Click Beetle Mitogenomics with the Definition of a New Subfamily Hapatesinae from Australasia (Coleoptera: Elateridae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Structure of Mitochondrial Genomes
3.2. Analyses of Compositional Heterogeneity, Sequence Heterogeneity, and Sequence Completeness
3.3. Phylogenetic Relationships
3.4. Taxonomy
Hapatesinae, New Subfamily
4. Discussion
4.1. Mitogenomics of Elateridae
4.2. Phylogenetic Relationships of Click Beetle Subfamilies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, C.; Lawrence, J.F.; Rosa, S.P. Elateridae Leach, 1815. In Handbook of Zoology, Vol. 2, Coleoptera, Beetles; Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Walter de Gruyter GmbH & Co.: Berlin, Germany; New York, NY, USA, 2010; pp. 75–103. [Google Scholar]
- Bocakova, M.; Bocak, L.; Hunt, T.; Teräväinen, M.; Vogler, A.P. Molecular phylogenetics of Elateriformia (Coleoptera): Evolution of bioluminescence and neoteny. Cladistics 2007, 23, 477–496. [Google Scholar] [CrossRef]
- Ribak, G.; Weihs, D. Jumping without Using Legs: The jump of the click-beetles (Elateridae) is morphologically constrained. PLoS ONE 2011, 6, e20871. [Google Scholar] [CrossRef] [PubMed]
- Bolmin, O.; Wei, L.H.; Hazel, A.M.; Dunn, A.C.; Wissa, A.; Alleyne, M. Latching of the click beetle (Coleoptera: Elateridae) thoracic hinge enabled by the morphology and mechanics of conformal structures. J. Exp. Biol. 2019, 222, jeb196683. [Google Scholar] [CrossRef] [PubMed]
- Whalley, P.E.S. The systematics and palaeogeography of the lower Jurassic insects of Dorset, England. Bull. Brit. Mus. Nat. Hist. 1985, 39, 107–189. [Google Scholar]
- Doludenko, M.P.; Ponomarenko, A.G.; Sakulina, G.V. La Géologie du Gisement Unique de la Faune et de la Flore du Jurassique Supérieur d’Aulié (Karatau, Kazakhstan du Sud); Académie des Sciences de l’URSS, Institut Géologique: Moscow, Russia, 1990. [Google Scholar]
- Chang, H.L.; Kirejtshuk, A.G.; Ren, D.; Shih, C.K. First fossil click beetles from the Middle Jurassic of Inner Mongolia, China (Coleoptera: Elateridae). Ann. Zool. 2009, 59, 7–14. [Google Scholar] [CrossRef]
- Sohn, J.C.; Nam, G.S.; Choi, S.W.; Ren, D. New fossils of Elateridae (Insecta, Coleoptera) from Early Cretaceous Jinju Formation (South Korea) with their implications to evolutionary diversity of extinct Protagrypninae. PLoS ONE 2019, 14, e0225502. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Ślipiński, S.A.; et al. Family-group names in Coleoptera (Insecta). Zookeys 2011, 88, 1–972. [Google Scholar] [CrossRef] [PubMed]
