F)) a point-connection line graph to illustrate the survival over time and locomotion.
![Insects 12 01024 g005 Insects 12 01024 g005]()
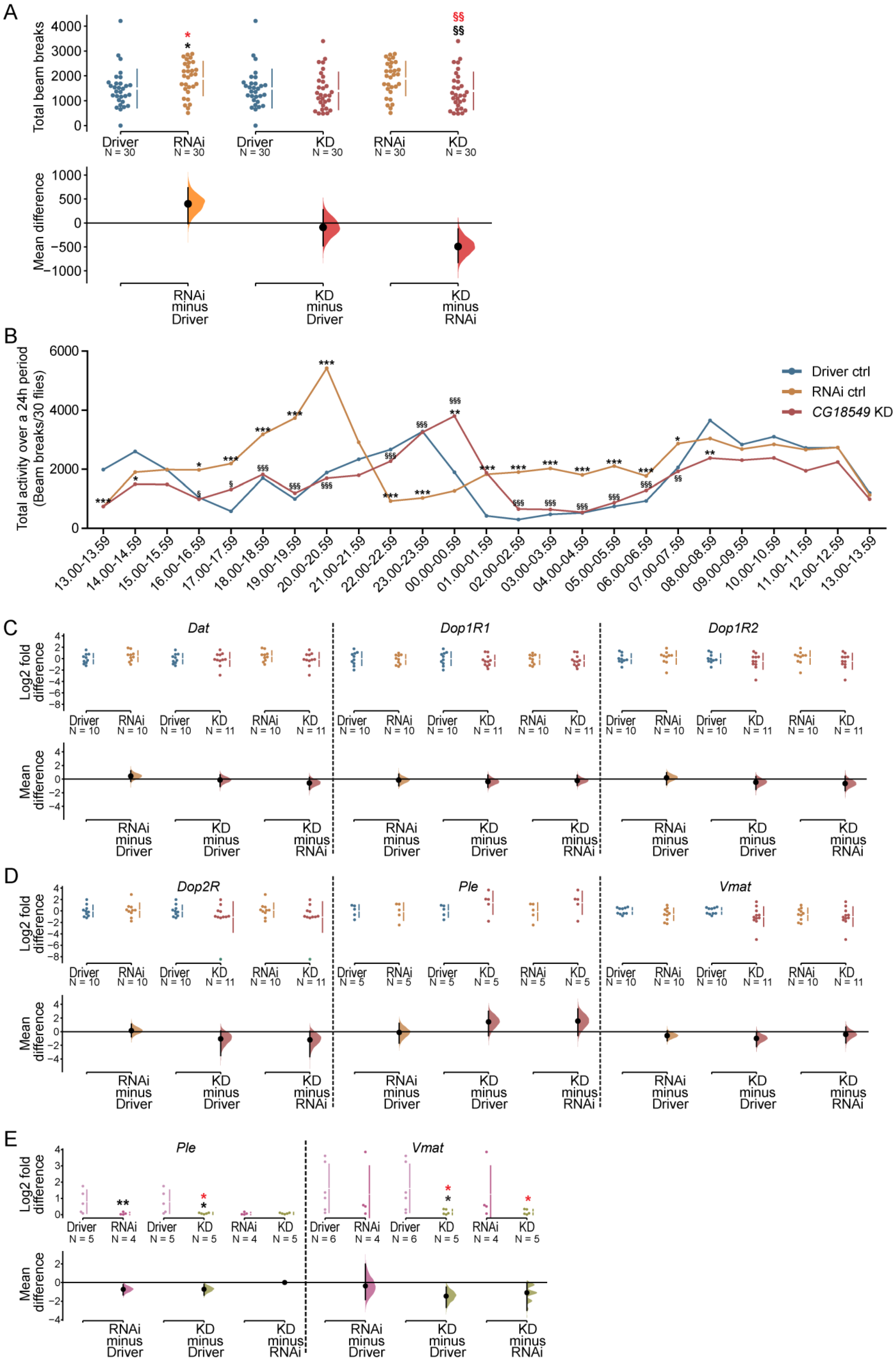
Figure 6.
Ubiquitous knockdown of CG18549 and locomotion monitoring and transcriptional measurements of Dopamine signal transmission genes and the vesicular monoamine transporter in CG18549 knockdown flies. The CG18549 RNAi line 1 was used in this setup. F1 progenies from Driver control, RNAi control and CG18549 knockdown (n = 30) were transferred to glass tubes with enriched Jazz mix standard food and contained in the DAMS for 24 h to record the activity level (locomotion, beam breaks). Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney (MW) as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences *= significant difference against the Driver control, § = significant difference against the RNAi control). (A) Mean differences for 3 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. Kruskal–Wallis p = 0.0112; RNAi ctrl mins Driver ctrl: MW p = 0.0225, effect size 4 × 102 [95.0% CI −12.8, 7.37 × 102] p = 0.04; CG18549 knockdown minus Driver ctrl: MW p = 0.3061, effect size −91.4 [95.0% CI −4.76 × 102, 2.78 × 102] p = 0.654; CG18549 knockdown minus RNAi ctrl: MW p = 0.0075, effect size is −4.92 × 102 [95.0% CI −8.27 × 102, −1.24 × 102] p = 0.0124). (B) Total activity over time. (C–E) The ubiquitous da-GAL4 driver, w1118 and CG18549 RNAi line 1 and 2 were used. Gene expressions of Dat, Dop1R1, Dop1R2, Dop2R, Ple and Vmat were measured using qPCR, log2 fold changes were calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49) for CG18549 RNAi line 1 and controls, while one reference gene (Actin42a) was used for CG18549 RNAi line 2 and controls. The Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * p < 0.0491, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (C,D) Data of CG18549 RNAi line 1. The mean differences for 18 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. (C) Dat: Kruskal–Wallis p = 0.5425, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.439 [95.0% CI −0.333, 1.21] p = 0.295; CG18549 knockdown minus Driver ctrl: −0.128 [95.0% CI −1.09, 0.64] p = 0.786; CG18549 knockdown minus RNAi ctrl: −0.567 [95.0% CI −1.52, 0.239] p = 0.247); Dop1R1: Kruskal–Wallis p = 0.7009, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.142 [95.0% CI −0.984, 0.707] p = 0.76; CG18549 knockdown minus Driver ctrl −0.363 [95.0% CI −1.2, 0.582] p = 0.43; CG18549 knockdown minus RNAi ctrl: is −0.221 [95.0% CI−0.922, 0.574] p = 0.582); Dop1R2: Kruskal–Wallis p = 0.3470, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.192 [95.0% CI −0.822, 0.929] p = 0.672; CG18549 knockdown minus Driver ctrl −0.455 [95.0% CI −1.52, 0.369] p = 0.392; CG18549 knockdown minus RNAi ctrl: −0.647 [95.0% CI −1.7, 0.413] p = 0.269). (D) Dop2R: Kruskal–Wallis p = 0.2889, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.141 [95.0% CI −0.768, 1.09] p = 0.782; CG18549 knockdown minus Driver ctrl −1.06 [95.0% CI −3.5, 0.0975] p = 0.278; CG18549 knockdown minus RNAi ctrl: −1.2 [95.0% CI −3.65, 0.0492] p = 0.222); Ple: Kruskal–Wallis p = 0.2276, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.115 [95.0% CI −1.7, 1.23] p = 0.927; CG18549 knockdown minus Driver ctrl 1.43 [95.0% CI −0.615, 2.98] p = 0.188; CG18549 knockdown minus RNAi ctrl: 1.55 [95.0% CI −0.584, 3.33] p = 0.22); Vmat: Kruskal–Wallis p = 0.1780, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.585 [95.0% CI −1.35, 0.106] p = 0.145; CG18549 knockdown minus Driver ctrl −0.991 [95.0% CI −2.19, −0.115] p = 0.0804; CG18549 knockdown minus RNAi ctrl: −0.406 [95.0% CI −1.67, 0.658] p = 0.549). (E) Data of CG18549 RNAi line 2. Cummings estimation plot with same setup as in B and C: Ple (Kruskal–Wallis p = 0.0212; MW p = 0.0061, effect size RNAi ctrl minus Driver ctrl: MW −0.725 [95.0% CI −1.33, −0.215] p = 0.0568; CG18549 knockdown minus Driver ctrl: MW p = 0.0101, effect size −0.716 [95.0% CI −1.35, −0.2] p = 0.0252; CG18549 knockdown minus RNAi ctrl: MW p = 1, effect size 0.00867 [95.0% CI −0.0823, 0.0645] p = 0.883); Vmat (RNAi ctrl minus Driver ctrl: MW p = 0.0823, effect size −0.362 [95.0% CI −1.84, 1.98] p = 0.687; CG18549 knockdown minus Driver ctrl: MW p = 0.0411, effect size −1.45 [95.0% CI −2.68, −0.497] p = 0.0394; CG18549 knockdown minus RNAi ctrl: MW p = 0.0823, effect size−1.09 [95.0% CI −2.99, −0.11] p = 0.035.
