Diversity and Regulation of S-Adenosylmethionine Dependent Methyltransferases in the Anhydrobiotic Midge
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SAM-Dependent Methyltransferases
2.2. Differential Expression Analysis and Clustering
2.3. Modelling of PIMTs and SAM Interactions
2.4. Recombinant Protein Purification
2.5. Protein Identification
2.6. Enzymatic Activity Assay
3. Results
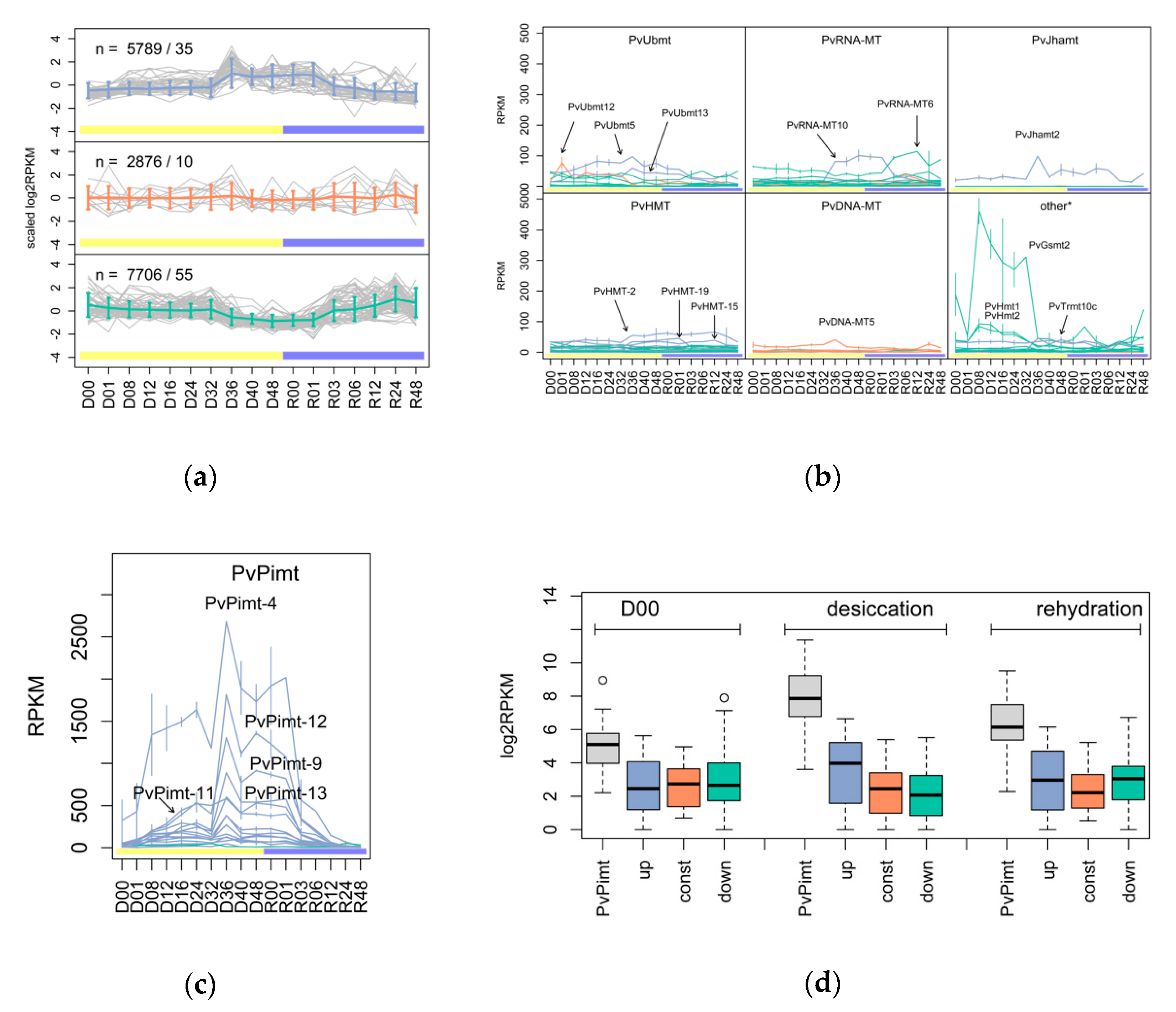
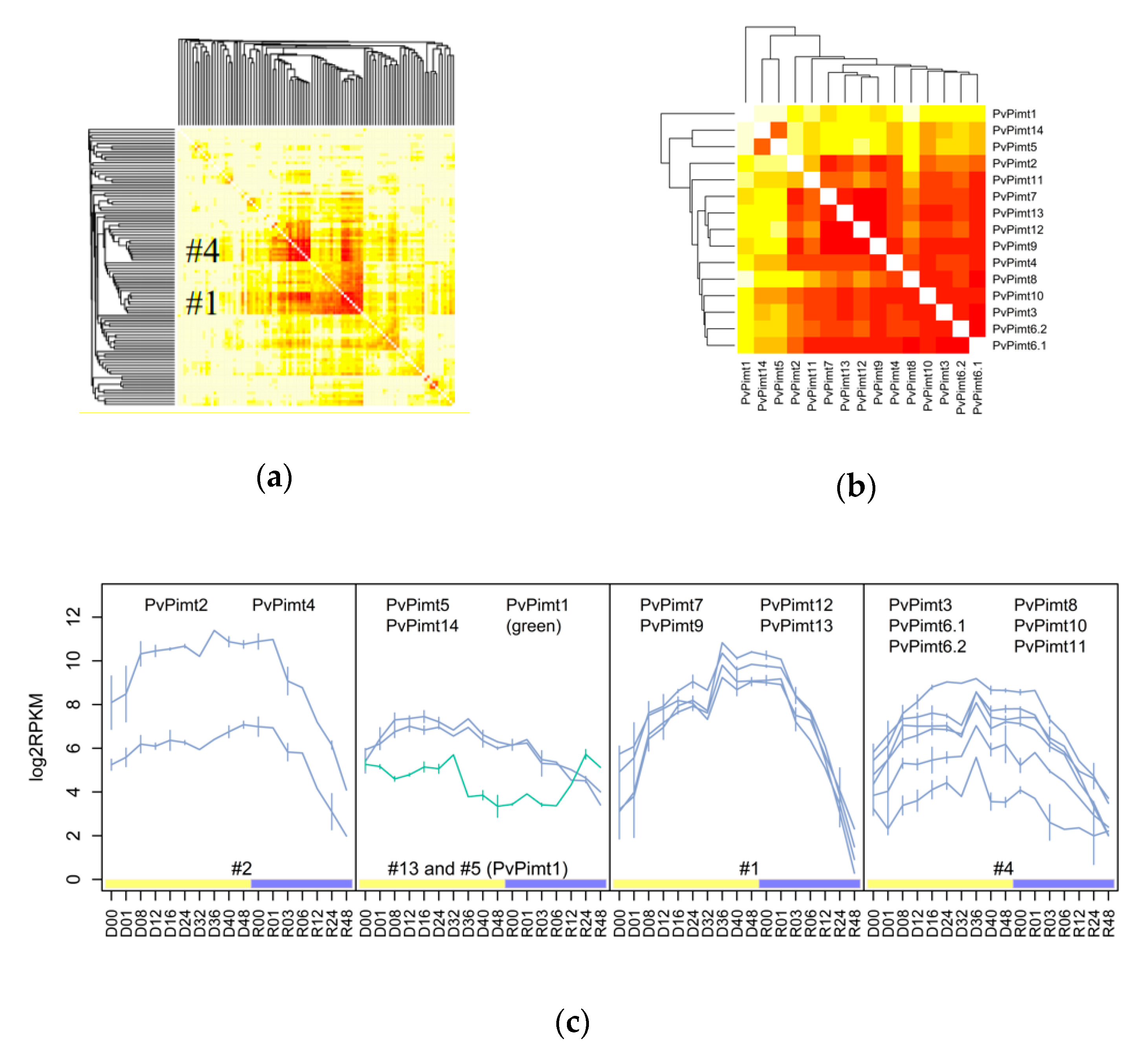
3.1. The Variety and Differential Expression of SAM-Dependent Methyltransferases in Dipteran Insects
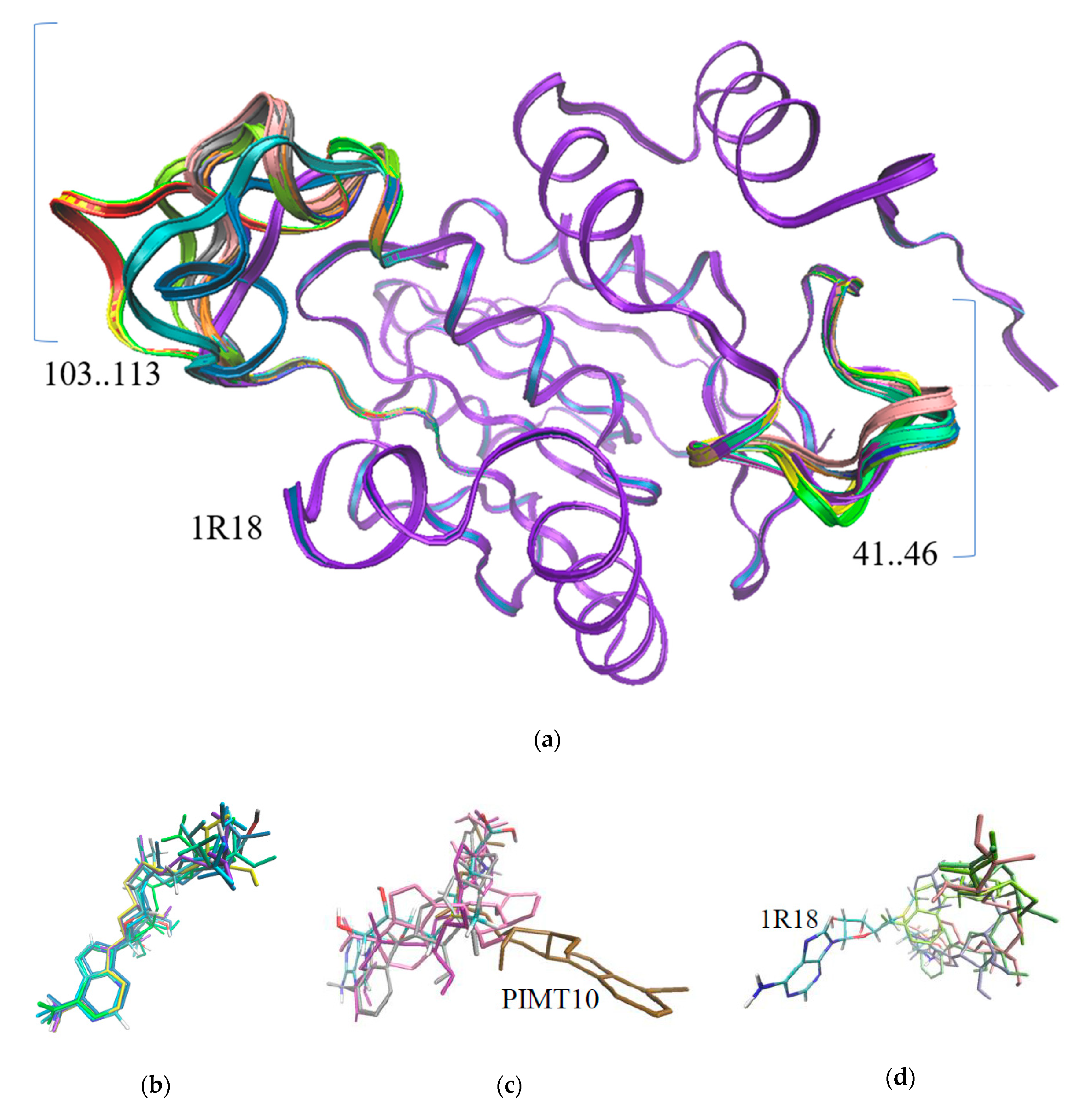
3.2. The Structure Prediction of Anhydrobiosis Specific PIMTs
3.3. Mass Spectrometry and Enzymatic Activity of Recombinant PIMTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watanabe, M.; Kikawada, T.; Okuda, T. Increase of internal ion concentration triggers trehalose synthesis associated with cryptobiosis in larvae of Polypedilum vanderplanki. J. Exp. Biol. 2003, 206, 2281–2286. [Google Scholar] [CrossRef]
- Clegg, J.S. Cryptobiosis—A peculiar state of biological organization. Comp. Biochem. Physiol. Part B 2001, 128, 613–624. [Google Scholar] [CrossRef]
- Cornette, R.; Kikawada, T. The induction of anhydrobiosis in the sleeping chironomid: Current status of our knowledge. IUBMB Life 2011, 63, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Gusev, O.; Suetsugu, Y.; Cornette, R.; Kawashima, T.; Logacheva, M.D.; Kondrashov, A.S.; Penin, A.A.; Hatanaka, R.; Kikuta, S.; Shimura, S.; et al. Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat. Commun. 2014, 5, 4784. [Google Scholar] [CrossRef] [PubMed]
