A Novel Allele Specific Polymerase Chain Reaction (AS-PCR) Assay to Detect the V1016G Knockdown Resistance Mutation Confirms Its Widespread Presence in Aedes albopictus Populations from Italy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
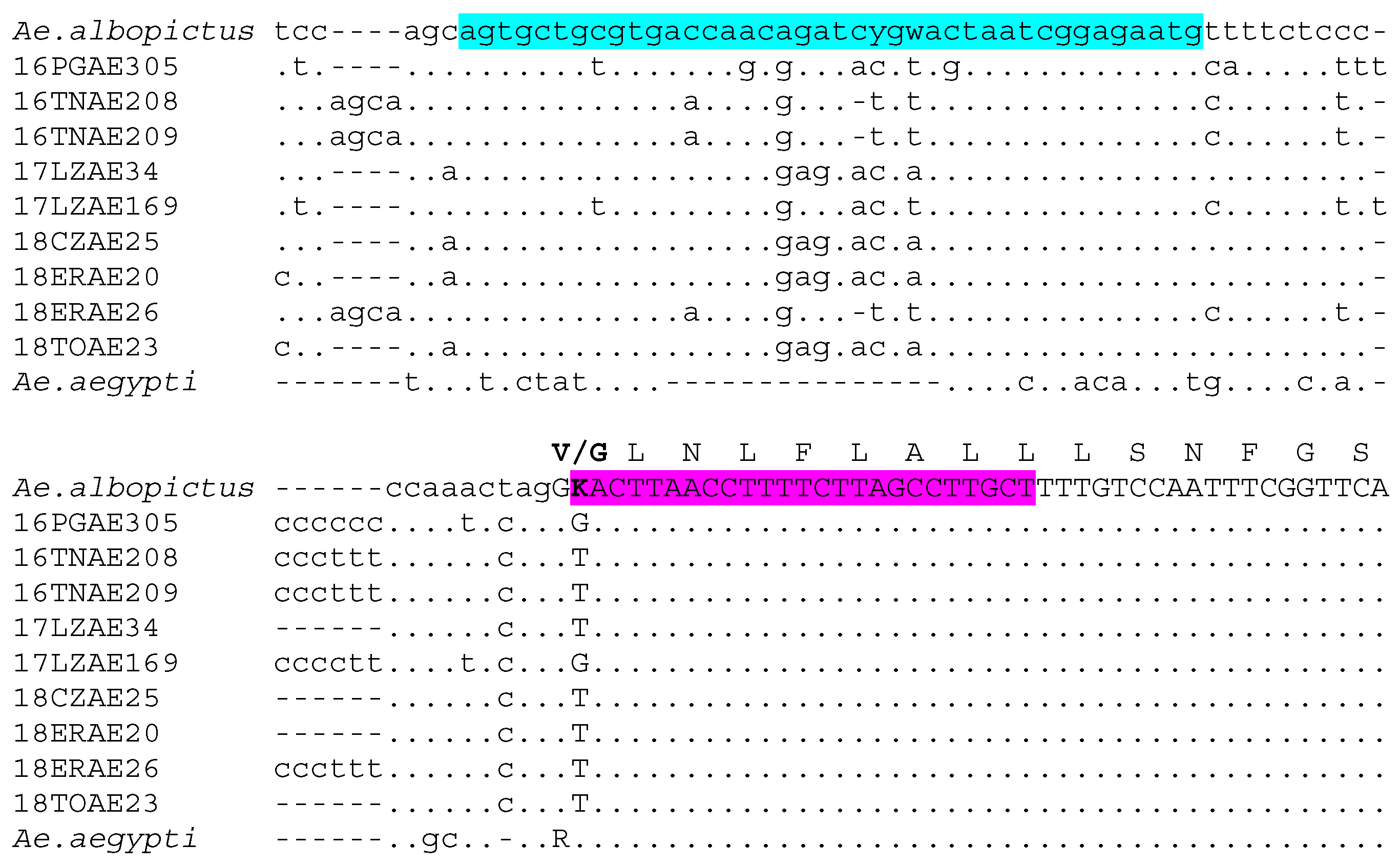
2.1. Design of Aedes Albopictus V1016G AS-PCR Assay
2.2. Validation of the Novel AS-PCR Assay on Field Samples from Italy
3. Results
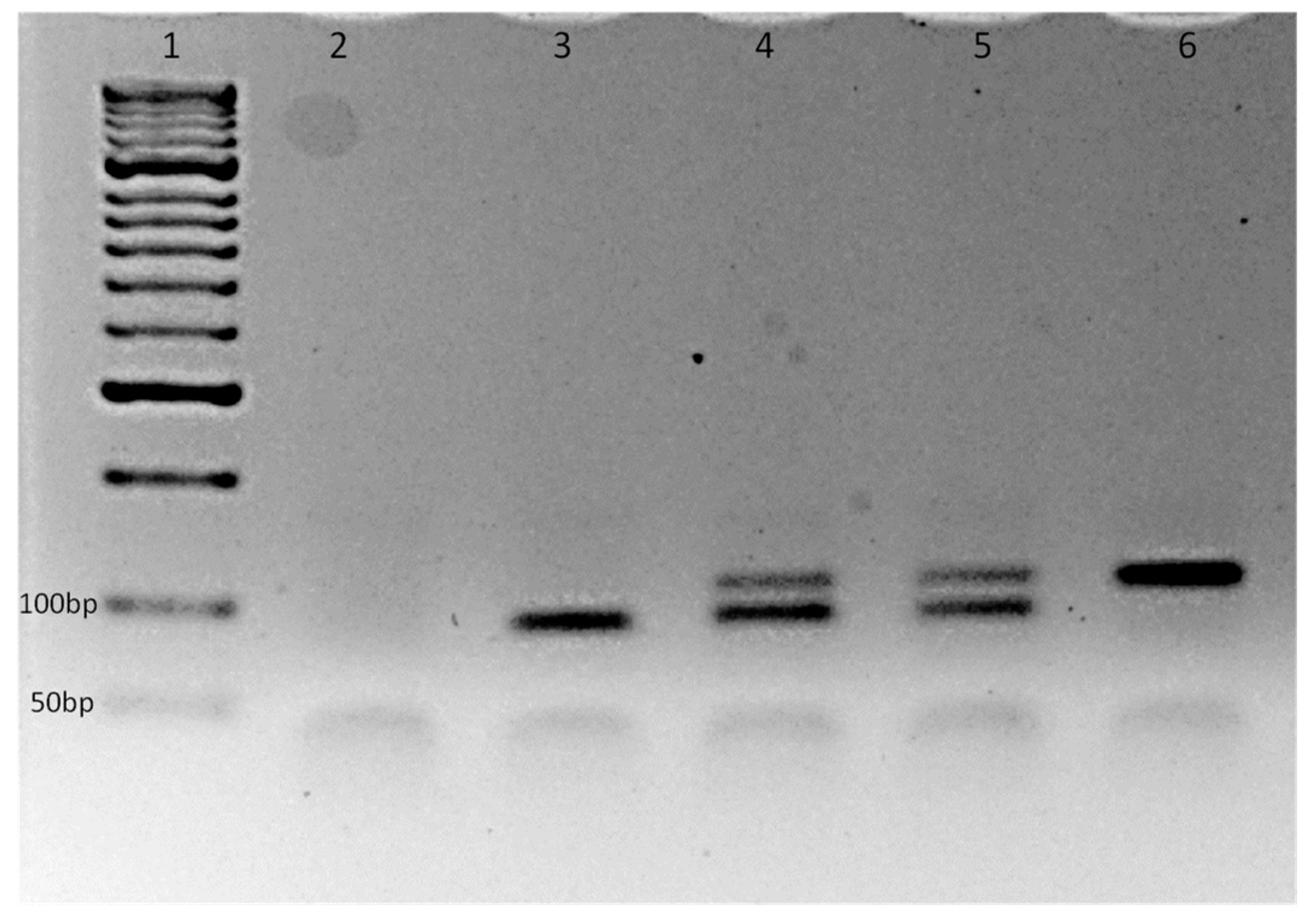
3.1. The Novel Aedes Albopictus V1016G AS-PCR Assay
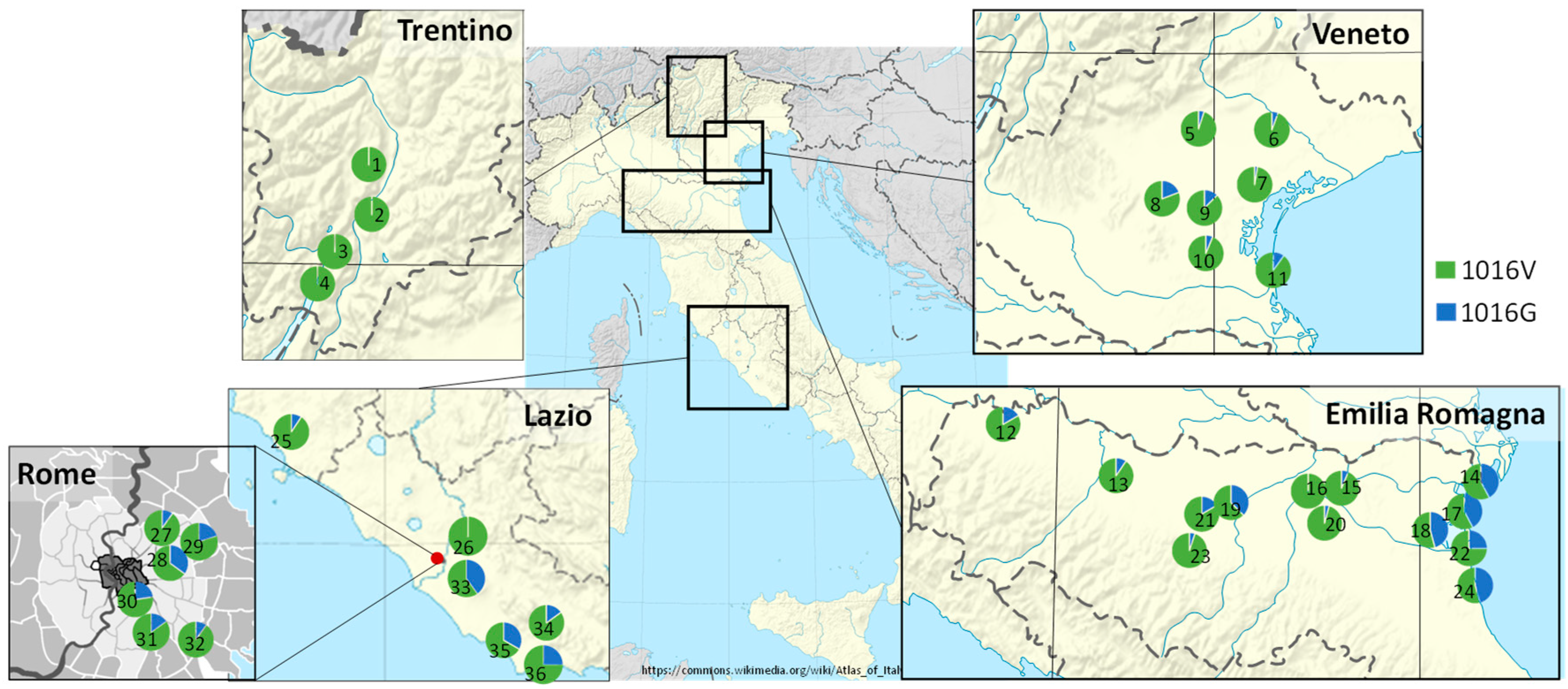
3.2. Frequency of 1016G kdr Allele in Aedes Albopictus Field Populations from Italy
4. Discussion
4.1. Aedes Albopictus V1016G AS-PCR Assay
4.2. Presence of 1016G kdr Allele in Aedes Albopictus Field Populations from Italy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Vector Control Repsonse 2017–2030; WHO: Geneva, Switzerland, 2017; p. 2030. [Google Scholar]
- WHO. A Global Brief on Vector-Borne Diseases; WHO: Geneva, Switzerland, 2014; pp. 1–56. [Google Scholar]
- Rezza, G. Aedes albopictus and the reemergence of Dengue. BMC Public Health 2012, 12, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caminade, C.; Medlock, J.M.; Ducheyne, E.; McIntyre, K.M.; Leach, S.; Baylis, M.; Morse, A.P. Suitability of European climate for the Asian tiger mosquito Aedes albopictus: Recent trends and future scenarios. J. R. Soc. Interface 2012, 9, 2708–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhold, J.M.; Lazzari, C.R.; Lahondère, C. Effects of the Environmental Temperature on Aedes aegypti and Aedes albopictus Mosquitoes: A Review. Insects 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Chen, Y.; Yan, H.; Zhang, P.; Xu, X.; Tang, B.; Zhao, P.; Ren, R. A survey of the 2014 dengue fever epidemic in Guangzhou, China. Emerg. Microbes Infect. 2015, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Gjenero-Margan, I.; Aleraj, B.; Krajcar, D.; Lesnikar, V.; Klobučar, A.; Pem-Novosel, I.; Kurečić-Filipović, S.; Komparak, S.; Martićз, R.; Duričić, S.; et al. Autochthonous dengue fever in Croatia, August-September 2010. Euro Surveill. 2011, 16, 19805. [Google Scholar] [PubMed]
- Succo, T.; Leparc-Goffart, I.; Ferré, J.; Roiz, D.; Broche, B.; Maquart, M.; Noel, H.; Catelinois, O.; Entezam, F.; Caire, D.; et al. Autochthonous dengue outbreak in Nimes, South of France, July to September 2015. Eurosurveillance 2016, 21, 1–7. [Google Scholar] [CrossRef]
- Lazzarini, L.; Barzon, L.; Foglia, F.; Manfrin, V.; Pacenti, M.; Pavan, G.; Rassu, M.; Montarsi, F.; Martini, S.; Zanella, F.; et al. First autochthonous dengue outbreak in Italy, August 2020. Eurosurveillance 2020, 1, 8–11. [Google Scholar]
- Tomasello, D.; Schlagenhauf, P. Chikungunya and dengue autochthonous cases in Europe, 2007–2012. Travel Med. Infect. Dis. 2013, 11, 274–284. [Google Scholar] [CrossRef]
- ECDC. Autochthonous Cases of Dengue in Spain and France; ECDC: Stockholm, Sweden, 2019; pp. 1–8. [Google Scholar]
