Genetic Parameters of Honey Bee Colonies Traits in a Canadian Selection Program
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Pedigree Determination
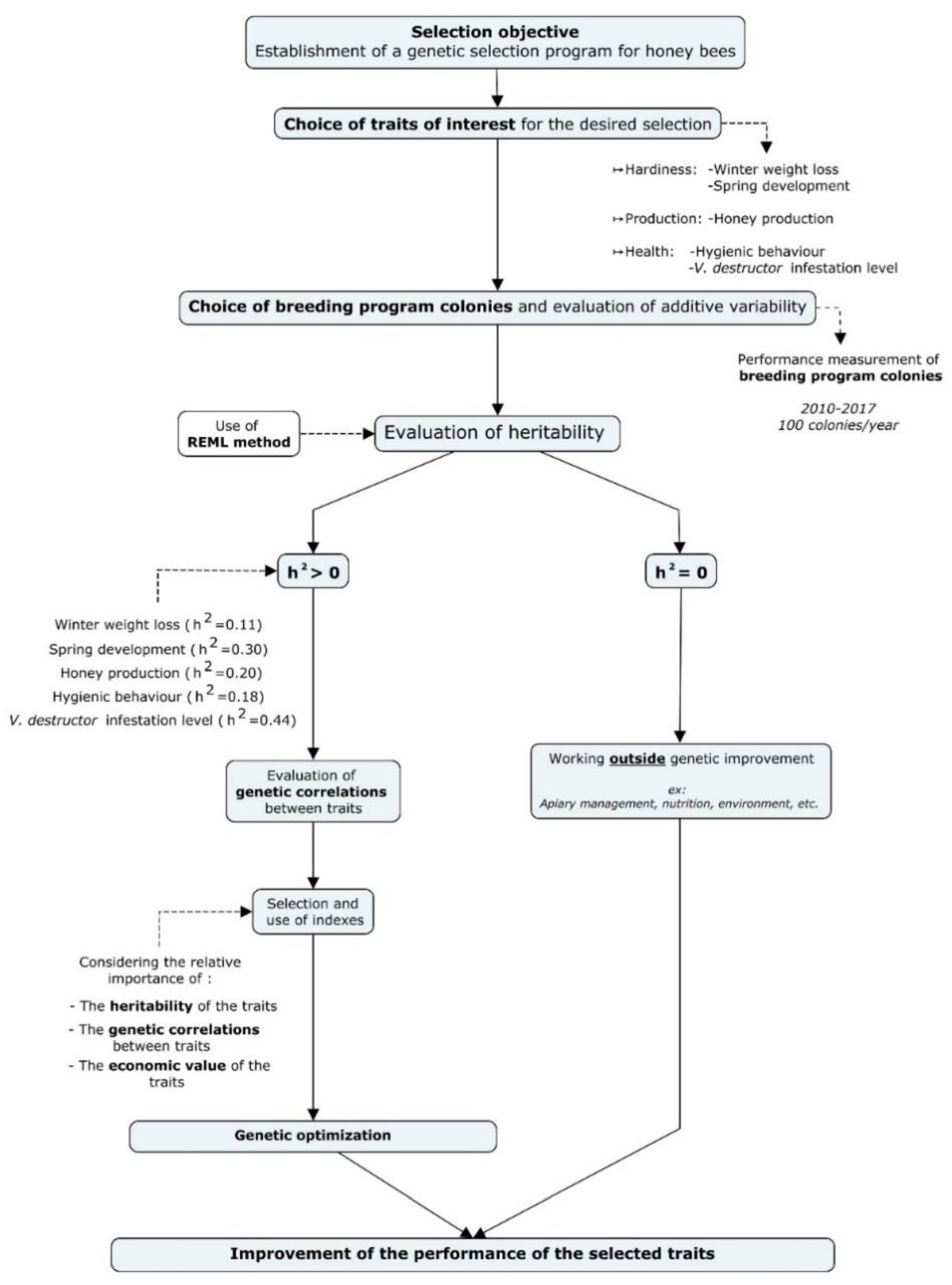
2.3. Traits Measured to Evaluate Colony Performance
2.3.1. Productivity of Honey Production
2.3.2. Health
2.3.3. Hardiness
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- CCSI, Canadian Centre for Swine Improvement. Canadian Centre for Swine Improvement—2019 Annual Report. Available online: https://www.ccsi.ca/meetings/annual/2019AnnualReportEnglish.pdf (accessed on 15 May 2020).
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef] [PubMed]
- CDN, Canadian Dairy Network. Breed Improvement Programs: Critical to Success and Profitability. Canadian Dairy Network. Available online: https://www.cdn.ca/articles.php?_postback_=1&_formname_=form1&fromdate=01%2F01%2F1999&todate=11%2F21%2F2019&keywords=genetic+improvement&search=Search&_returnurl_=https%3A%2F%2Fwww.google.com%2F (accessed on 15 May 2020).