- Furuchi, T.; Sakurako, K.; Katane, M.; Sekine, M.; Homma, H. The role of protein L-isoaspartyl/D-aspartyl O-methyltransferase (PIMT) in intracellular signal transduction. Chem. Biodivers. 2010, 7, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S. Aging as war between chemical and biochemical processes: Protein methylation and the recognition of age-damaged proteins for repair. Ageing Res. Rev. 2003, 2, 263–285. [Google Scholar] [CrossRef]
- Shimizu, T.; Matsuoka, Y.; Shirasawa, T. Biological significance of isoaspartate and its repair system. Biol. Pharm. Bull. 2005, 28, 1590–1596. [Google Scholar] [CrossRef]
- Wakankar, A.A.; Borchardt, R.T. Formulation considerations for proteins susceptible to asparagine deamidation and aspartate isomerization. J. Pharm. Sci. 2006, 95, 2321–2336. [Google Scholar] [CrossRef]
- Kagan, R.M.; McFadden, H.J.; McFadden, P.N.; O’Connor, C.; Clarke, S. Molecular phylogenetics of a protein repair methyltransferase. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1997, 117, 379–385. [Google Scholar] [CrossRef]
- Mudgett, M.B.; Lowenson, J.D.; Clarke, S. Protein repair L-isoaspartyl methyltransferase in plants (Phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds). Plant Physiol. 1997, 115, 1481–1489. [Google Scholar] [CrossRef]
- Bennett, E.J.; Bjerregaard, J.; Knapp, J.E.; Chavous, D.A.; Friedman, A.M.; Royer, W.E., Jr.; O’Connor, C.M. Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis. Biochemistry 2003, 42, 12844–12853. [Google Scholar] [CrossRef] [PubMed]