- Douglas, H. Phylogenetic relationships of Elateridae inferred from adult morphology, with special reference to the position of Cardiophorinae. Zootaxa 2011, 2900, 1–45. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Ślipiński, S.A.; Seago, A.E.; Thayer, M.K.; Newton, A.F.; Marvaldi, A.E. Phylogeny of the Coleoptera based on morphological characters of adults and larvae. Ann. Zool. 2011, 61, 1–217. [Google Scholar] [CrossRef]
- Crowson, R.A. A review of the classification of Cantharoidea (Coleoptera), with definition of two new families Cneoglossidae and Omethidae. Rev. Univ. Madrid 1972, 21, 35–77. [Google Scholar]
- Kundrata, R.; Bocak, L. The phylogeny and limits of Elateridae (Insecta, Coleoptera): Is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool. Scr. 2011, 40, 364–378. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocakova, M.; Bocak, L. The comprehensive phylogeny of the superfamily Elateroidea (Coleoptera: Elateriformia). Mol. Phylogenet. Evol. 2014, 76, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Bocak, L.; Motyka, M.; Bocek, M.; Bocakova, M. Incomplete sclerotization and phylogeny: The phylogenetic classification of Plastocerus (Coleoptera: Elateroidea). PLoS ONE 2018, 13, e0194026. [Google Scholar] [CrossRef] [PubMed]
- Sagegami-Oba, R.; Oba, Y.; Ôhira, H. Phylogenetic relationships of click-beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: Insights into the evolution of bioluminescence in Elateridae. Mol. Phylogenet. Evol. 2007, 42, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, A.V.; Kirejtshuk, A.G.; Shapovalov, A.M. Drilorhinus, a new genus of the family Drilidae Lacordaire, 1857 (Coleoptera: Elateroidea) from Iran. Zootaxa 2019, 4577, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Muona, J.; Taräväinen, M. A re-evaluation of the Eucnemidae larval characters. Papeis Avul. Zool. Spec. Issue 2020, 60. [Google Scholar] [CrossRef]
- Bocak, L.; Kusy, D.; Motyka, M.; Bocek, M. Drilidae Blanchard, 1845: Multi-gene molecular phylogenies versus morphological similarity. An answer to Kovalev et al. Zootaxa 2019, 4674, 142–146. [Google Scholar] [CrossRef]
- Kundrata, R.; Gunter, N.L.; Douglas, H.; Bocak, L. Next step toward a molecular phylogeny of click-beetles (Coleoptera: Elateridae): Redefinition of Pityobiinae, with a description of a new subfamily, Parablacinae, from the Australasian Region. Austral Entomol. 2016, 55, 291–302. [Google Scholar] [CrossRef]