- Angelini, R.; Finarelli, A.C.; Angelini, P.; Po, C.; Petropulacos, K.; Macini, P.; Fiorentini, C.; Fortuna, C.; Venturi, G.; Romi, R.; et al. Un’epidemia di febbre chikungunya nella provincia di Ravenna. Eurosurveillance 2007, 12. [Google Scholar] [CrossRef]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; et al. Infection with chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- Venturi, G.; Di Luca, M.; Fortuna, C.; Remoli, M.E.; Riccardo, F.; Severini, F.; Toma, L.; Del Manso, M.; Benedetti, E.; Caporali, M.G.; et al. Detection of a chikungunya outbreak in Central Italy, August to September 2017. Eurosurveillance 2017, 22, 17–00646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldacchino, F.; Caputo, B.; Chandre, F.; Drago, A.; della Torre, A.; Montarsi, F.; Rizzoli, A. Control methods against invasive Aedes mosquitoes in Europe: A review. Pest Manag. Sci. 2015, 71, 1471–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vontas, J.; Kioulos, E.; Pavlidi, N.; Morou, E.; Della Torre, A.; Ranson, H. Insecticide resistance in the major dengue vectors Aedes albopictus and Aedes aegypti. Pestic. Biochem. Physiol. 2012, 104, 126–131. [Google Scholar] [CrossRef]
- Ranson, H.; Abdallah, H.; Badolo, A.; Guelbeogo, W.M.; Kerah-Hinzoumbé, C.; Yangalbé-Kalnoné, E.; Sagnon, N.; Simard, F.; Coetzee, M. Insecticide resistance in Anopheles gambiae: Data from the first year of a multi-country study highlight the extent of the problem. Malar. J. 2009, 8, 299. [Google Scholar] [CrossRef] [Green Version]
- Moyes, C.L.; Vontas, J.; Martins, A.J.; Ng, L.C.; Koou, S.Y.; Dusfour, I.; Raghavendra, K.; Pinto, J.; Corbel, V.; David, J.-P.; et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 2017, 7, e0005625. [Google Scholar] [CrossRef]
- Fonseca, D.M.; Unlu, I.; Crepeau, T.; Farajollahi, A.; Healy, S.P.; Bartlett-Healy, K.; Strickman, D.; Gaugler, R.; Hamilton, G.; Kline, D.; et al. Area-wide management of Aedes albopictus. Part 2: Gauging the efficacy of traditional integrated pest control measures against urban container mosquitoes. Pest Manag. Sci. 2013, 69, 1351–1361. [Google Scholar] [CrossRef]
- Manica, M.; Cobre, P.; Rosà, R.; Caputo, B. Not in my backyard: Effectiveness of outdoor residual spraying from and-held sprayers against the mosquito Aedes albopictus in Rome, Italy. Pest Manag. Sci. 2017, 73, 138–145. [Google Scholar] [CrossRef]
- Carrieri, M.; Bellini, R.; MacCaferri, S.; Gallo, L.; Maini, S.; Celli, G. Tolerance thresholds for Aedes albopictus and Aedes caspius in Italian Urban areas. J. Am. Mosq. Control Assoc. 2008, 24, 377–386. [Google Scholar] [CrossRef]
- Diabate, A.; Brengues, C.; Baldet, T.; Dabiré, K.R.; Hougard, J.M.; Akogbeto, M.; Kengne, P.; Simard, F.; Guillet, P.; Hemingway, J.; et al. The spread of the Leu-Phe kdr mutation through Anopheles gambiae complex in Burkina Faso: Genetic introgression and de novo phenomena. Trop. Med. Int. Health 2004, 9, 1267–1273. [Google Scholar] [CrossRef]