- Mazin, P.V.; Shagimardanova, E.; Kozlova, O.; Cherkasov, A.; Sutormin, R.; Stepanova, V.V.; Stupnikov, A.; Logacheva, M.; Penin, A.; Sogame, Y.; et al. Cooption of heat shock regulatory system for anhydrobiosis in the sleeping chironomid Polypedilum vanderplanki. Proc. Natl. Acad. Sci. USA 2018, 115, E2477–E2486. [Google Scholar] [CrossRef] [PubMed]
- Furuchi, T.; Kosugi, S.; Ohno, K.; Egawa, T.; Sekine, M.; Katane, M.; Homma, H. High-performance liquid chromatographic method to measure protein L-isoaspartyl/D-aspartyl O-methyltransferase activity in cell lysates. Anal. Biochem. 2009, 384, 207–212. [Google Scholar] [CrossRef]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef]
- Martin, J.L.; McMillan, F.M. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Ogé, L.; Bourdais, G.; Bove, J.; Collet, B.; Godin, B.; Granier, F.; Boutin, J.P.; Job, D.; Jullien, M.; Grappin, P. Protein repair L-isoaspartyl methyltransferasel is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 2008, 20, 3022–3037. [Google Scholar] [CrossRef]
- Hatanaka, R.; Gusev, O.; Cornette, R.; Shimura, S.; Kikuta, S.; Okada, J.; Okuda, T.; Kikawada, T. Diversity of the expression profiles of late embryogenesis abundant (LEA) protein encoding genes in the anhydrobiotic midge Polypedilum vanderplanki. Planta 2015, 242, 451–459. [Google Scholar] [CrossRef]
- Mizobuchi, M.; Murao, K.; Takeda, R.; Kakimoto, Y. Tissue-specific expression of isoaspartyl protein carboxyl methyltransferase gene in rat brain and testis. J. Neurochem. 1994, 62, 322–328. [Google Scholar] [CrossRef]
- Verma, P.; Kaur, H.; Petla, B.P.; Rao, V.; Saxena, S.C.; Majee, M. Protein L-isoaspartyl-O-methyltransferase2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol. 2013, 161, 1141–1157. [Google Scholar] [CrossRef]
