The Identification of Boll Weevil, Anthonomus grandis grandis (Coleoptera: Curculionidae), Genes Involved in Pheromone Production and Pheromone Biosynthesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Boll Weevil Collections and Experimental Design
2.2. RNA Extraction and Sequencing
2.3. Transcriptome Assembly and Annotation
2.4. RNA-Seq Analysis
3. Results and Discussion
3.1. Transcriptome
3.2. RNA-Seq Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paula, D.P.; Claudino, D.; Timbó, R.V.; Miranda, J.E.; Bemquerer, M.; Ribeiro, A.; Sujii, E.; Fontes, E.; Pires, C. Reproductive dormancy in boll-weevil from populations of the Midwest of Brazil. J. Econ. Ѐntomol. 2013, 106, 86–96. [Google Scholar] [CrossRef]
- Smith, J.W. Boll weevil eradication: Area-wide pest management. Ann. Ѐntomol. Soc. Am. 1998, 91, 239–247. [Google Scholar] [CrossRef]
- Carter, F.L.; Nelson, T.C.; Jordan, A.G.; Smith, J.R. US cotton declares war on the boll weevil. In Boll Weevil Eradication in the United States through 1999; The Cotton Foundation: Memphis, TN, USA, 1999; pp. 25–54. [Google Scholar]
- El-Lissy, O.A.; Grefenstette, W.J. Progress of boll weevil eradication in the U.S., 2005. In Proceedings of the Beltwide Cotton Conferences, San Antonio, TX, USA, 3–6 January 2006; pp. 1266–1276. [Google Scholar]
- Texas Boll Weevil Eradication Foundation. Trapping. Available online: https://www.txbollweevil.org/trapping.html (accessed on 7 May 2021).
- Tumlinson, J.H.; Hardee, D.D.; Gueldner, R.C.; Thompson, A.C.; Hedin, P.A.; Minyard, J.P. Sex pheromones produced by male boll weevil: Isolation, identification, and synthesis. Science 1969, 166, 1010–1012. [Google Scholar] [CrossRef]
- Hardee, D.D.; McKibben, G.H.; Gueldner, R.C.; Mitchell, E.B.; Tumlinson, J.H.; Cross, W.H. Boll weevils in nature respond to grandlure, a synthetic pheromone. J. Econ. Ѐntomol. 1972, 65, 97–100. [Google Scholar]
- Rogers, C.; Oakes, S.; Rummel, D. Evaluation of infield pheromone traps for boll weevil suppression in the Texas Rolling Plains. Res. Monogr -Tex. Agric. Exp. Station 1976, 8, 45–52. [Google Scholar]
- Rummel, D.R.; Bottrell, D.G. Seasonally related decline in response of boll weevils to pheromone traps during mid-season. Environ. Ѐntomol. 1976, 5, 783–787. [Google Scholar] [CrossRef]
- Wolfenbarger, D.A.; Graham, H.M.; Parker, R.D.; Davis, J.W. Boll weevil: Seasonal patterns of response to traps baited with grandlure in the Lower Rio Grande Valley. Environ. Ѐntomol. 1976, 5, 403–408. [Google Scholar] [CrossRef]
- Suh, C.P.-C.; Westbrook, J.K. Failure of pheromone traps in detecting incipient populations of boll weevils (Coleoptera: Curculiondae): Investigation of two potential contributing factors. J. Ѐntomol. Sci. 2014, 49, 211–214. [Google Scholar] [CrossRef]