- Soderlund, D.M. Molecular mechanisms of pyrethroid insecticide neurotoxicity: Recent advances. Arch. Toxicol. 2012, 86, 165–181. [Google Scholar] [CrossRef] [Green Version]
- IRAC. Prevention and Management of Insecticide Resistance in Vectors of Public Health Importance; Insecticide Resistance Action Committee: Brussels, Belgium, 2011. [Google Scholar]
- Hemingway, J.; Ranson, H. Insecticide Resistance in Insect Vectors of Human Disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Nikou, D.; Vontas, J.; Donnelly, M.J.; Williamson, M.S.; Field, L.M. The Vector Population Monitoring Tool (VPMT): High-Throughput DNA-Based Diagnostics for the Monitoring of Mosquito Vector Populations. Malar. Res. Treat. 2010, 2010, 190434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenhouse, S.A.; Plernsub, S.; Yanola, J.; Lumjuan, N.; Dantrakool, A. Detection of the V1016G mutation in the voltage-gated sodium channel gene of Aedes aegypti (Diptera: Culicidae) by allele-specific PCR assay, and its distribution and effect on deltamethrin resistance in Thailand. Parasites Vectors 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saavedra-Rodriguez, K.; Urdaneta-Marquez, L.; Rajatileka, S.; Moulton, M.; Flores, A.E.; Fernandez-Salas, I.; Bisset, J.; Rodriguez, M.; Mccall, P.J.; Donnelly, M.J.; et al. A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol. Biol. 2007, 16, 785–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuaycharoensuk, T.; Juntarajumnong, W.; Boonyuan, W.; Bangs, M.J.; Akratanakul, P.; Thammapalo, S.; Jirakanjanakit, N.; Tanasinchayakul, S.; Chareonviriyaphap, T. Frequency of pyrethroid resistance in Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in Thailand. J. Vector Ecol. 2011, 36, 204–212. [Google Scholar] [CrossRef]
- Ishak, I.H.; Kamgang, B.; Ibrahim, S.S.; Riveron, J.M.; Irving, H.; Wondji, C.S. Pyrethroid Resistance in Malaysian Populations of Dengue Vector Aedes aegypti Is Mediated by CYP9 Family of Cytochrome P450 Genes. PLoS Negl. Trop. Dis. 2017, 11, e0005302. [Google Scholar] [CrossRef] [Green Version]
- Ngoagouni, C.; Kamgang, B.; Brengues, C.; Yahouedo, G.; Paupy, C.; Nakouné, E.; Kazanji, M.; Chandre, F. Susceptibility profile and metabolic mechanisms involved in Aedes aegypti and Aedes albopictus resistant to DDT and deltamethrin in the Central African Republic. Parasites Vectors 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Lee RM, L.; Choong CT, H.; Goh, B.P.L.; Ng, L.C.; Lam-Phua, S.G. Bioassay and biochemical studies of the status of pirimiphos-methyl and cypermethrin resistance in Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus (Diptera: Culicidae) in Singapore. Trop. Biomed. 2014, 31, 670–679. [Google Scholar]