Figure 6.
Ubiquitous knockdown of CG18549 and locomotion monitoring and transcriptional measurements of Dopamine signal transmission genes and the vesicular monoamine transporter in CG18549 knockdown flies. The CG18549 RNAi line 1 was used in this setup. F1 progenies from Driver control, RNAi control and CG18549 knockdown (n = 30) were transferred to glass tubes with enriched Jazz mix standard food and contained in the DAMS for 24 h to record the activity level (locomotion, beam breaks). Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney (MW) as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences *= significant difference against the Driver control, § = significant difference against the RNAi control). (A) Mean differences for 3 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. Kruskal–Wallis p = 0.0112; RNAi ctrl mins Driver ctrl: MW p = 0.0225, effect size 4 × 102 [95.0% CI −12.8, 7.37 × 102] p = 0.04; CG18549 knockdown minus Driver ctrl: MW p = 0.3061, effect size −91.4 [95.0% CI −4.76 × 102, 2.78 × 102] p = 0.654; CG18549 knockdown minus RNAi ctrl: MW p = 0.0075, effect size is −4.92 × 102 [95.0% CI −8.27 × 102, −1.24 × 102] p = 0.0124). (B) Total activity over time. (C–E) The ubiquitous da-GAL4 driver, w1118 and CG18549 RNAi line 1 and 2 were used. Gene expressions of Dat, Dop1R1, Dop1R2, Dop2R, Ple and Vmat were measured using qPCR, log2 fold changes were calculated according to the delta Ct method using three reference genes (Actin42a, Rpl11, Rp49) for CG18549 RNAi line 1 and controls, while one reference gene (Actin42a) was used for CG18549 RNAi line 2 and controls. The Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * p < 0.0491, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (C,D) Data of CG18549 RNAi line 1. The mean differences for 18 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. (C) Dat: Kruskal–Wallis p = 0.5425, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.439 [95.0% CI −0.333, 1.21] p = 0.295; CG18549 knockdown minus Driver ctrl: −0.128 [95.0% CI −1.09, 0.64] p = 0.786; CG18549 knockdown minus RNAi ctrl: −0.567 [95.0% CI −1.52, 0.239] p = 0.247); Dop1R1: Kruskal–Wallis p = 0.7009, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.142 [95.0% CI −0.984, 0.707] p = 0.76; CG18549 knockdown minus Driver ctrl −0.363 [95.0% CI −1.2, 0.582] p = 0.43; CG18549 knockdown minus RNAi ctrl: is −0.221 [95.0% CI−0.922, 0.574] p = 0.582); Dop1R2: Kruskal–Wallis p = 0.3470, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.192 [95.0% CI −0.822, 0.929] p = 0.672; CG18549 knockdown minus Driver ctrl −0.455 [95.0% CI −1.52, 0.369] p = 0.392; CG18549 knockdown minus RNAi ctrl: −0.647 [95.0% CI −1.7, 0.413] p = 0.269). (D) Dop2R: Kruskal–Wallis p = 0.2889, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: 0.141 [95.0% CI −0.768, 1.09] p = 0.782; CG18549 knockdown minus Driver ctrl −1.06 [95.0% CI −3.5, 0.0975] p = 0.278; CG18549 knockdown minus RNAi ctrl: −1.2 [95.0% CI −3.65, 0.0492] p = 0.222); Ple: Kruskal–Wallis p = 0.2276, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.115 [95.0% CI −1.7, 1.23] p = 0.927; CG18549 knockdown minus Driver ctrl 1.43 [95.0% CI −0.615, 2.98] p = 0.188; CG18549 knockdown minus RNAi ctrl: 1.55 [95.0% CI −0.584, 3.33] p = 0.22); Vmat: Kruskal–Wallis p = 0.1780, effect size unpaired mean difference RNAi ctrl minus Driver ctrl: −0.585 [95.0% CI −1.35, 0.106] p = 0.145; CG18549 knockdown minus Driver ctrl −0.991 [95.0% CI −2.19, −0.115] p = 0.0804; CG18549 knockdown minus RNAi ctrl: −0.406 [95.0% CI −1.67, 0.658] p = 0.549). (E) Data of CG18549 RNAi line 2. Cummings estimation plot with same setup as in B and C: Ple (Kruskal–Wallis p = 0.0212; MW p = 0.0061, effect size RNAi ctrl minus Driver ctrl: MW −0.725 [95.0% CI −1.33, −0.215] p = 0.0568; CG18549 knockdown minus Driver ctrl: MW p = 0.0101, effect size −0.716 [95.0% CI −1.35, −0.2] p = 0.0252; CG18549 knockdown minus RNAi ctrl: MW p = 1, effect size 0.00867 [95.0% CI −0.0823, 0.0645] p = 0.883); Vmat (RNAi ctrl minus Driver ctrl: MW p = 0.0823, effect size −0.362 [95.0% CI −1.84, 1.98] p = 0.687; CG18549 knockdown minus Driver ctrl: MW p = 0.0411, effect size −1.45 [95.0% CI −2.68, −0.497] p = 0.0394; CG18549 knockdown minus RNAi ctrl: MW p = 0.0823, effect size−1.09 [95.0% CI −2.99, −0.11] p = 0.035.
![Insects 12 01024 g006 Insects 12 01024 g006]()
Figure 7.