- McKenna, D.D.; Wild, A.L.; Kanda, K.; Bellamy, C.L.; Beutel, R.G.; Caterino, M.S.; Farnum, C.W.; Hawks, D.C.; Ivie, M.A.; Jameson, M.L.; et al. The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015, 40, 835–880. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Che, L.H.; Li, Y.; Dan, L.; Pang, H.; Ślipiński, A.; Zhang, P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018, 9, 205. [Google Scholar] [CrossRef]
- Kusy, D.; Motyka, M.; Andújar, C.; Bocek, M.; Masek, M.; Sklenarova, K.; Kokas, F.; Bocakova, M.; Vogler, A.P.; Bocak, L. Genome sequencing of Rhinorhipus Lawrence exposes an early branch of the Coleoptera. Front. Zool. 2018, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef] [PubMed]
- Kusy, D.; Motyka, M.; Bocek, M.; Vogler, A.P.; Bocak, L. Genome sequences identify three families of Coleoptera as morphologically derived click beetles (Elateridae). Sci. Rep. 2018, 8, 17084. [Google Scholar] [CrossRef] [PubMed]
- Kusy, D.; He, J.W.; Bybee, S.M.; Motyka, M.; Bi, W.X.; Podsiadlowski, L.; Li, X.Y.; Bocak, L. Phylogenomic relationships of bioluminescent elateroids define the ‘lampyroid’ clade with clicking Sinopyrophoridae as its earliest member. Syst. Entomol. 2021. [Google Scholar] [CrossRef]
- Davis, A.L.V.; Scholtz, C.H.; Philips, T.K. Historical biogeography of scarabaeine dung beetles. J. Biogeogr. 2002, 29, 1217–1256. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucl. Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucl. Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Dodsworth, S.; Culverwell, C.L.; Bocak, L.; Ahrens, D.; Littlewood, D.T.J.; Pons, J.; Vogler, A.P. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucl. Acids Res. 2010, 38, e197. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Ahrens, D.; Ollikainen, A.; Culverwell, L.C.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Linard, B.; Crampton-Platt, A.; Moriniere, J.; Timmermans, M.J.T.N.; Andujar, C.; Arribas, P.; Miller, K.E.; Lipecki, J.; Favreau, E.; Hunter, A.; et al. The contribution of mitochondrial metagenomics to large-scale data mining and phylogenetic analysis of Coleoptera. Mol. Phylogenet. Evol. 2018, 128, 1–11. [Google Scholar] [CrossRef] [PubMed]
- He, J.W.; Bi, W.; Dong, Z.; Liu, G.; Zhao, R.; Wang, W.; He, X.L. The mitochondrial genome of the first luminous click-beetle (Coleoptera: Elateridae) recorded in Asia. Mitochondr. DNA Part B 2019, 4, 565–567. [Google Scholar] [CrossRef]