- Thanispong, K.; Sathantriphop, S.; Malaithong, N.; Bangs, M.J.; Chareonviriyaphap, T. Establishment of Diagnostic Doses of Five Pyrethroids for Monitoring Physiological Resistance in Aedes albopictus in Thailand. J. Am. Mosq. Control Assoc. 2015, 31, 346–352. [Google Scholar] [CrossRef]
- Chen, H.; Li, K.; Wang, X.; Yang, X.; Lin, Y.; Cai, F.; Zhong, W.; Lin, C.; Lin, Z.; Ma, Y. First identification of kdr allele F1534S in VGSC gene and its association with resistance to pyrethroid insecticides in Aedes albopictus populations from Haikou City, Hainan Island, China. Infect. Dis. Poverty 2016, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Liu, L.; Cheng, P.; Yang, L.; Chen, J.; Lu, Y.; Wang, H.; Chen, X.G.; Gong, M. Bionomics and insecticide resistance of Aedes albopictus in Shandong, a high latitude and high-risk dengue transmission area in China. Parasites Vectors 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Chen, H.M.; Shi, H.; Peng, H.; Ma, Y.J. Correlation between adult pyrethroid resistance and knockdown resistance (kdr) mutations in Aedes albopictus (Diptera: Culicidae) field populations in China. Infect. Dis. Poverty 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Guo, Y.; Deng, J.; Xu, J.; Zhou, G.; Zhou, T.; Li, Y.; Zhong, D.; Kong, L.; Wang, X.; et al. Fast emerging insecticide resistance in Aedes albopictus in Guangzhou, China: Alarm to the dengue epidemic. PLoS Negl. Trop. Dis. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Kamgang, B.; Marcombe, S.; Chandre, F.; Nchoutpouen, E.; Nwane, P.; Etang, J.; Corbel, V.; Paupy, C. Insecticide susceptibility of Aedes aegypti and Aedes albopictus in Central Africa. Parasites Vectors 2011, 4, 79. [Google Scholar] [CrossRef] [Green Version]
- Kamgang, B.; Yougang, A.P.; Tchoupo, M.; Riveron, J.M.; Wondji, C. Temporal distribution and insecticide resistance profile of two major arbovirus vectors Aedes aegypti and Aedes albopictus in Yaoundé, the capital city of Cameroon. Parasites Vectors 2017, 10. [Google Scholar] [CrossRef]
- Bengoa, M.; Eritja, R.; Delacour, S.; Miranda, M.Á.; Sureda, A.; Lucientes, J. First data on resistance to pyrethroids in wild populations of Aedes albopictus from Spain. J. Am. Mosq. Control Assoc. 2017, 33, 246–249. [Google Scholar] [CrossRef]
- Pichler, V.; Bellini, R.; Veronesi, R.; Arnoldi, D.; Rizzoli, A.; Lia, R.P.; Otranto, D.; Montarsi, F.; Carlin, S.; Ballardini, M.; et al. First evidence of resistance to pyrethroid insecticides in Italian Aedes albopictus populations 26 years after invasion. Pest Manag. Sci. 2018, 74, 1319–1327. [Google Scholar] [CrossRef] [Green Version]
- Pichler, V.; Malandruccolo, C.; Paola, S.; Bellini, R.; Severini, F.; Toma, L.; Di Luca, M.; Montarsi, F.; Ballardini, M.; Manica, M.; et al. Phenotypic and genotypic pyrethroid resistance of Aedes albopictus, with focus on the 2017 chikungunya outbreak in Italy. Pest Manag. Sci. 2019. [Google Scholar] [CrossRef]