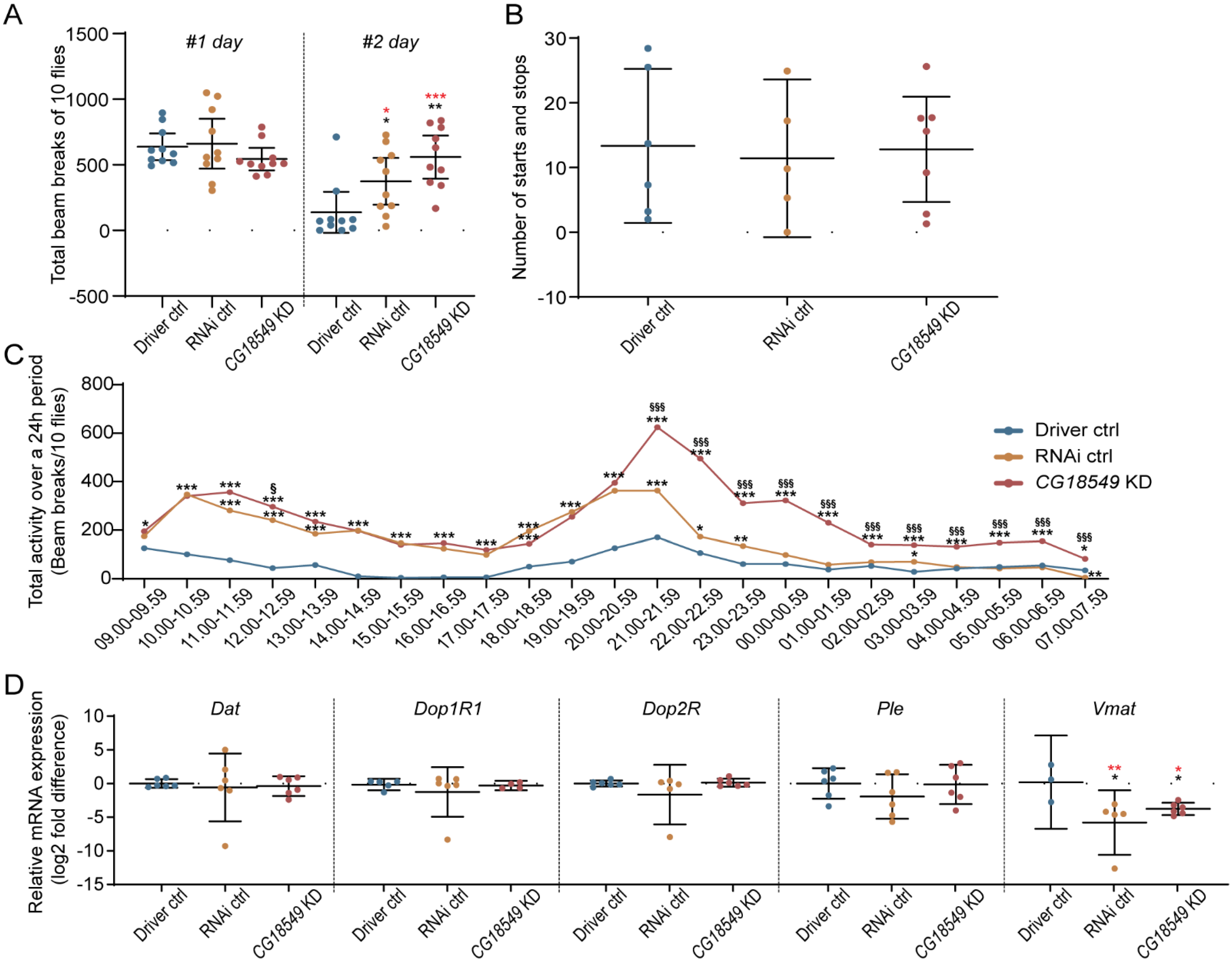
Locomotion of CG18549 RNAi line 1 using the elav-GAL4:GAL80 driver. Conditional (onset one day post-eclosion) brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Five- to seven-days-old male flies (n = 10) were transferred to glass tubes prepared with standard Jazz mix food for 48 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Scatter plot displaying the total beam breaks of run 1 (Kruskal–Wallis p = 0.1792) and run 2 (Kruskal–Wallis p = 0.0023, Mann–Whitney CG18549 knockdown * p = 0.0017, RNAi ctrl * p = 0.0211, Mean difference for 3 comparisons were calculated with 95% confidence intervals. (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control): day #1 unpaired mean difference RNAi ctrl minus Driver ctrl: 22.3 [95.0% CI −1.54 × 102, 1.97 × 102] p = 0.811; CG18549 knockdown minus Driver ctrl: −94.7 [95.0% CI −2.02 × 102, 14.8] p = 0.127; CG18549 knockdown minus RNAi ctrl: −1.17 × 102 [95.0% CI −2.97 × 102, 52.8] p = 0.225), day #2 unpaired mean difference RNAi ctrl minus Driver ctrl: 2.36 × 102 [95.0% CI 4.7, 3.98 × 102] p = 0.039; CG18549 knockdown minus Driver ctrl: 4.21 × 102 [95.0% CI 1.89 × 102, 5.71 × 102] p = 0.001; CG18549 knockdown minus RNAi ctrl: 1.85 × 102 [95.0% CI −18.6, 3.79 × 102] p = 0.0908. (B) Number of starts and stops for each genotype. (C) Total activity plotted over time (* = statistical difference against Driver control, § = statistical difference against RNAi control, * p < 0.05, ** p < 0.001, *** p < 0.0001). (D) Gene expressions of Dat, Dop1R1, Dop2R, Ple and Vmat were measured using qPCR, log2 fold changes were calculated according to the delta Ct method using one reference genes (Actin42a), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * p < 0.0491, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, §= significant difference against the RNAi control. No differences were measured for Dat, Dop1R1, Dop2R and Ple, while Vmat was found to be downregulated in the RNAi ctrl (Mann–Whitney * p = 0.0357) and CG18549 knockdown (Mann–Whitney * p = 0.0476) flies. Dat (Kruskal–Wallis p = 0.9107, unpaired mean difference RNAi ctrl minus Driver ctrl: −0.591 [95.0% CI −5.12, 2.22] p = 0.84; CG18549 knockdown minus Driver ctrl: is −0.387 [95.0% CI −1.57, 0.631] p = 0.547; CG18549 knockdown minus RNAi ctrl: 0.205 [95.0% CI −2.63, 4.82] p = 0.938), Dop1R1 (Kruskal–Wallis p = 0.6546, unpaired mean difference RNAi ctrl minus Driver ctrl: is −1.1 [95.0% CI −5.33, 0.647] p = 0.86; CG18549 knockdown minus Driver ctrl: is −0.148 [95.0% CI −0.683, 0.644] p = 0.741; CG18549 knockdown minus RNAi ctrl: 0.948 [95.0% CI −0.771, 4.06] p = 0.945), Dop2R (Kruskal–Wallis p = 0.7604, unpaired mean difference RNAi ctrl minus Driver ctrl: −1.66 [95.0% CI −6.45, 0.109] p = 0.234; CG18549 knockdown minus Driver ctrl: 0.125 [95.0% CI −0.346, 0.686] p = 0.676; CG18549 knockdown minus RNAi ctrl: 1.78 [95.0% CI −0.0278, 6.58] p = 0.178), Ple (Kruskal–Wallis p = 0.4785, unpaired mean difference RNAi ctrl minus Driver ctrl: −1.93 [95.0% CI −4.51, 1.13] p = 0.228; CG18549 knockdown minus Driver ctrl: is −0.14 [95.0% CI −2.69, 2.43] p = 0.941; CG18549 knockdown minus RNAi ctrl: 1.79 [95.0% CI −1.25, 4.86] p = 0.311) and Vmat (Kruskal–Wallis * p = 0.0351; Driver ctrl vs RNAi ctrl MW * p = 0.0357, Driver ctrl vs. CG18549 knockdown MW * p = 0.0476; unpaired mean difference RNAi ctrl minus Driver ctrl: −6.01 [95.0% CI −10.5, −2.67] p = 0.005; CG18549 knockdown minus Driver ctrl: −3.97 [95.0% CI −6.27, −0.931] p = 0.0188; CG18549 knockdown minus RNAi ctrl: 2.04 [95.0% CI −0.0492, 7.1] p = 0.188).