- Rummel, D.; Bottrell, D. Relationship of overwintered boll weevil response to pheromone traps and natural entry into cotton. Res. Monogr.-Tex. Agric. Exp. Stn. 1976, 8, 26–31. [Google Scholar]
- Hardee, D.D.; Cross, W.H.; Huddleston, P.M.; Davich, T.B. Survey and control of the boll weevil in West Texas with traps baited with males. J. Econ. Ѐntomol. 1970, 63, 1041–1048. [Google Scholar] [CrossRef]
- Boyd, F., Jr.; Brazzel, J.; Helms, W.; Moritz, R.; Edwards, R. Spring destruction of overwintered boll weevils in West Texas with wing traps. J. Econ. Entomol. 1973, 66, 507–510. [Google Scholar] [CrossRef]
- Scott, W.P.; Lloyd, E.P.; Bryson, J.O.; Davich, T.B. Trap plots for suppression of low-density overwintered populations of boll weevils. J. Econ. Ѐntomol. 1974, 67, 281–283. [Google Scholar] [CrossRef]
- Cherry, E.T. Monitoring boll weevil movement with pheromone traps. Tenn. Farm Home Sci. Prog. Rep. 1974, 90, 27–29. [Google Scholar]
- Segers, J.; Urban, T.; George, D.; Benedict, J.; Walmsley, M.; Pieters, E. Seasonal numbers, sex diapause states of boll weevils captured in pheromone traps in the Lower Gulf Coast of Texas. Southwest. Entomol. 1988, 12, 311–316. [Google Scholar]
- White, J.R.; Rummel, D.R. Emergence profile of overwintered boll weevils and entry into cotton. Environ. Ѐntomol. 1978, 7, 7–14. [Google Scholar] [CrossRef]
- Spurgeon, D.W. Age dependence of pheromone production by the boll weevil (Coleoptera: Curculionidae). Environ. Ѐntomol. 2003, 32, 31–38. [Google Scholar] [CrossRef]
- Spurgeon, D.W.; Suh, C.P.-C. Pheromone production by the boll weevil (Coleoptera: Curculionidae) fed cotton squares and bolls. J. Ѐntomol. Sci. 2009, 44, 209–221. [Google Scholar] [CrossRef][Green Version]
- Suh, C.-C.; Spurgeon, D. Continued pheromone release by boll weevils (Coleoptera: Curculionidae) following host removal. J. Ѐntomol. Sci. 2016, 51, 332–335. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B.J.B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Waterhouse, R.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. Bioinformatics 2019, 1962, 227–245. [Google Scholar]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.; Smyth, G. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Keeling, C.I.; Blomquist, G.J.; Tittiger, C. Coordinated gene expression for pheromone biosynthesis in the pine engraver beetle, Ips pini (Coleoptera: Scolytidae). Naturwissenschaften 2004, 91, 324–328. [Google Scholar] [CrossRef]
- Bellés, X.; Martín, D.; Piulachs, M.-D. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu. Rev. Ѐntomol. 2005, 50, 181–199. [Google Scholar] [CrossRef]