- Richards, S.L.; Balanay JA, G.; Fields, M.; Vandock, K. Baseline Insecticide Susceptibility Screening Against Six Active Ingredients for Culex and Aedes (Diptera: Culicidae) Mosquitoes in the United States. J. Med. Entomol. 2017, 54, 1–14. [Google Scholar] [CrossRef]
- Richards, S.L.; Anne, J.; Balanay, G.; White, A.V.; Hope, J.; Vandock, K.; Byrd, B.D.; Reiskind, M.H. Insecticide Susceptibility Screening Against Culex and Aedes (Diptera: Culicidae) Mosquitoes From the United States. J. Med. Entomol. 2018, 55, 398–407. [Google Scholar] [CrossRef]
- Balaska, S.; Fotakis, E.A.; Kioulos, I.; Grigoraki, L.; Mpellou, S.; Chaskopoulou, A.; Vontas, J. Bioassay and molecular monitoring of insecticide resistance status in Aedes albopictus populations from Greece, to support evidence based vector control. Parasites Vectors 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Bonizzoni, M.; Zhong, D.; Zhou, G.; Cai, S.; Yan, G.; Chen, X. Multi-country Survey Revealed Prevalent and Novel F1534S Mutation in Voltage-Gated Sodium Channel (VGSC) Gene in Aedes albopictus. PLoS Negl. Trop. Dis. 2016, 10, e0004696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasai, S.; Ng, L.C.; Lam-phua, S.G.; Tang, C.S. First Detection of a Putative Knockdown Resistance Gene in Major Mosquito Vector, Aedes albopictus. Jpn. J. Infect. Dis. 2011, 64, 217–221. [Google Scholar] [PubMed]
- Marcombe, S.; Farajollahi, A.; Healy, S.P.; Clark, G.G.; Fonseca, D.M. Insecticide resistance status of United States populations of Aedes albopictus and mechanisms involved. PLoS ONE 2014, 9, e101992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasai, S.; Caputo, B.; Tsunoda, T.; Cuong, T.C.; Maekawa, Y.; Lam-phua, S.G.; Pichler, V.; Itokawa, K.; Murota, K.; Komagata, O.; et al. Eurosurveillance First detection of a Vssc allele V1016G conferring a high level of insecticide resistance in Aedes albopictus collected from Europe (Italy) and Asia (Vietnam), 2016: A new emerging threat to controlling arboviral diseases. Eurosurveillance 2019, 24, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tancredi, A.; Papandrea, D.; Marconcini, M.; Carballar-Lejarazu, R.; Casas-Martinez, M.; Lo, E.; Chen, X.; Malacrida, R.; Bonizzoni, M. Tracing temporal and geographic distribution of resistance to pyrethroids in the arboviral vector Aedes albopictus. PloS Negl. Trop. Dis. 2020, 14, e0008350. [Google Scholar] [CrossRef]
- Zhu, C.Y.; Zhao, C.C.; Wang, Y.G.; Ma, D.L.; Song, X.P.; Wang, J. Establishment of an innovative and sustainable PCR technique for 1534 locus mutation of the knockdown resistance (kdr) gene in the dengue vector Aedes albopictus. Parasites Vectors 2019, 12, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yang, C.; Liu, N.; Li, M.; Tong, Y.; Zeng, X.; Qiu, X. Knockdown resistance (kdr) mutations within seventeen field populations of Aedes albopictus from Beijing China: First report of a novel V1016G mutation and evolutionary origins of kdr haplotypes. Parasites Vectors 2019, 12, 1–16. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Giraldo-Calderón, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Emrich, S.; Collins, F.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; et al. VectorBase: An updated Bioinformatics Resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef]