- Desrosiers, R.R.; Fanélus, I. Damaged proteins bearing L-isoaspartyl residues and aging: A dynamic equilibrium between generation of isomerized forms and repair by PIMT. Curr. Aging Sci. 2011, 4, 8–18. [Google Scholar] [CrossRef]
- Ryabova, A.; Cornette, R.; Cherkasov, A.; Watanabe, M.; Okuda, T.; Shagimardanova, E.; Kikawada, T.; Gusev, O. Combined metabolome and transcriptome analysis reveals key components of complete desiccation tolerance in an anhydrobiotic insect. Proc. Natl. Acad. Sci. USA 2020, 117, 19209–19220. [Google Scholar] [CrossRef] [PubMed]
- Gomez, T.A.; Banfield, K.L.; Clarke, S.G. The protein L-isoaspartyl-O-methyltransferase functions in the Caenorhabditis elegans stress response. Mech. Ageing Dev. 2008, 129, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Kindrachukt, J.; Parent, J.; Davies, G.F.; Dinsmore, M.; Attah-Poku, S.; Napper, S. Overexpression of L-isoaspartate O-methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. J. Biol. Chem. 2003, 278, 50880–50886. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Qi, C.; Cao, W.Q.; Yeldandi, A.V.; Rao, M.S.; Reddy, J.K. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl. Acad. Sci. USA 2001, 98, 10380–10385. [Google Scholar] [CrossRef] [PubMed]
- Thapar, N.; Kim, A.K.; Clarke, S. Distinct patterns of expression but similar biochemical properties of protein L-isoaspartyl methyltransferase in higher plants. Plant Physiol. 2001, 125, 1023–1035. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Reinikainen, T.; Leisola, M. Characterization of glycine sarcosine N-methyltransferase and sarcosine dimethylglycine N-methyltransferase. Appl. Environ. Microbiol. 2001, 67, 2044–2050. [Google Scholar] [CrossRef]