- González-Caballero, N.; Rodríguez-Vega, A.; Dias-Lopes, G.; Valenzuela, J.G.; Ribeiro, J.M.; Carvalho, P.C.; Valente, R.H.; Brazil, R.P.; Cuervo, P. Expression of the mevalonate pathway enzymes in the Lutzomyia longipalpis (Diptera: Psychodidae) sex pheromone gland demonstrated by an integrated proteomic approach. J. Proteom. 2014, 96, 117–132. [Google Scholar] [CrossRef][Green Version]
- Beedle, A.; Walton, M.; Goodwin, T. Isoprenoid biosynthesis in aseptic larvae of Calliphora erythrocephala. Insect Biochem. 1975, 5, 465–472. [Google Scholar] [CrossRef]
- Tittiger, C.; Blomquist, G. Pheromone production in pine bark beetles. Adv. Insect Physiol. 2016, 50, 235–263. [Google Scholar]
- Tillman, J.A.; Holbrook, G.L.; Dallara, P.L.; Schal, C.; Wood, D.L.; Blomquist, G.J.; Seybold, S.J. Endocrine regulation of de novo aggregation pheromone biosynthesis in the pine engraver, Ips pini (Say) (Coleoptera: Scolytidae). Insect Biochem. Mol. Biol. 1998, 28, 705–715. [Google Scholar] [CrossRef]
- Stroumbakis, N.D.; Li, Z.; Tolias, P.P. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic central nervous system. Mol. Cell. Biol. 1996, 16, 192–201. [Google Scholar] [CrossRef][Green Version]
- Goodman, C.S. The likeness of being: Phylogenetically conserved molecular mechanisms of growth cone guidance. Cell 1994, 78, 353–356. [Google Scholar] [CrossRef]
- Yamaguchi, M. The transcriptional regulation of regucalcin gene expression. Mol. Cell. Biochem. 2011, 346, 147–171. [Google Scholar] [CrossRef]
- Barrett, A.; Rawlings, N. Families and Clans of Serine Peptidases. Arch. Biochem. Biophys. 1995, 318, 247–250. [Google Scholar] [CrossRef]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar] [CrossRef]
- Perkin, L.; Elpidina, E.; Oppert, B. RNA interference and dietary inhibitors induce a similar compensation response in Tribolium castaneum larvae. Insect Mol. Biol. 2017, 26, 35–45. [Google Scholar] [CrossRef]
- Oliveira-Neto, O.B.; Batista, J.A.; Rigden, D.J.; Fragoso, R.R.; Silva, R.O.; Gomes, E.A.; Franco, O.L.; Dias, S.C.; Cordeiro, C.M.; Monnerat, R.G.; et al. A diverse family of serine proteinase genes expressed in cotton boll weevil (Anthonomus grandis): Implications for the design of pest-resistant transgenic cotton plants. Insect Biochem. Mol. Biol. 2004, 34, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.; Li, C.-F.; He, Z.; Lu, Y.; Liu, X.-S.; Wang, Y.-F.; Ip, Y.T.; Strand, M.R.; Yu, X.-Q. Toll family members bind multiple Spätzle proteins and activate antimicrobial peptide gene expression in Drosophila. J. Biol. Chem. 2019, 294, 10172–10181. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Swevers, L.; Iatrou, K.; Huvenne, H.; Smagghe, G. Bombyx mori DNA/RNA non-specific nuclease: Expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 2012, 58, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.G.; Jones, T.M. The role of chemical communication in mate choice. Biol. Rev. 2007, 82, 265–289. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.J.; Jokinen, I.; Kortet, R.; Vainikka, A.; Suhonen, J. Do pheromones reveal male immunocompetence? Proc. Biol. Sci. 2002, 269, 1681–1685. [Google Scholar] [CrossRef] [PubMed]
- Sugumaran, M. Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigment. Cell Res. 2002, 15, 2–9. [Google Scholar] [CrossRef]
- Stoehr, A. Costly melanin ornaments: The importance of taxon? Func. Ecol. 2006, 20, 276–281. [Google Scholar] [CrossRef]
- Arakane, Y.; Muthukrishnan, S.; Beeman, R.W.; Kanost, M.R.; Kramer, K.J. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. USA 2005, 102, 11337–11342. [Google Scholar] [CrossRef]
- Arakane, Y.; Lomakin, J.; Beeman, R.W.; Muthukrishnan, S.; Gehrke, S.H.; Kanost, M.R.; Kramer, K.J. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J. Biol. Chem. 2009, 284, 16584–16594. [Google Scholar] [CrossRef]
- Lukacsovich, T.; Yuge, K.; Awano, W.; Asztalos, Z.; Kondo, S.; Juni, N.; Yamamoto, D. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch. Insect Biochem. Physiol. 2003, 54, 77–94. [Google Scholar] [CrossRef] [PubMed]



| Raw sequences | 352,189,343 |
| Total transcripts | 195,128 |
| Average sequence length | 922.89 bp |
| N50 | 2040 bp |
| BUSCO | |
| Complete | 99.27% |
| Complete single copy | 97.81% |
| Complete duplicated | 1.46% |
| Fragmented | 0.59% |
| Missing | 0.15% |
| Sequence Name | Sequence Description | BLAST Top Hit | Fold Change | UP/DOWN | p-Value | FDR |
|---|---|---|---|---|---|---|
| Mevalonate pathway or associated with pheromone production and biosynthesis | ||||||
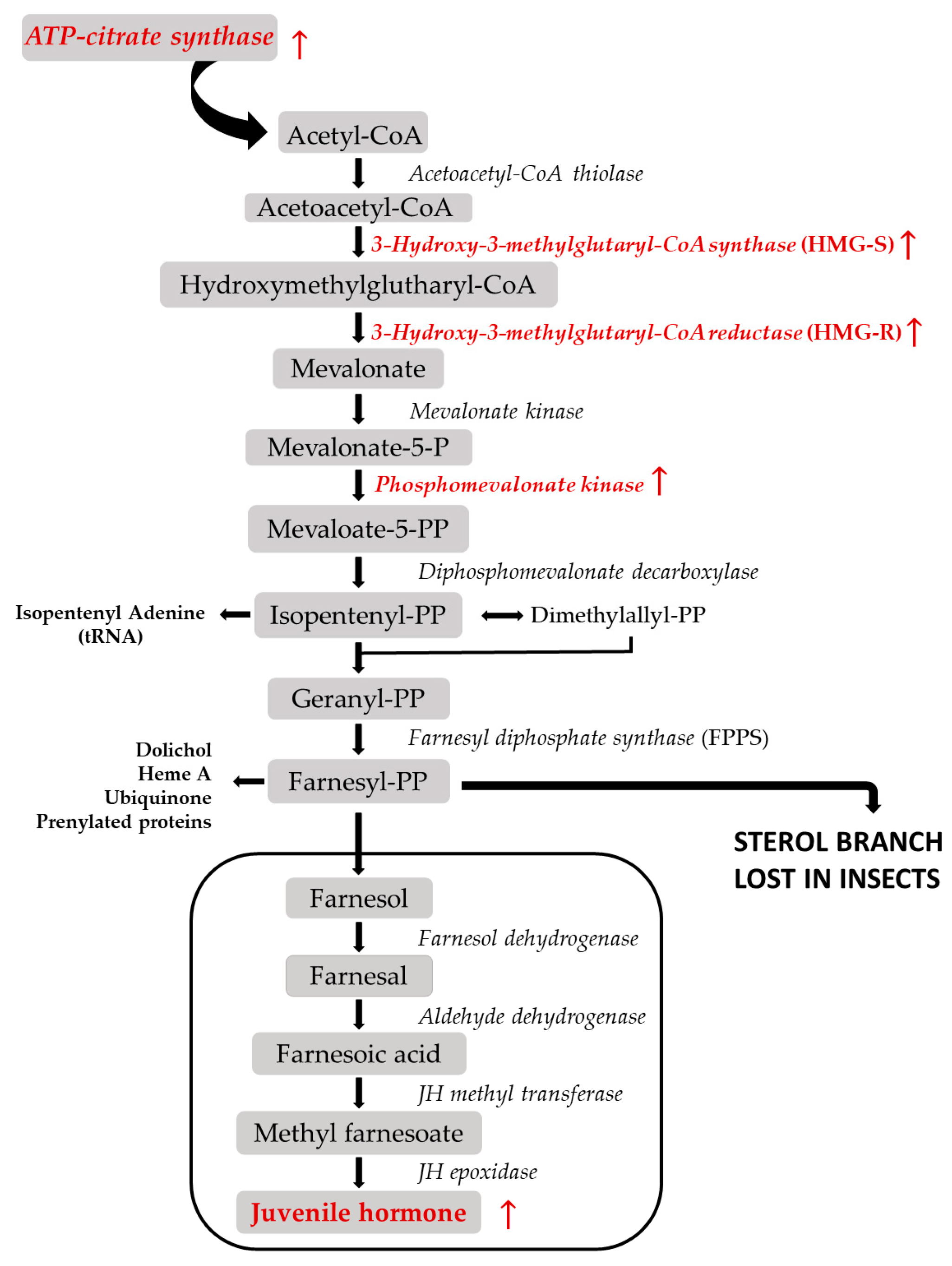
| TRINITY_DN11108 | hydroxymethylglutaryl-CoA synthase 1 (HMG-S) | D. ponderosae | 23.08 | ↑ | 1.45 × 10−37 | 2.67 × 10−34 |
| TRINITY_DN13668 | dolichyl pyrophosphate Man9GlcNAc2 α-1,3-glucosyltransferase | D. ponderosae | 2.01 | ↑ | 4.44 × 10−05 | 1.82 × 10−03 |
| TRINITY_DN5050 | farnesyl pyrophosphate synthase-like | I. pini | 11.77 | ↑ | 1.30 × 10−04 | 4.59 × 10−03 |
| TRINITY_DN5422 | phosphomevalonate kinase | D. ponderosae | 3.05 | ↑ | 9.80 × 10−12 | 1.51 × 10−09 |
| TRINITY_DN8104 | farnesyl pyrophosphate synthase | I. pini | 34.05 | ↑ | 1.10 × 10−05 | 5.33 × 10−04 |
| TRINITY_DN8499 | isopentenyl-diphosphate Delta-isomerase 1 | A. glabripennis | 37.64 | ↑ | 1.57 × 10−09 | 1.69 × 10−07 |
| TRINITY_DN873 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase-like (HMG-R) | I. paraconfusus | 16.57 | ↑ | 9.18 × 10−29 | 9.18 × 10−26 |