- Papp, A.C.; Pinsonneault, J.K.; Cooke, G.; Sadée, W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques 2003, 34, 1068–1072. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.C.; Berninger, M.S.; Hartley, J.L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 1990, 93, 125–128. [Google Scholar] [CrossRef]
- Rider, M.A.; Byrd, B.D.; Keating, J.; Wesson, D.M.; Caillouet, K.A. PCR detection of malaria parasites in desiccated Anopheles mosquitoes is uninhibited by storage time and temperature. Malar. J. 2012, 11, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imyanitov, E.N.; Buslov, K.G.; Suspitsin, E.N.; Kuligina, E.S.; Belogubova, E.V.; Grigoriev, M.Y.; Togo, A.V.; Hanson, K.P. Improved reliability of allele-specific PCR. Biotechniques 2002, 33, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Pichler, V.; Kotsakiozi, P.; Caputo, B.; Serini, P.; Caccone, A.; Della Torre, A. Complex interplay of evolutionary forces shaping population genomic structure of invasive Aedes albopictus in southern Europe. PLoS Negl. Trop. Dis. 2019, 13, e0007554. [Google Scholar] [CrossRef]
- Manni, M.; Guglielmino, C.R.; Scolari, F.; Vega-Rùa, A.; Failloux, A.B.; Somboon, P.; Lisa, A.; Savini, G.; Bonizzoni, M.; Gomulski, L.M.; et al. Genetic evidence for a worldwide chaotic dispersion pattern of the arbovirus vector, Aedes albopictus. PLoS Negl. Trop. Dis. 2017, 11, e0005332. [Google Scholar] [CrossRef]
- Battaglia, V.; Gabrieli, P.; Brandini, S.; Capodiferro, M.R.; Javier, P.A.; Chen, X.G.; Achilli, A.; Semino, O.; Gomulski, L.M.; Malacrida, A.R.; et al. The worldwide spread of the tiger mosquito as revealed by mitogenome haplogroup diversity. Front. Genet. 2016, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, R.; Gentile, G.; Carrieri, M.; Maccagnani, B.; Stermieri, L.; Bellini, R. Seasonal pattern of daily activity of Aedes caspius, Aedes detritus, Culex modestus, and Culex pipiens in the Po Delta of northern Italy and significance for vector-borne disease risk assessment. J. Vector Ecol. 2012, 37, 49–61. [Google Scholar] [CrossRef]
- Martinez-Torres, D.E.A. Molecular characterization of pyrethroid knockdown (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol. Biol. 1998, 7, 179–184. [Google Scholar] [CrossRef] [Green Version]
| Site | Coordinates | Sampling Year | Treatment | N | Genotype Frequency | Freq. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lat | Long | GG | VG | VV | 1016G | ||||||
| Trentino | 1 | Mezzolombardo | 46°12′44.03″ N | 11°5′45.77″ E | 2020 | NA | 10 | - | - | 1.000 | 0.000 |