- Amaral, D.T.; Mitani, Y.; Ohmiya, Y.; Viviani, V.R. Organization and comparative analysis of the mitochondrial genomes of bioluminescent Elateroidea (Coleoptera: Polyphaga). Gene 2016, 586, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Bininda-Emonds, O. TransAlign: Using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinform. 2005, 6, 156. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef]
- Kück, P.; Meid, S.A.; Gross, C.; Wägele, J.W.; Misof, B. AliGROOVE—Visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Jermiin, L.S.; Jayaswal, V.; Ababneh, F.; Robinson, J. Phylogenetic model evaluation. In Bioinformatics, Data, Sequence Analysis and Evolution; Keith, J., Ed.; Humana Press: Totowa, NJ, USA, 2008; Volume 1, pp. 331–363. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Meth. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H. An Approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H.; Hasegawa, M. Multiple Comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999, 16, 1114. [Google Scholar] [CrossRef]
- Kishino, H.; Hasegawa, M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989, 29, 170–179. [Google Scholar] [CrossRef]
- Strimmer, K.; Rambaut, A. Inferring confidence sets of possibly misspecified gene trees. Proc. Biol. Sci. 2002, 269, 137–142. [Google Scholar] [CrossRef]
- Kishino, H.; Miyata, T.; Hasegawa, M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 1990, 31, 151–160. [Google Scholar] [CrossRef]
- Calder, A.A. Coleoptera: Elateroidea. In Zoological Catalogue of Australia; Wells, A., Ed.; CSIRO Publishing: Melbourne, Australia, 1998; Volume 29.6, p. 248. [Google Scholar]
- Neboiss, A. The genera Hapatesus Candèze and Toorongus, gen. nov. (Coleoptera: Elateridae). Austr. J. Zool. 1957, 5, 496–520. [Google Scholar] [CrossRef]
- Tarnawski, D. A world catalogue of Ctenicerini Fleutiaux, 1936. Part II (Coleoptera: Elateridae: Athoinae). Genus 2001, 12, 277–323. [Google Scholar]
- Neboiss, A. Genus Hapatesus from Austro-Malayan sub-region (Coleoptera: Elateridae). Proc. R. Soc. Vict. 1958, 70, 169–174. [Google Scholar]
- Girard, C. Les Coléoptères Elateridae de Lamto (Côte d’Ivoire). Bull. l’Inst. Fond. Afr. Noire 1971, 23, 449–650. [Google Scholar]
- Girard, C. Sept nouvelles espèces afrotropicales de Dicronychidae du genre Eudicronychus Méquignon (Coleoptera). Bull. Soc. Entomol. Fr. 1991, 96, 145–154. [Google Scholar]