| TRINITY_DN4352 | ATP-citrate synthase | D. ponderosae | 6.66 | ↑ | 1.89 × 10−25 | 3.07 × 10−32 |
| Juvenile hormone | ||||||
| TRINITY_DN11992 | juvenile hormone inducible protein | D. ponderosae | 7.75 | ↑ | 7.40 × 10−13 | 1.39 × 10−10 |
| TRINITY_DN4315 | juvenile hormone esterase-like | D. ponderosae | 2.98 | ↑ | 5.14 × 10−11 | 7.24 × 10−09 |
| TRINITY_DN874 | juvenile hormone esterase-like | D. ponderosae | 2.71 | ↑ | 1.99 × 10−08 | 1.79 × 10−06 |
| TRINITY_DN4184 | juvenile hormone binding protein | R. ferrugineus | 5.64 | ↑ | 2.69 × 10−08 | 2.34 × 10−06 |
| TRINITY_DN9052 | juvenile hormone inducible protein | D. ponderosae | 4.53 | ↑ | 7.72 × 10−07 | 4.94 × 10−05 |
| TRINITY_DN115965 | putative juvenile hormone inducible protein | D. ponderosae | 3.26 | ↑ | 1.10 × 10−05 | 5.33 × 10−04 |
| TRINITY_DN11903 | juvenile hormone acid O-methyltransferase | D. ponderosae | 2.89 | ↑ | 5.87 × 10−05 | 2.31 × 10−03 |
| Fatty acid metabolism | ||||||
| TRINITY_DN58 | fatty acid synthase | D. ponderosae | 4.92 | ↑ | 2.20 × 10−26 | 1.74 × 10−23 |
| TRINITY_DN5986 | glycerol-3-phosphate phosphatase-like | S. oryzae | 7.50 | ↑ | 3.68 × 10−26 | 2.89 × 10−23 |
| Neurological and hormonal regulation | ||||||
| TRINITY_DN4411 | protein shuttle craft like | D. ponderosae | 5.38 | ↑ | 3.60 × 10−29 | 3.64 × 10−26 |
| TRINITY_DN72610 | regucalcin-like | D. ponderosae | 5.28 | ↑ | 1.45 × 10−25 | 1.11 × 10−22 |
| Sequence Name | Sequence Description | Blast Top Hit | Fold Change | UP/DOWN | p-Value | FDR |
|---|---|---|---|---|---|---|
| Peptidase activity | ||||||
| TRINITY_DN62502 | transmembrane protease serine 9-like | Z. cucurbitae | 27.18 | ↑ | 1.17 × 10−53 | 7.28 × 10−50 |
| TRINITY_DN35065 | cathepsin L1-like | R. ferrugineus | 14.42 | ↑ | 3.42 × 10−51 | 1.73 × 10−47 |
| TRINITY_DN2409 | carboxypeptidase B-like | D. ponderosae | 12.74 | ↑ | 9.29 × 10−41 | 2.28 × 10−37 |
| TRINITY_DN5051 | venom serine carboxypeptidase-like | S. oryzae | 26.91 | ↑ | 2.87 × 10−31 | 3.46 × 10−28 |
| TRINITY_DN13263 | trypsin-like serine prtoease | A. grandis | 15.38 | ↑ | 4.09 × 10−26 | 3.18 × 10−23 |
| TRINITY_DN4893 | venom serine protease-like | D. ponderosae | 10.49 | ↑ | 2.70 × 10−22 | 1.45 × 10−19 |
| TRINITY_DN34699 | trypsin alpha 3-like (Agser2p)—induced by eating | A. grandis | 3.83 | ↑ | 6.99 × 10−15 | 1.76 × 10−12 |
| TRINITY_DN35978 | brachyurin-like (Agser9p) | A. grandis | 3.36 | ↑ | 9.32 × 10−12 | 1.46 × 10−09 |
| TRINITY_DN4825 | brachyurin-like (Agser5p)—induced by eating | A. grandis | 2.26 | ↑ | 5.60 × 10−06 | 2.93 × 10−04 |
| TRINITY_DN49721 | brachyurin-like (Agser9p) | A. grandis | 3.56 | ↑ | 9.43 × 10−13 | 1.74 × 10−10 |