- Obata, F.; Miura, M. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 2015, 18, 8332. [Google Scholar] [CrossRef]
- Shinoda, T.; Itoyama, K. Juvenile hormone acid methyltransferase: A key regulatory enzyme for insect metamorphosis. Proc. Natl. Acad. Sci. USA 2003, 100, 11986–11991. [Google Scholar] [CrossRef]
- Mandal, S.S.; Chu, C.; Wada, T.; Handa, H.; Shatkin, A.J.; Reinberg, D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. USA 2004, 101, 7572–7577. [Google Scholar] [CrossRef]
| Insect Species | D. mel | P. van | P. nub | B. ant |
|---|---|---|---|---|
| Homocysteine S-methyltransferase EC 2.1.1.10 (Hmt) | 2 | 2 | 1 | 1 |
| Histone-arginine N-methyltransferase EC 2.1.1.125 (HMT) | 10 | 6 | 8 | 5 |
| DNA adenine-specific MTases EC 2.1.1.72 | 7 | 5 | 7 | 6 |
| RNA MTases | 31 | 24 | 26 | 18 |
| Histone-lysine methyltransferase EC 2.1.1.43 (HMT) | 18 | 21 | 18 | 19 |
| PIMT EC 2.1.1.77 | 1 | 15 | 1 | 1 |
| Diphthamide methyltransferase EC 2.1.1.98 (Dph) | 1 | 1 | 1 | 1 |
| Ubiquinone biosynthesis O-methyltransferases (Ubmt) | 4 | 13 | 13 | 2 |
| Protein-S-isoprenylcysteine O-methyltransferase EC 2.1.1.100 (Icmt) | 1 | 1 | 1 | 1 |
| Glycine/Sarcosine N-methyltransferase EC 2.1.1.156 (Gsmt) | 1 | 2 | 1 | 1 |
| Diphthamide methyltransferase EC 2.1.1.98 (Dph) | 1 | 1 | 1 | 1 |
| DNA (cytosine-C5) MTase EC 2.1.1.37 | 1 | 1 | 1 | 1 |
| Juvenile hormone acid methyltransferase EC 2.1.1.325 (Jhamt) | 1 | 4 | 4 | 2 |
| Protein N-terminal methyltransferase EC 2.1.1.244 (NMT) | 1 | 1 | 1 | 1 |
| Spermidine/spermine synthases family EC 2.5.1.22 (Sps) | 1 | 1 | 1 | 1 |
| DNA (cytosine-C5) MTase EC 2.1.1.37 | 1 | 1 | 1 | 1 |