Figure 7.
Locomotion of CG18549 RNAi line 1 using the elav-GAL4:GAL80 driver. Conditional (onset one day post-eclosion) brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Five- to seven-days-old male flies (n = 10) were transferred to glass tubes prepared with standard Jazz mix food for 48 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Scatter plot displaying the total beam breaks of run 1 (Kruskal–Wallis p = 0.1792) and run 2 (Kruskal–Wallis p = 0.0023, Mann–Whitney CG18549 knockdown * p = 0.0017, RNAi ctrl * p = 0.0211, Mean difference for 3 comparisons were calculated with 95% confidence intervals. (in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control): day #1 unpaired mean difference RNAi ctrl minus Driver ctrl: 22.3 [95.0% CI −1.54 × 102, 1.97 × 102] p = 0.811; CG18549 knockdown minus Driver ctrl: −94.7 [95.0% CI −2.02 × 102, 14.8] p = 0.127; CG18549 knockdown minus RNAi ctrl: −1.17 × 102 [95.0% CI −2.97 × 102, 52.8] p = 0.225), day #2 unpaired mean difference RNAi ctrl minus Driver ctrl: 2.36 × 102 [95.0% CI 4.7, 3.98 × 102] p = 0.039; CG18549 knockdown minus Driver ctrl: 4.21 × 102 [95.0% CI 1.89 × 102, 5.71 × 102] p = 0.001; CG18549 knockdown minus RNAi ctrl: 1.85 × 102 [95.0% CI −18.6, 3.79 × 102] p = 0.0908. (B) Number of starts and stops for each genotype. (C) Total activity plotted over time (* = statistical difference against Driver control, § = statistical difference against RNAi control, * p < 0.05, ** p < 0.001, *** p < 0.0001). (D) Gene expressions of Dat, Dop1R1, Dop2R, Ple and Vmat were measured using qPCR, log2 fold changes were calculated according to the delta Ct method using one reference genes (Actin42a), the Driver control was set as control. Kruskal–Wallis with Dunn’s comparison against the control was used to initially analyze differences, exact p-values were calculated using multiple corrected Mann–Whitney as a post hoc test * p < 0.0491, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, §= significant difference against the RNAi control. No differences were measured for Dat, Dop1R1, Dop2R and Ple, while Vmat was found to be downregulated in the RNAi ctrl (Mann–Whitney * p = 0.0357) and CG18549 knockdown (Mann–Whitney * p = 0.0476) flies. Dat (Kruskal–Wallis p = 0.9107, unpaired mean difference RNAi ctrl minus Driver ctrl: −0.591 [95.0% CI −5.12, 2.22] p = 0.84; CG18549 knockdown minus Driver ctrl: is −0.387 [95.0% CI −1.57, 0.631] p = 0.547; CG18549 knockdown minus RNAi ctrl: 0.205 [95.0% CI −2.63, 4.82] p = 0.938), Dop1R1 (Kruskal–Wallis p = 0.6546, unpaired mean difference RNAi ctrl minus Driver ctrl: is −1.1 [95.0% CI −5.33, 0.647] p = 0.86; CG18549 knockdown minus Driver ctrl: is −0.148 [95.0% CI −0.683, 0.644] p = 0.741; CG18549 knockdown minus RNAi ctrl: 0.948 [95.0% CI −0.771, 4.06] p = 0.945), Dop2R (Kruskal–Wallis p = 0.7604, unpaired mean difference RNAi ctrl minus Driver ctrl: −1.66 [95.0% CI −6.45, 0.109] p = 0.234; CG18549 knockdown minus Driver ctrl: 0.125 [95.0% CI −0.346, 0.686] p = 0.676; CG18549 knockdown minus RNAi ctrl: 1.78 [95.0% CI −0.0278, 6.58] p = 0.178), Ple (Kruskal–Wallis p = 0.4785, unpaired mean difference RNAi ctrl minus Driver ctrl: −1.93 [95.0% CI −4.51, 1.13] p = 0.228; CG18549 knockdown minus Driver ctrl: is −0.14 [95.0% CI −2.69, 2.43] p = 0.941; CG18549 knockdown minus RNAi ctrl: 1.79 [95.0% CI −1.25, 4.86] p = 0.311) and Vmat (Kruskal–Wallis * p = 0.0351; Driver ctrl vs RNAi ctrl MW * p = 0.0357, Driver ctrl vs. CG18549 knockdown MW * p = 0.0476; unpaired mean difference RNAi ctrl minus Driver ctrl: −6.01 [95.0% CI −10.5, −2.67] p = 0.005; CG18549 knockdown minus Driver ctrl: −3.97 [95.0% CI −6.27, −0.931] p = 0.0188; CG18549 knockdown minus RNAi ctrl: 2.04 [95.0% CI −0.0492, 7.1] p = 0.188).
![Insects 12 01024 g007 Insects 12 01024 g007]()
Figure 8.
Locomotion on CG18549 RNAi line 1 using the elav-GAL4 driver. Brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Five- to seven-days-old male flies (n = 10) were transferred to glass tubes prepared with standard Jazz mix food for 72 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (*= significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Mean differences for 9 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. Kruskal–Wallis Run 1 p = 0.0093, Run 2 p ≤ 0.0001, Run 3 p ≤ 0.0001, Driver ctrl and RNAi Run 1: 4.39 × 102 [95.0% CI 1.14 × 102, 8.14 × 102] p = 0.0222, Run 2: 8.52 × 102 [95.0% CI 5.58 × 102, 1.22 × 103] p = 0.0, Run 3: 9.81 × 102 [95.0% CI 6.67 × 102, 1.27 × 103] p = 0.0; CG18549 knockdown and Driver ctrl Run 1: 9.69 × 102 [95.0% CI 4.81 × 102, 1.67 × 103] p = 0.002, Run 2: 1.48 × 103 [95.0% CI 1.08 × 103, 2.01 × 103] p = 0.0, Run 3: 1.1 × 103 [95.0% CI 7.78 × 102, 1.41 × 103] p = 0.0; CG18549 knockdown and RNAi ctrl Run 1: 5.3 × 102 [95.0% CI −12.5, 1.25 × 103] p = 0.131, Run 2: 6.28 × 102 [95.0% CI 1.47 × 102, 1.2 × 103] p = 0.035, Run 3: 1.19 × 102 [95.0% CI −2.57 × 102, 5.1 × 102] p = 0.579. Mann–Whitney post hoc test Run 1: CG18549 knockdown p = 0.0029, RNAi ctrl p = 0.0355; Run 2; CG18549 knockdown * p ≤0.0001 § p = 0.0312, RNAi ctrl * p = 0.0006; Run 3: CG18549 knockdown * p ≤ 0.0001, RNAi ctrl * p ≤ 0.0001. (B) Total activity plotted over time, * p < 0.05, ** p < 0.001, *** p < 0.001 (*= significant difference against the Driver control, § = significant difference against the RNAi control. (C–F) Brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Male flies were aged to 13 (Driver ctrl n = 9, RNAi ctrl n = 12, CG18549 knockdown n = 24) and 21 (n = 30) days. They were transferred to glass tubes prepared with standard Jazz mix food for 72 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * p < 0.0492, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control. (C,D) Scatter plot displaying the total beam breaks, (C) 13 days old flies (Kruskal–Wallis p = 0.0063, Mann–Whitney CG18549 knockdown * p = 0.0129, RNAi ctrl * p = 0.0040; unpaired mean difference between Driver ctrl and RNAi ctrl is −1.35 × 103 [95.0% CI −2.43 × 103, −6.14 × 102] p = 0.0008, Driver ctrl and CG18549 knockdown is −1.08 × 103 [95.0% CI −2.23 × 103, −3.91 × 102] p = 0.0052, RNAi ctrl and CG18549 knockdown is 2.69 × 102 [95.0% CI −20.0, 5.5 × 102] p = 0.131) and (D) 21 days old flies (no differences). (E,F) Total activity plotted over time, (E) 13 days old flies and (F) 21 days old flies.