| 2 | Trento | 46°4′8.56″ N | 11°7′16.98″ E | 2020 | NA | 30 | - | - | 1.000 | 0.000 | |
| 3 | Arco | 45°55′2.49″ N | 10°54′36.52″ E | 2020 | NA | 10 | - | - | 1.000 | 0.000 | |
| 4 | Riva del Garda | 45°51′51.46″ N | 10°50′37.80″ E | 2020 | NA | 10 | - | - | 1.000 | 0.000 | |
| Total | 60 | - | - | 1.000 | 0.000 | ||||||
| Veneto | 5 | Castel Franco | 45°40′56.19″ N | 11°55′23.99″ E | 2019 | NA | 20 | - | 0.100 | 0.900 | 0.050 |
| 6 | Treviso | 45°39′41.48″ N | 12°15′32.61″ E | 2019 | Low | 18 | - | 0.111 | 0.889 | 0.056 | |
| 7 | Mestre | 45°28′12.36″ N | 12°13′28.64″ E | 2019 | NA | 20 | - | 0.050 | 0.950 | 0.025 | |
| 8 | Padova | 45°23′58.41″ N | 11°50′20.42″ E | 2019 | High | 13 | - | 0.385 | 0.615 | 0.192 | |
| 9 | Legnaro | 45°21′16.09″ N | 11°57′4.04″ E | 2019 | Low | 12 | - | 0.250 | 0.750 | 0.125 | |
| 10 | Brugine | 45°17′5.82″ N | 11°59′52.53″ E | 2019 | Low | 16 | - | 0.125 | 0.875 | 0.063 | |
| 11 | Chioggia | 45°12′13.93″ N | 12°17′16.21″ E | 2019 | High | 20 | - | 0.200 | 0.800 | 0.100 | |
| Total | 119 | - | 0.160 | 0.840 | 0.080 | ||||||
| Emilia Romagna | 12 | Piacenza | 45°03′08″ N | 9°41′36″ E | 2019 | Low | 12 | - | 0.333 | 0.667 | 0.167 |
| 13 | Parma | 44°47′57″ N | 10°19′34″ E | 2019 | low | 21 | - | 0.190 | 0.810 | 0.095 | |
| 14 | Lido di Volano | 44°47′44″ N | 12°15′46″ E | 2019 | Yes | 13 | - | 0.846 | 0.154 | 0.423 | |
| 15 | Malalbergo | 44°43′09.17″ N | 11°31′53.54″ E | 2019 | High | 15 | - | 0.133 | 0.867 | 0.067 | |
| 16 | Ponticelli | 44°41′55.15″ N | 11°28′24.20″ E | 2019 | Low | 16 | - | - | 1.000 | 0.000 | |
| 17 | Comacchio | 44°41′41″ N | 12°10′54″ E | 2019 | High | 12 | - | 0.833 | 0.167 | 0.417 | |
| 18 | Porto Garibaldi | 44°40′40″ N | 12°14′40″ E | 2019 | High | 12 | - | 0.917 | 0.083 | 0.458 | |
| 19 | Modena Nord | 44°40′0.08″ N | 10°54′47.42″ E | 2019 | Low | 12 | 0.083 | 0.583 | 0.333 | 0.375 | |
| 20 | Altedo | 44°39′48.35″ N | 11°30′11.02″ E | 2019 | Low | 12 | - | 0.083 | 0.917 | 0.042 | |
| 21 | Modena Nord-Ovest | 44°39′25.72″ N | 10°56′54.87″ E | 2019 | Low | 12 | - | 0.333 | 0.667 | 0.167 | |
| 22 | Lido di Spina | 44°39′05″ N | 12°14′56″ E | 2019 | High | 12 | 0.083 | 0.333 | 0.583 | 0.250 | |
| 23 | Maranello | 44°31′51″ N | 10°52′07″ E | 2019 | Low | 10 | - | 0.100 | 0.900 | 0.050 | |
| 24 | Marina Romea | 44°29′00″ N | 12°16′00″ E | 2019 | High | 11 | 0.182 | 0.545 | 0.273 | 0.455 | |
| Total | 170 | 0.024 | 0.382 | 0.594 | 0.215 | ||||||
| Toscana | 25 | Grosseto | 42°45′23.8″ N | 11°05′53.7″ E | 2020 | NA | 16 | - | 0.187 | 0.813 | 0.094 |
| Total | 16 | - | 0.187 | 0.813 | 0.094 | ||||||
| Lazio | 26 | Guidonia | 41°56′04.75″ N | 12°40′00.02″ E | 2020 | NA | 10 | - | - | 1.000 | 0.000 |
| 27 | Roma Pertini | 41°55′17.14″ N | 12°32′30.11″ E | 2020 | NA | 10 | - | 0.200 | 0.800 | 0.100 | |
| 28 | Roma-Villa Mirafiori | 41°55′08.1″ N | 12°31′02″ E | 2020 | NA | 10 | 0.100 | 0.500 | 0.400 | 0.350 | |