- Girard, C. Sept nouvelles espèces afrotropicales du genre Eudicronychus Méquignon, 1931 (Coleoptera, Eudicronychidae). Bull. Soc. Entomol. Fr. 2011, 116, 135–146. [Google Scholar]
- Girard, C. Deux espeèces nouvelles du genre Anisomerus Schwarz, 1897 (Coleoptera, Eudicronychidae). Bull. Soc. Entomol. Fr. 2017, 122, 475–478. [Google Scholar] [CrossRef]
- Schwarz, O.C.E. Über die Systematische der Elateriden-Gattungen Dicronychus Castelnau, und Tarsalgus Candèze. Dtsch. Entomol. Zeit. 1897, 1897, 9–16. [Google Scholar]
- Dolin, V.G. Zhilkovanie kry’lev zhukov-shchelkunov (Coleoptera, Elateridae) i ego znachenie dlya sistematiki semeistva. Zool. Zhurnal 1975, 54, 1618–1633. [Google Scholar]
- Bocak, L.; Barton, C.; Crampton-Platt, A.; Chesters, D.; Ahrens, D.; Vogler, A.P. Building the Coleoptera tree-of-life for >8000 species: Composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014, 39, 97–110. [Google Scholar] [CrossRef]
- Stibick, J.N.L. Classification of the Elateridae (Coleoptera). Relation- ships and classification of the subfamilies and tribes. Pacif. Insects 1979, 20, 145–186. [Google Scholar]
- Dolin, V. A role of larval and wing venation characters in the systematics of Elateroidea (Coleoptera). In Meetings in Memory of N.A. Cholodovsky; Lecture at the 52nd Annual Meeting; 1 April 1999; Zoological Institute RAS: St. Peterburg, Russian, 2000; p. 50. [Google Scholar]
- Schwarz, O.C.E. Coleoptera, fam. Elateridae. In Genera Insectorum; P Wytsman: Brussels, Belgium, 1906; Volume 46, pp. 1–224. [Google Scholar]
- Dolin, W.G.; Girard, C. Zur Kenntnis der Schnellkäfer-Gattung Diplophoenicus Candèze, 1893, aus Madagascar (Coleoptera, Elateridae). Bull. Soc. Entomol. Fr. 2003, 108, 55–60. [Google Scholar]
- Liu, Y.; Song, F.; Jiang, P.; Wilson, J.J.; Cai, W.; Li, H. Compositional heterogeneity in true bug mitochondrial phylogenomics. Mol. Phylogenet. Evol. 2018, 118, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, T.; Dang, K.; Bu, W. Compositional and mutational rate heterogeneity in mitochondrial genomes and its effect on the phylogenetic inferences of Cimicomorpha (Hemiptera: Heteroptera). BMC Genom. 2018, 19, 264. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.-G.; Liu, J.-P.; Sun, C.-H.; Vogler, A.P.; Cai, W.-Z. Capturing the phylogeny of holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef] [PubMed]
- Nie, R.; Vogler, A.P.; Yang, X.-K.; Lin, M. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst. Entomol. 2021. [Google Scholar] [CrossRef]