Figure 8.
Locomotion on CG18549 RNAi line 1 using the elav-GAL4 driver. Brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Five- to seven-days-old male flies (n = 10) were transferred to glass tubes prepared with standard Jazz mix food for 72 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (*= significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Mean differences for 9 comparisons are shown in the above Cumming estimation plot. The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. Kruskal–Wallis Run 1 p = 0.0093, Run 2 p ≤ 0.0001, Run 3 p ≤ 0.0001, Driver ctrl and RNAi Run 1: 4.39 × 102 [95.0% CI 1.14 × 102, 8.14 × 102] p = 0.0222, Run 2: 8.52 × 102 [95.0% CI 5.58 × 102, 1.22 × 103] p = 0.0, Run 3: 9.81 × 102 [95.0% CI 6.67 × 102, 1.27 × 103] p = 0.0; CG18549 knockdown and Driver ctrl Run 1: 9.69 × 102 [95.0% CI 4.81 × 102, 1.67 × 103] p = 0.002, Run 2: 1.48 × 103 [95.0% CI 1.08 × 103, 2.01 × 103] p = 0.0, Run 3: 1.1 × 103 [95.0% CI 7.78 × 102, 1.41 × 103] p = 0.0; CG18549 knockdown and RNAi ctrl Run 1: 5.3 × 102 [95.0% CI −12.5, 1.25 × 103] p = 0.131, Run 2: 6.28 × 102 [95.0% CI 1.47 × 102, 1.2 × 103] p = 0.035, Run 3: 1.19 × 102 [95.0% CI −2.57 × 102, 5.1 × 102] p = 0.579. Mann–Whitney post hoc test Run 1: CG18549 knockdown p = 0.0029, RNAi ctrl p = 0.0355; Run 2; CG18549 knockdown * p ≤0.0001 § p = 0.0312, RNAi ctrl * p = 0.0006; Run 3: CG18549 knockdown * p ≤ 0.0001, RNAi ctrl * p ≤ 0.0001. (B) Total activity plotted over time, * p < 0.05, ** p < 0.001, *** p < 0.001 (*= significant difference against the Driver control, § = significant difference against the RNAi control. (C–F) Brain CG18549 knockdown was performed, and activity was measured in the DAMS system. Male flies were aged to 13 (Driver ctrl n = 9, RNAi ctrl n = 12, CG18549 knockdown n = 24) and 21 (n = 30) days. They were transferred to glass tubes prepared with standard Jazz mix food for 72 h. Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * p < 0.0492, ** p < 0.0099, *** p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control. (C,D) Scatter plot displaying the total beam breaks, (C) 13 days old flies (Kruskal–Wallis p = 0.0063, Mann–Whitney CG18549 knockdown * p = 0.0129, RNAi ctrl * p = 0.0040; unpaired mean difference between Driver ctrl and RNAi ctrl is −1.35 × 103 [95.0% CI −2.43 × 103, −6.14 × 102] p = 0.0008, Driver ctrl and CG18549 knockdown is −1.08 × 103 [95.0% CI −2.23 × 103, −3.91 × 102] p = 0.0052, RNAi ctrl and CG18549 knockdown is 2.69 × 102 [95.0% CI −20.0, 5.5 × 102] p = 0.131) and (D) 21 days old flies (no differences). (E,F) Total activity plotted over time, (E) 13 days old flies and (F) 21 days old flies.
![Insects 12 01024 g008 Insects 12 01024 g008]()
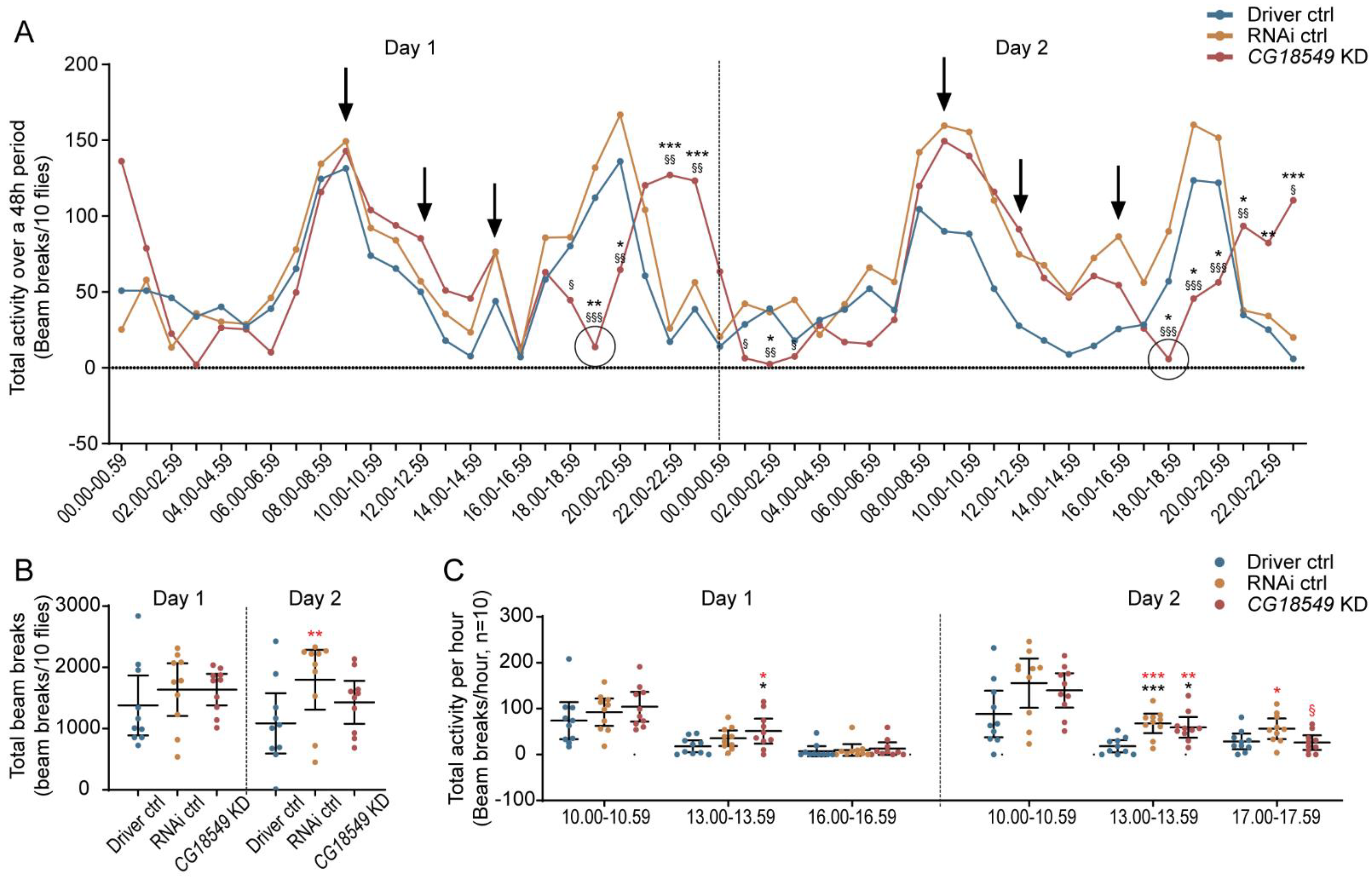
Figure 9.