| 29 | Roma Pietralata | 41°55′02.22″ N | 12°33′18.83″ E | 2020 | NA | 10 | - | 0.400 | 0.600 | 0.200 | |
| 30 | Roma CTO | 41°51′29.82″ N | 12°29′13.85″ E | 2020 | NA | 11 | - | 0.455 | 0.545 | 0.227 | |
| 31 | Roma Appio Latino | 41°51′14.51″ N | 12°30′15.85″ E | 2020 | NA | 10 | 0.100 | 0.100 | 0.800 | 0.150 | |
| 32 | Roma-Appia Pignatelli | 41°50′48.20″ N | 12°32′55.40″ E | 2020 | NA | 10 | - | 0.200 | 0.800 | 0.100 | |
| 33 | Ciampino | 41°47′56,24″ N | 12°36′16,54″ E | 2020 | NA | 10 | 0.300 | 0.200 | 0.500 | 0.400 | |
| 34 | Fondi | 41°21′26.51″ N | 13°25′37.70″ E | 2020 | NA | 10 | - | 0.300 | 0.700 | 0.150 | |
| 35 | Sabaudia | 41°18′07.56″ N | 13°01′25.59″ E | 2020 | NA | 9 | 0.111 | 0.444 | 0.444 | 0.333 | |
| 36 | Terracina | 41°17′05.45″ N | 13°14′44.33″ E | 2020 | NA | 14 | - | 0.500 | 0.500 | 0.250 | |
| Total | 114 | 0.053 | 0.307 | 0.640 | 0.206 | ||||||
| TOTAL | 479 | 0.021 | 0.255 | 0.724 | 0.148 | ||||||
| AS-PCR | |||||
|---|---|---|---|---|---|
| GG | VG | VV | TOT | ||
| Sequencing | GG | 5 | 5 | ||
| VG | 1 | 13 | 14 | ||
| VV | 1 | 19 | 20 | ||
| TOT | 6 | 14 | 19 | 39 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pichler, V.; Mancini, E.; Micocci, M.; Calzetta, M.; Arnoldi, D.; Rizzoli, A.; Lencioni, V.; Paoli, F.; Bellini, R.; Veronesi, R.; et al. A Novel Allele Specific Polymerase Chain Reaction (AS-PCR) Assay to Detect the V1016G Knockdown Resistance Mutation Confirms Its Widespread Presence in Aedes albopictus Populations from Italy. Insects 2021, 12, 79. https://doi.org/10.3390/insects12010079
Pichler V, Mancini E, Micocci M, Calzetta M, Arnoldi D, Rizzoli A, Lencioni V, Paoli F, Bellini R, Veronesi R, et al. A Novel Allele Specific Polymerase Chain Reaction (AS-PCR) Assay to Detect the V1016G Knockdown Resistance Mutation Confirms Its Widespread Presence in Aedes albopictus Populations from Italy. Insects. 2021; 12(1):79. https://doi.org/10.3390/insects12010079
Chicago/Turabian StylePichler, Verena, Emiliano Mancini, Martina Micocci, Maria Calzetta, Daniele Arnoldi, Annapaola Rizzoli, Valeria Lencioni, Francesca Paoli, Romeo Bellini, Rodolfo Veronesi, and et al. 2021. "A Novel Allele Specific Polymerase Chain Reaction (AS-PCR) Assay to Detect the V1016G Knockdown Resistance Mutation Confirms Its Widespread Presence in Aedes albopictus Populations from Italy" Insects 12, no. 1: 79. https://doi.org/10.3390/insects12010079
APA StylePichler, V., Mancini, E., Micocci, M., Calzetta, M., Arnoldi, D., Rizzoli, A., Lencioni, V., Paoli, F., Bellini, R., Veronesi, R., Martini, S., Drago, A., De Liberato, C., Ermenegildi, A., Pinto, J., della Torre, A., & Caputo, B. (2021). A Novel Allele Specific Polymerase Chain Reaction (AS-PCR) Assay to Detect the V1016G Knockdown Resistance Mutation Confirms Its Widespread Presence in Aedes albopictus Populations from Italy. Insects, 12(1), 79. https://doi.org/10.3390/insects12010079