| Leucine carboxyl methyltransferase EC 2.1.1.233 (Lcmt) | 1 | 1 | 1 | 1 |
| Methylene-fatty-acyl-phospholipid synthase EC 2.1.1.16 (Mfap) | - | 1 | 1 | 1 |
| Methyltransferase type 11 (Mt11) | 3 | 2 | 2 | 2 |
| Unchr_MeTrfase_Williams-Beuren Family (WBS_methylT) | 1 | 1 | 1 | 1 |
| PIMT | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1R18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Energy, kcal/mol | −7.6 | −8.9 | −7.1 | −7.8 | −7.9 | −7.7 | −6.8 | −7.1 | −8.3 | −7.0 | −8.3 | −8.3 | −7.4 | −8.1 | −8.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deviatiiarov, R.; Ayupov, R.; Laikov, A.; Shagimardanova, E.; Kikawada, T.; Gusev, O. Diversity and Regulation of S-Adenosylmethionine Dependent Methyltransferases in the Anhydrobiotic Midge. Insects 2020, 11, 634. https://doi.org/10.3390/insects11090634
Deviatiiarov R, Ayupov R, Laikov A, Shagimardanova E, Kikawada T, Gusev O. Diversity and Regulation of S-Adenosylmethionine Dependent Methyltransferases in the Anhydrobiotic Midge. Insects. 2020; 11(9):634. https://doi.org/10.3390/insects11090634
Chicago/Turabian StyleDeviatiiarov, Ruslan, Rustam Ayupov, Alexander Laikov, Elena Shagimardanova, Takahiro Kikawada, and Oleg Gusev. 2020. "Diversity and Regulation of S-Adenosylmethionine Dependent Methyltransferases in the Anhydrobiotic Midge" Insects 11, no. 9: 634. https://doi.org/10.3390/insects11090634
APA StyleDeviatiiarov, R., Ayupov, R., Laikov, A., Shagimardanova, E., Kikawada, T., & Gusev, O. (2020). Diversity and Regulation of S-Adenosylmethionine Dependent Methyltransferases in the Anhydrobiotic Midge. Insects, 11(9), 634. https://doi.org/10.3390/insects11090634