Brain knockdown of CG18549 affects the stress response after stimulation with bright light. Brain CG18549 knockdown using the elav-GAL4 driver and the CG18549 RNAi line 1 was performed, and flies (n = 10) were placed in a DAMS for 48 h). Stress was induced by exposing the flies to bright light (LED, 1500 lumens) for 10 min three times (9.00, 12.00 and 15.00/16.00) per day, Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Total activity over the 48 h period. Black arrows illustrate the stimulation, and the two circulated points show the large dip in activity that the CG18549 knockdown flies experienced post-stimulation. (B) Total beam breaks per genotype divided into day 1 (Kruskal–Wallis p = 0.3682, unpaired mean difference between Driver ctrl and RNA ctrl 2.56 × 102 [95.0% CI −3.51 × 102, 7.15 × 102] p = 0.379, Driver ctrl and CG18549 knockdown 2.57 × 102 [95.0% CI −2.8 × 102, 6.53 × 102] p = 0.301, RNAi ctrl and CG18549 knockdown 0.8 [95.0% CI −3.76 × 102, 4.42 × 102] p = 0.997) and day 2 (Kruskal–Wallis p = 0.0599, unpaired mean difference between Driver ctrl and RNA ctrl 7.11 × 102 [95.0% CI 69.2, 1.22 × 103] p = 0.0384, Driver ctrl and CG18549 knockdown 3.43 × 102 [95.0% CI −1.68 × 102, 8.17 × 102] p = 0.202, RNAi ctrl and CG18549 knockdown −3.68 × 102 [95.0% CI −8 × 102, 1.93 × 102] p = 0.169). (C) Total activity one-hour post-stimulation [10.00–10.59 day 1: Kruskal–Wallis p = 0.2975, effect size RNAi ctrl minus Driver ctrl: 18.3 [95.0% CI −31.5, 53.7] p = 0.427, CG18549 knockdown minus Driver ctrl: 30.1 [95.0% CI −17.7, 70.2] p = 0.203, CG18549 knockdown minus RNAi ctrl: 11.8 [95.0% CI −21.0, 51.2] p = 0.548; day 2: Kruskal–Wallis p = 0.0849, effect size RNAi ctrl minus Driver ctrl: 67.3 [95.0% CI −0.9, 1.22 × 102] p = 0.0576, CG18549 knockdown minus Driver ctrl: 51.5 [95.0% CI −4.3, 99.3] p = 0.077, CG18549 knockdown minus RNAi ctrl: is −15.8 [95.0% CI −66.2, 41.1] p = 0.578; 13.00–13.59 day 1: Kruskal–Wallis p = 0.0664, Mann–Whitney * p = 0.0444, effect size RNAi ctrl minus Driver ctrl: 17.6 [95.0% CI 0.7, 35.5] p = 0.0766, CG18549 knockdown minus Driver ctrl: 33.1 [95.0% CI 9.9, 59.9]. p = 0.0246, CG18549 knockdown mins RNAi ctrl: 15.5 [95.0% CI −9.6, 43.0] p = 0.285; day 2: Kruskal–Wallis p = 0.0012, * p = 0.0009 and 0.0136, effect size RNAi ctrl minus Driver ctrl: 49.6 [95.0% CI 27.7, 68.0] p = 0.0004, CG18549 knockdown minus Driver ctrl: 41.2 [95.0% CI 21.9, 65.0] p = 0.0012, CG18549 knockdown minus RNAi ctrl: −8.4 [95.0% CI −30.6, 21.2] p = 0.53; 16.00–16.59 Kruskal–Wallis p = 0.3540, effect size RNAi ctrl minus Driver ctrl: 2.6 [95.0% CI −10.0, 18.1] p = 0.64, CG18549 knockdown minus Driver ctrl: 5.6 [95.0% CI −8.2, 21.0] p = 0.469, CG18549 knockdown minus RNAi ctrl: 3.0 [95.0% CI −12.8, 18.2] p = 0.692; 17.00–17.59 Kruskal–Wallis p = 0.3682, effect size RNAi ctrl minus Driver ctrl: 27.8 [95.0% CI 4.0, 50.6] p = 0.0414, CG18549 knockdown minus Driver ctrl: −2.4 [95.0% CI −22.7, 15.8] p = 0.808, CG18549 knockdown minus RNAi ctrl: −30.2 [95.0% CI −51.8, −6.9] p = 0.0244].
Figure 9.
Brain knockdown of CG18549 affects the stress response after stimulation with bright light. Brain CG18549 knockdown using the elav-GAL4 driver and the CG18549 RNAi line 1 was performed, and flies (n = 10) were placed in a DAMS for 48 h). Stress was induced by exposing the flies to bright light (LED, 1500 lumens) for 10 min three times (9.00, 12.00 and 15.00/16.00) per day, Differences were calculated using Kruskal–Wallis with Dunn’s comparison and/or Mann–Whitney as a post hoc test to calculate exact p-values * or § p < 0.0492, ** or §§ p < 0.0099, *** or §§§ p < 0.0001 (* = significant difference against the Driver control, § = significant difference against the RNAi control, in red effect size differences * = significant difference against the Driver control, § = significant difference against the RNAi control). (A) Total activity over the 48 h period. Black arrows illustrate the stimulation, and the two circulated points show the large dip in activity that the CG18549 knockdown flies experienced post-stimulation. (B) Total beam breaks per genotype divided into day 1 (Kruskal–Wallis p = 0.3682, unpaired mean difference between Driver ctrl and RNA ctrl 2.56 × 102 [95.0% CI −3.51 × 102, 7.15 × 102] p = 0.379, Driver ctrl and CG18549 knockdown 2.57 × 102 [95.0% CI −2.8 × 102, 6.53 × 102] p = 0.301, RNAi ctrl and CG18549 knockdown 0.8 [95.0% CI −3.76 × 102, 4.42 × 102] p = 0.997) and day 2 (Kruskal–Wallis p = 0.0599, unpaired mean difference between Driver ctrl and RNA ctrl 7.11 × 102 [95.0% CI 69.2, 1.22 × 103] p = 0.0384, Driver ctrl and CG18549 knockdown 3.43 × 102 [95.0% CI −1.68 × 102, 8.17 × 102] p = 0.202, RNAi ctrl and CG18549 knockdown −3.68 × 102 [95.0% CI −8 × 102, 1.93 × 102] p = 0.169). (C) Total activity one-hour post-stimulation [10.00–10.59 day 1: Kruskal–Wallis p = 0.2975, effect size RNAi ctrl minus Driver ctrl: 18.3 [95.0% CI −31.5, 53.7] p = 