| TRINITY_DN63273 | trypsin alpha-3-like (Agser2p)—induced by feeding | A. grandis | 3.57 | ↑ | 2.63 × 10−13 | 5.21 × 10−11 |
| TRINITY_DN69718 | serine protease (Agser12p) | A. grandis | 2.27 | ↑ | 2.86 × 10−05 | 1.24 × 10−03 |
| Immune response | ||||||
| TRINITY_DN4819 | protein spaetzle 3 | D. ponderosae | 25.78 | ↑ | 3.30 × 10−45 | 1.11 × 10−41 |
| TRINITY_DN3567 | DNA/RNA non-specific nuclease 1 | A. grandis | 13.54 | ↑ | 3.13 × 10−39 | 6.34 × 10−36 |
| Sexual maturity | ||||||
| TRINITY_DN10824 | transcription factor Ken | 7.86 | ↓ | 1.35 × 10−44 | 4.39 × 10−41 | |
| Cuticle tanning and sclerotization | ||||||
| TRINITY_DN16013 | pupal cuticle protein 36-like | D. ponderosae | 89.53 | ↓ | 6.46 × 10−89 | 2.61 × 10−84 |
| TRINITY_DN11117 | cuticular protein 141 | D. ponderosae | 13.54 | ↓ | 1.68 × 10−68 | 2.72 × 10−64 |
| TRINITY_DN3947 | chitin deacetylase 4 | D. ponderosae | 20.48 | ↓ | 1.14 × 10−63 | 1.16 × 10−59 |
| TRINITY_DN4706 | chitin synthase 1 | A. grandis | 5.84 | ↓ | 5.48 × 10−35 | 8.37 × 10−32 |
| TRINITY_DN1999 | cuticle protein 19.8-like | S. oryzae | 15.15 | ↓ | 1.49 × 10−32 | 1.89 × 10−29 |
| TRINITY_DN4303 | cuticular protein analogous to peritrophins | D. ponderosae | 5.84 | ↓ | 1.53 × 10−32 | 1.91 × 10−29 |
| TRINITY_DN2312 | laccase 2 | A. eugenii | 18.71 | ↓ | 9.87 × 10−83 | 2.66 × 10−78 |
| TRINITY_DN22343 | dopamine D2-like receptor | S. oryzae | 4.85 | ↓ | 4.95 × 10−09 | 4.90 × 10−07 |
| TRINITY_DN6738 | dopamine N-acetyltransferase | D. ponderosae | 2.03 | ↓ | 4.40 × 10−04 | 1.30 × 10−02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perkin, L.C.; Perez, J.L.; Suh, C.P.-C. The Identification of Boll Weevil, Anthonomus grandis grandis (Coleoptera: Curculionidae), Genes Involved in Pheromone Production and Pheromone Biosynthesis. Insects 2021, 12, 893. https://doi.org/10.3390/insects12100893
Perkin LC, Perez JL, Suh CP-C. The Identification of Boll Weevil, Anthonomus grandis grandis (Coleoptera: Curculionidae), Genes Involved in Pheromone Production and Pheromone Biosynthesis. Insects. 2021; 12(10):893. https://doi.org/10.3390/insects12100893
Chicago/Turabian StylePerkin, Lindsey C., Jose L. Perez, and Charles P.-C. Suh. 2021. "The Identification of Boll Weevil, Anthonomus grandis grandis (Coleoptera: Curculionidae), Genes Involved in Pheromone Production and Pheromone Biosynthesis" Insects 12, no. 10: 893. https://doi.org/10.3390/insects12100893
APA StylePerkin, L. C., Perez, J. L., & Suh, C. P.-C. (2021). The Identification of Boll Weevil, Anthonomus grandis grandis (Coleoptera: Curculionidae), Genes Involved in Pheromone Production and Pheromone Biosynthesis. Insects, 12(10), 893. https://doi.org/10.3390/insects12100893