- Shen, X.X.; Li, Y.; Hittinger, C.T.; Chen, X.X.; Rokas, A. An investigation of irreproducibility in maximum likelihood phylogenetic inference. Nat. Commun. 2020, 11, 6090. [Google Scholar] [CrossRef]

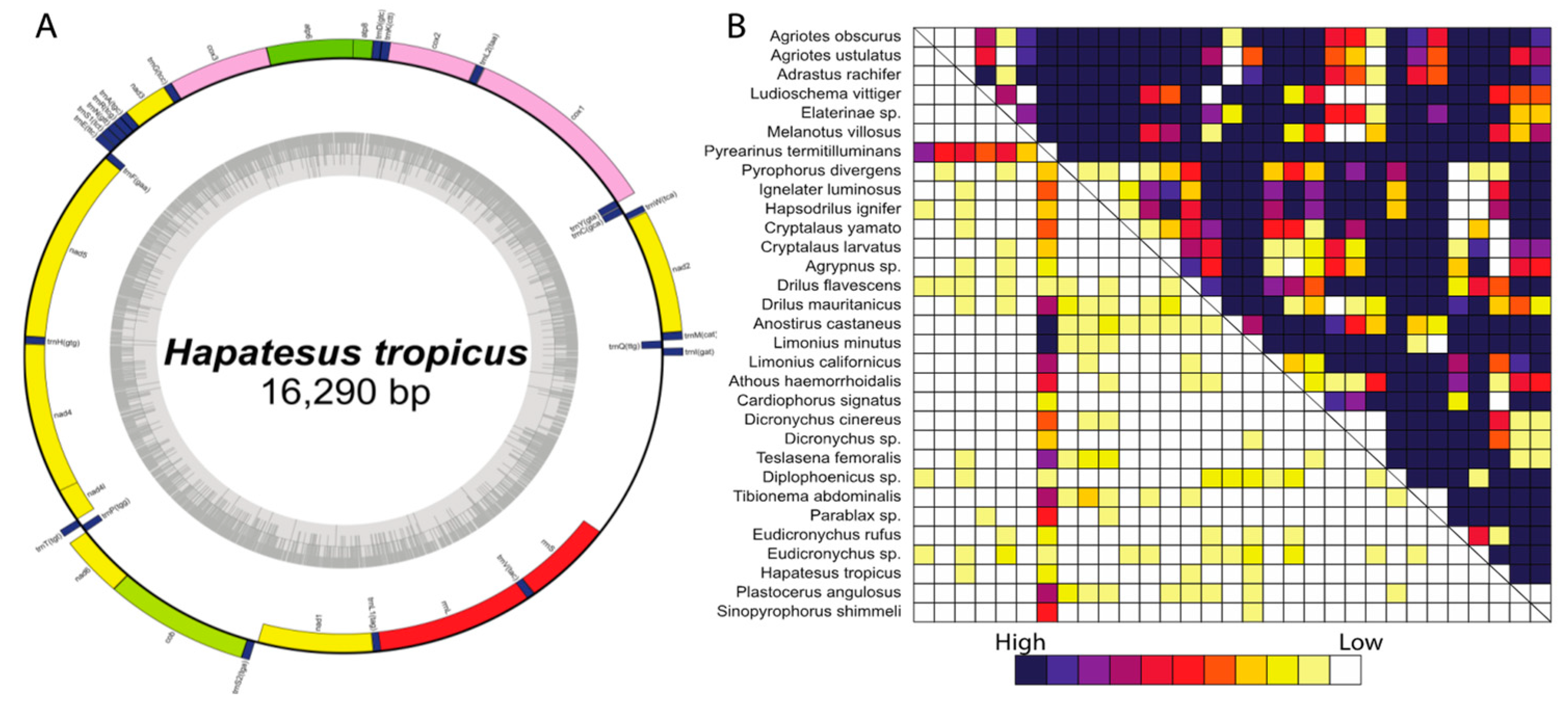


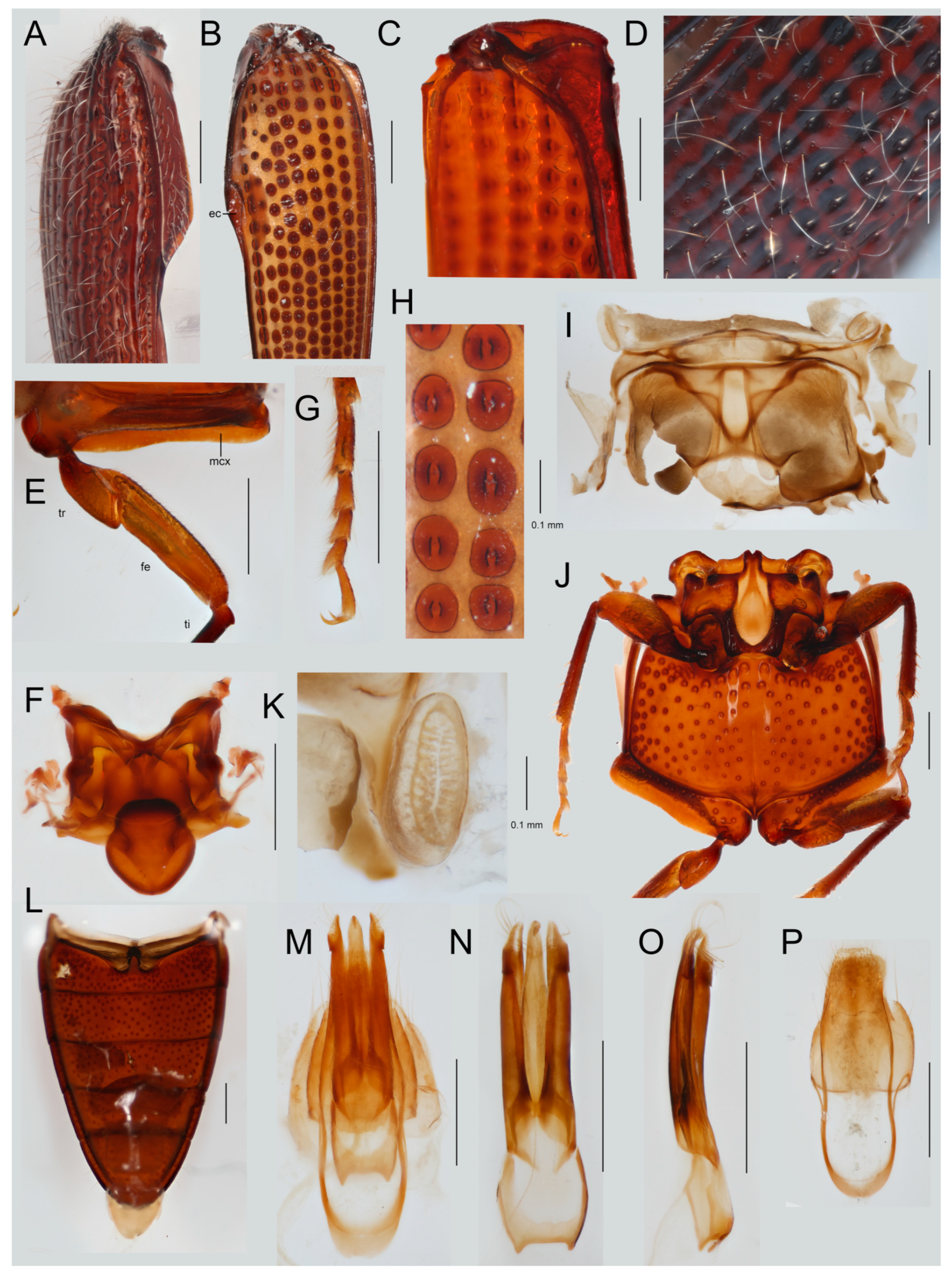

| Voucher Number | Genus, Species | Subfamily | Tribe | Geographic Origin |
|---|---|---|---|---|
| G18004 | Drilus mauritanicus | Agrypninae | Drilini | Spain |
| G19011 | Diplophoenicus sp. | Morostomatinae | Madagascar | |
| G20012 | Tibionema abdominalis | Pityobiinae | Chile | |
| G19006 | Parablax sp. | Parablacinae | Queensland | |
| G19007 | Eudicronychus rufus | Elaterinae | Eudicronychini | Zambia |
| G20004 | Eudicronychus sp. | Elaterinae | Eudicronychini | Zambia |
| G20007 | Hapatesus tropicus | Hapatesinae | New Guinea | |
| A01544 | Plastocerus angulosus | Dendrometrinae and/or Plastocerinae | Turkey |
| Topology | logL | Deltal | bp-RELL | p-KH | p-SH | p-WKH | p-WSH | c-ELW | p-AU | Validity |
|---|---|---|---|---|---|---|---|---|---|---|
| Unconstrained position | −195,557.3 | 0 | 0.987 | 0.994 | 1 | 0.984 | 0.998 | 0.987 | 0.992 | accepted |
| Sister to DMCA clade | −195,590.6 | 33.30 | 0.001 | 0.006 | 0.117 | 0.006 | 0.012 | 0.001 | 0.002 | rejected |
| Sister to Dendrometrinae | −195,594.0 | 36.68 | 0.012 | 0.016 | 0.073 | 0.016 | 0.039 | 0.012 | 0.012 | rejected |
| Terminal in Dendrometrinae | −195,726.3 | 169.0 | 0 | 0 | 0 | 0 | 0 | 2.83 × 10−25 | 2.84 × 10−39 | rejected |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusy, D.; Motyka, M.; Bocak, L. Click Beetle Mitogenomics with the Definition of a New Subfamily Hapatesinae from Australasia (Coleoptera: Elateridae). Insects 2021, 12, 17. https://doi.org/10.3390/insects12010017
Kusy D, Motyka M, Bocak L. Click Beetle Mitogenomics with the Definition of a New Subfamily Hapatesinae from Australasia (Coleoptera: Elateridae). Insects. 2021; 12(1):17. https://doi.org/10.3390/insects12010017
Chicago/Turabian StyleKusy, Dominik, Michal Motyka, and Ladislav Bocak. 2021. "Click Beetle Mitogenomics with the Definition of a New Subfamily Hapatesinae from Australasia (Coleoptera: Elateridae)" Insects 12, no. 1: 17. https://doi.org/10.3390/insects12010017
APA StyleKusy, D., Motyka, M., & Bocak, L. (2021). Click Beetle Mitogenomics with the Definition of a New Subfamily Hapatesinae from Australasia (Coleoptera: Elateridae). Insects, 12(1), 17. https://doi.org/10.3390/insects12010017