0.427, CG18549 knockdown minus Driver ctrl: 30.1 [95.0% CI −17.7, 70.2] p = 0.203, CG18549 knockdown minus RNAi ctrl: 11.8 [95.0% CI −21.0, 51.2] p = 0.548; day 2: Kruskal–Wallis p = 0.0849, effect size RNAi ctrl minus Driver ctrl: 67.3 [95.0% CI −0.9, 1.22 × 102] p = 0.0576, CG18549 knockdown minus Driver ctrl: 51.5 [95.0% CI −4.3, 99.3] p = 0.077, CG18549 knockdown minus RNAi ctrl: is −15.8 [95.0% CI −66.2, 41.1] p = 0.578; 13.00–13.59 day 1: Kruskal–Wallis p = 0.0664, Mann–Whitney * p = 0.0444, effect size RNAi ctrl minus Driver ctrl: 17.6 [95.0% CI 0.7, 35.5] p = 0.0766, CG18549 knockdown minus Driver ctrl: 33.1 [95.0% CI 9.9, 59.9]. p = 0.0246, CG18549 knockdown mins RNAi ctrl: 15.5 [95.0% CI −9.6, 43.0] p = 0.285; day 2: Kruskal–Wallis p = 0.0012, * p = 0.0009 and 0.0136, effect size RNAi ctrl minus Driver ctrl: 49.6 [95.0% CI 27.7, 68.0] p = 0.0004, CG18549 knockdown minus Driver ctrl: 41.2 [95.0% CI 21.9, 65.0] p = 0.0012, CG18549 knockdown minus RNAi ctrl: −8.4 [95.0% CI −30.6, 21.2] p = 0.53; 16.00–16.59 Kruskal–Wallis p = 0.3540, effect size RNAi ctrl minus Driver ctrl: 2.6 [95.0% CI −10.0, 18.1] p = 0.64, CG18549 knockdown minus Driver ctrl: 5.6 [95.0% CI −8.2, 21.0] p = 0.469, CG18549 knockdown minus RNAi ctrl: 3.0 [95.0% CI −12.8, 18.2] p = 0.692; 17.00–17.59 Kruskal–Wallis p = 0.3682, effect size RNAi ctrl minus Driver ctrl: 27.8 [95.0% CI 4.0, 50.6] p = 0.0414, CG18549 knockdown minus Driver ctrl: −2.4 [95.0% CI −22.7, 15.8] p = 0.808, CG18549 knockdown minus RNAi ctrl: −30.2 [95.0% CI −51.8, −6.9] p = 0.0244].
![Insects 12 01024 g009 Insects 12 01024 g009]()
Table 1.
Display the name and the forward and reverse sequence of primers. Housekeeping genes are marked in red.
Table 1.
Display the name and the forward and reverse sequence of primers. Housekeeping genes are marked in red.
| Primer | Forward | Reverse |
|---|
| Actin42A | acaacacttccgctcct | gaacacaatatggtttgcttatgc |
| Akh | tagtgctgtgttaattac | tcattctgagttctatg |
| AkhR | aggagcgactttgatgag | tccgtagcagtagatgaa |
| CG18549 | tgttcgtctttacggcattcc | gtgtagccctcacccttgaa |
| DAT | gcttcaaaccataagttctaa | tcggacttgatattatctacaa |
| Dilp2 | ctgcagtgaaaagctcaacga | tcattctgagttctatg |
| Dilp3 | tgaacgaactatcactcaacagtct | agagaactttggaccccgtgaa |
| Dilp5 | gaggcaccttgggcctattc | catgtggtgagattcggagct |
| Dilp6 | gtccaaagtcctgctagtcct | tctgttcgtattccgtgggtg |
| InR | caagccgttcgtccatc | tcattccaaagtcaggaa |
| Pale | tcaagaaatcctacagtat | cacaatgcaatcttccag |
| Rp49 | cacaccaaatcttacaaaatgtgtga | aatccggccttgcacatg |
| Rpl11 | ccatcggtatctatggtctgga | catcgtatttctggaacc |
| Vmat | gtgaccttcgggacgatag | actagagcgggaaaaccagc |
Table 2.
RNA sequencing log2 fold change as a validation of qPCRs run on the Driver control (
da-GAL4 >
w1118), the RNAi control (
w1118 >
CG18549 RNAi line 1) and the CG18549 knockdown (
da-GAL4 >
CG18549 RNAi line 1) flies. The table summarize the gene,
flybase.org (accessed on 16 March 2021) identity (FB ID) and the log2 fold change and Wald test
p-values.
Table 2.
RNA sequencing log2 fold change as a validation of qPCRs run on the Driver control (
da-GAL4 >
w1118), the RNAi control (
w1118 >
CG18549 RNAi line 1) and the CG18549 knockdown (
da-GAL4 >
CG18549 RNAi line 1) flies. The table summarize the gene,
flybase.org (accessed on 16 March 2021) identity (FB ID) and the log2 fold change and Wald test
p-values.
| Gene | FB ID | Wald Test |
|---|
| Driver vs. RNAi | Driver vs. Knockdown | RNAi vs. Knockdown |
|---|
| Log2 Fold Change | p-Value | Log2 Fold Change | p-Value | Log2 Fold Change | p-Value |
|---|
| CG18549 | FBgn0038053 | 0.27 | 0.57 | −0.47 | 0.32 | −0.74 | 0.15 |
| Akh | FBgn0004552 | 0.35 | 0.27 | 0.66 | 0.04 | 0.31 | 0.36 |
| AkhR | FBgn0025595 | −0.19 | 0.36 | −0.34 | 0.09 | −0.15 | 0.49 |
| Ilp2 | FBgn0036046 | 0.70 | 0.09 | 1.16 | 0.004 | 0.45 | 0.30 |
| Ilp3 | FBgn0044050 | 0.33 | 0.34 | 0.65 | 0.06 | 0.32 | 0.38 |
| Ilp5 | FBgn0044048 | 0.09 | 0.86 | 1.05 | 0.04 | 0.96 | 0.07 |
| Ilp6 | FBgn0044047 | −0.04 | 0.76 | −0.15 | 0.31 | −0.10 | 0.50 |
| InR | FBgn0283499 | N/A | N/A | N/A | N/A | N/A | N/A |
| Dat | FBgn0034136 | −0.29 | 0.27 | −0.27 | 0.30 | 0.02 | 0.96 |
| Dop1R1 | FBgn0011582 | −0.56 | 0.05 | −0.56 | 0.05 | −0.01 | 0.99 |
| Dop1R2 | FBgn0266137 | 0.13 | 0.66 | −0.31 | 0.29 | −0.44 | 0.16 |
| Dop2R | FBgn0053517 | −0.08 | 0.90 | −1.25 | 0.03 | −1.17 | 0.06 |
| Ple | FBgn0005626 | −0.45 | 0.17 | −0.93 | 0.005 | −0.48 | 0.18 |
| Vmat | FBgn0260964 | −0.14 | 0.60 | −0.74 | 0.007 | −0.60 | 0.05 |
Table 3.
Survival proportions of the Driver control (blue column, da-GAL4 > w1118), the RNAi control (yellow column, w1118 > CG18549 RNAi line 1) and the CG18549 knockdown (red column, da-GAL4 > CG18549 RNAi line 1) flies that were used in the starvation resistance assay. The percentage together with the upper and lower 95% CI for each line at specific hours are presented. A grey row indicates the hours that a line deceased of starvation.
Table 3.
Survival proportions of the Driver control (blue column, da-GAL4 > w1118), the RNAi control (yellow column, w1118 > CG18549 RNAi line 1) and the CG18549 knockdown (red column, da-GAL4 > CG18549 RNAi line 1) flies that were used in the starvation resistance assay. The percentage together with the upper and lower 95% CI for each line at specific hours are presented. A grey row indicates the hours that a line deceased of starvation.
| Survival Proportions |
|---|
| Hours | Driver Control | RNAi Control | CG18549 Knockdown |
|---|
| Percentage | +Error | −Error | Percentage | +Error | −Error | Percentage | +Error | −Error |
|---|
| 0.00 | 100 | | | 100 | | | 100 | | |
| 27.36 | 96.43 | 3.06 | 19.18 | | | | | | |
| 27.91 | | | | 96.67 | 2.86 | 18.06 | | | |
| 27.95 | | | | 93.33 | 4.96 | 17.45 | | | |
| 28.00 | | | | 90.00 | 6.66 | 17.88 | | | |
| 28.20 | | | | 86.67 | 8.11 | 18.39 | | | |
| 28.48 | | | | | | | 96.67 | 2.86 | 18.06 |
| 28.53 | | | | 83.33 | 9.37 | 18.84 | | | |
| 29.11 | | | | | | | 93.33 | 4.96 | 17.45 |
| 29.50 | | | | | | | 90.00 | 6.66 | 17.88 |
| 29.75 | | | | 80.00 | 10.48 | 19.20 | | | |
| 30.07 | | | | 76.67 | 11.46 | 19.46 | | | |
| 30.37 | 92.86 | 5.31 | 18.51 | | | | | | |
| 30.53 | | | | 73.33 | 12.34 | 19.64 | | | |
| 31.38 | | | | | | | 86.67 | 8.12 | 18.39 |
| 31.53 | | | | 70.00 | 13.12 | 19.74 | | | |
| 31.82 | 89.29 | 7.13 | 18.93 | | | | | | |
| 31.87 | | | | 66.67 | 13.81 | 19.75 | | | |
| 33.23 | | | | 63.33 | 14.42 | 19.69 | | | |
| 33.4 | | | | 60.00 | 14.95 | 19.55 | | | |
| 34.1 | 85.71 | 8.67 | 19.42 | | | | | | |
| 34.13 | | | | | | | 83.33 | 9.37 | 18.84 |
| 34.23 | | | | 56.67 | 15.41 | 19.34 | | | |
| 34.98 | | | | | | | 80 | 10.48 | 19.20 |
| 35.17 | | | | | | | 76.67 | 11.46 | 19.46 |
| 35.18 | | | | | | | 73.33 | 12.34 | 19.64 |
| 35.20 | | | | | | | 70.00 | 13.12 | 19.74 |
| 35.50 | | | | 53.33 | 15.80 | 19.05 | | | |
| 35.60 | 82.14 | 10.01 | 19.85 | | | | | | |
| 35.73 | | | | | | | 66.67 | 13.81 | 19.75 |
| 35.77 | | | | | | | 63.33 | 14.42 | 19.69 |
| 35.80 | | | | | | | 60.00 | 14.95 | 19.55 |
| 35.82 | | | | 50 | 16.12 | 18.70 | | | |
| 36.87 | | | | 46.67 | 16.38 | 18.27 | | | |
| 36.95 | | | | 43.33 | 16.56 | 17.77 | | | |
| 37.50 | 78.57 | 11.18 | 20.17 | | | | | | |
| 37.67 | | | | | | | 56.67 | 15.41 | 19.34 |
| 37.72 | | | | 40.00 | 16.67 | 17.20 | | | |
| 37.92 | 75.00 | 12.21 | 20.40 | | | | | | |
| 38.20 | 71.43 | 13.13 | 20.51 | | | | | | |
| 38.38 | | | | 36.67 | 16.71 | 16.54 | | | |
| 38.43 | | | | 33.33 | 16.67 | 15.81 | | | |
| 38.72 | | | | 30.00 | 16.55 | 14.98 | | | |
| 38.93 | | | | 26.67 | 16.35 | 14.06 | | | |
| 39.02 | | | | | | | 53.33 | 15.80 | 19.05 |
| 39.6 | 67.86 | 13.94 | 20.54 | | | | | | |
| 39.87 | | | | | | | 50.00 | 16.12 | 18.70 |
| 40.20 | | | | | | | 46.67 | 16.38 | 18.27 |
| 40.40 | 64.29 | 14.65 | 20.48 | | | | | | |
| 41.27 | | | | 23.33 | 16.05 | 13.03 | | | |
| 41.62 | 60.71 | 15.27 | 20.32 | | | | | | |
| 42.17 | | | | | | | 43.33 | 16.56 | 17.77 |
| 42.40 | 57.14 | 15.80 | 20.09 | | | | | | |
| 42.68 | | | | 20.00 | 15.64 | 11.88 | | | |
| 42.95 | | | | | | | 40.00 | 16.67 | 17.20 |
| 43.40 | | | | 16.67 | 15.11 | 10.59 | | | |
| 44.03 | 53.57 | 16.25 | 19.76 | | | | | | |
| 44.98 | 50.00 | 16.62 | 19.36 | | | | | | |
| 45.1 | 46.43 | 16.90 | 18.87 | | | | | | |
| 45.93 | | | | | | | 36.67 | 16.71 | 16.54 |
| 46.23 | 42.86 | 17.10 | 18.29 | | | | | | |
| 46.55 | | | | | | | 33.33 | 16.67 | 15.81 |
| 46.73 | | | | 13.33 | 14.45 | 9.13 | | | |
| 47.92 | | | | | | | 30.00 | 16.55 | 14.98 |
| 48.60 | 39.29 | 17.22 | 17.62 | | | | | | |
| 49.65 | 35.71 | 17.26 | 16.86 | | | | | | |
| 49.82 | | | | 10.00 | 13.58 | 7.45 | | | |
| 50.22 | 32.14 | 17.20 | 16.00 | | | | | | |
| 50.55 | 28.57 | 17.04 | 15.03 | | | | | | |
| 50.75 | 25.00 | 16.78 | 13.94 | | | | | | |
| 50.82 | | | | 6.67 | 12.51 | 5.49 | | | |
| 51.00 | | | | | | | 26.67 | 16.35 | 14.06 |
| 51.3 | | | | | | | 23.33 | 16.05 | 13.03 |
| 52.18 | | | | | | | 20.00 | 15.64 | 11.88 |
| 52.40 | | | | | | | 16.67 | 15.11 | 10.59 |
| 52.45 | | | | | | | 13.33 | 14.44 | 9.13 |
| 52.73 | 21.43 | 16.40 | 12.72 | | | | | | |
| 55.38 | 17.86 | 15.89 | 11.35 | | | | | | |
| 55.77 | | | | 3.33 | 11.18 | 3.08 | | | |
| 55.8 | | | | 0 | 0 | 0 | | | |
| 56.53 | | | | | | | 10.00 | 13.58 | 7.45 |
| 56.75 | 14.29 | 15.21 | 9.79 | | | | | | |
| 57.12 | 10.71 | 14.35 | 7.99 | | | | | | |
| 58.08 | | | | | | | 6.67 | 12.51 | 5.49 |
| 62.87 | | | | | | | 3.33 | 11.18 | 3.08 |
| 63.67 | 7.14 | 13.23 | 5.89 | | | | | | |
| 64.12 | 3.57 | 11.84 | 3.31 | | | | | | |
| 68.28 | | | | | | | 0 | 0 | 0 |
| 74.02 | 0 | 0 | 0 | | | | | | |