- Minvielle, F. Principes d’Amélioration Génétique des Animaux Domestiques, 1st ed.; Les presses de l’université Laval: Québec, QC, Canada, 1990; pp. 15–68. [Google Scholar]
- De Rochambeau, H. Les bases de la génétique quantitative: Le progrès génétique et sa réalisation dans les expériences de sélection. In Elément de Génétique Quantitative et Application aux Populations Animales, special ed.; INRA Production Animales: Paris, France, 1992; Volume 1, pp. 83–86. [Google Scholar]
- Page, R.E.; Laidlaw, H.H. Honey bee genetics and breeding. In Hive and the Honey Bee, 3rd ed.; Graham, J., Ed.; Dadant & Sons: Hamilton, IL, USA, 2008; pp. 235–267. [Google Scholar]
- Sauvager, B. Hérédité chez l’Abeille et les Colonies d’Abeilles, 1st ed.; Anercea, au cœur de l’élevage: Surgères, France, 2019; pp. 28–30. [Google Scholar]
- Bienefeld, K.; Pirchner, F. Genetic correlations among several colony characters in the honey bee (Hymenoptera: Apidae) taking queen and worker effects into account. Ann. Entomol. Soc. Am. 1991, 84, 324–331. [Google Scholar] [CrossRef]
- Oldroyd, B.P.; Goodman, R.D. On the relative importance of queens and workers to honey production. Apidologie 1990, 21, 153–159. [Google Scholar] [CrossRef]
- Oxley, P.R.; Oldroyd, B.P. The Genetic architecture of honeybee breeding. In Advances in Insect Physiology, 1st ed.; Simpson, S.J., Casas, J., Eds.; Elsevier Ltd. Academic Press: Burlington, Australia, 2010; Volume 39, pp. 83–118. [Google Scholar]
- Bienefeld, K.; Pirchner, F. Heritabilities for several colony traits in the honeybee (Apis Mellifera Carnica). Apidologie 1990, 21, 175–183. [Google Scholar] [CrossRef]
- Smartbees, Sustainable Management of Resilient Bee Populations. Performance Testing Protocol A Guide for European Honey Bee Breeders. Available online: http.//www.smartbees-fp7.eu%0APerformance (accessed on 15 May 2020).
- Leveaux, G.; Van Laere, O. Beebreed.Eu Programme de Sélection. Abeille Cie. 2014, 2, 33–34. [Google Scholar]
- Bienefeld, K.; Ehrhardt, K.; Reinhardt, F. Genetic evaluation in the honey bee considering queen and worker effects—A BLUP-animal model approach. Apidologie 2007, 38, 77–85. [Google Scholar] [CrossRef]
- Bienefeld, K.; Ehrhardt, K.; Reinhardt, F. Bee breeding around the world—Noticeable success in honey bee selection after the introduction of genetic evaluation using BLUP. Am. Bee J. 2008, 8, 739–742. [Google Scholar]
- Jamieson, C.A. Fact about beekeeping in Canada. Bee World 1958, 39, 232–236. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee, 1st ed.; Harvard University Press: Campbridge, MA, USA, 1987; pp. 250–254. [Google Scholar]
- Boucher, C.; Desjardins, F.; Giovenazzo, P.; Marceau, J.; Pettigrew, A.; Tremblay, H.; Tremblay, N. Gestion Optimale du Rucher, 2nd ed.; CRAAQ: Québec, QC, Canada, 2011; pp. 15–18. [Google Scholar]
- Canadian Honey Council. Industry Overview. Available online: http://honeycouncil.ca/archive/cdn_apiculture_overview.php (accessed on 15 May 2020).
- Parker, R.; Melathopoulos, A.P.; White, R.; Pernal, S.F.; Guarna, M.; Foster, L. Ecological adaptation of diverse honey bee (Apis Mellifera) populations. PLoS ONE 2010, 5, e11096. [Google Scholar] [CrossRef]
- Canadian Honey Council. A national bee health action plan. HiveLights 2015, 28, 6–8. [Google Scholar]
- Furgala, B.; McCutcheon, D.M. Wintering productive colonies. In Hive and the Honey bee, 3rd ed.; Graham, J., Ed.; Dadant & Sons: Hamilton, IL, USA, 2008; pp. 829–868. [Google Scholar]
- Currie, R.W.; Pernal, S.F.; Guzmán-Novoa, E. Honey Bee colony losses in Canada. J. Apic. Res. 2010, 49, 104–106. [Google Scholar] [CrossRef]
- CAPA, Canadian Association of Professional Apiculturists, National Survey Committee and Provincial Apiculturists. Statement on honey bee wintereing losses in Canada. HiveLights 2019, 32, 13–20. [Google Scholar]
- Kevan, P.; Guzman, E.; Skinner, A.; van Englesdorp, D. Colony collapse disorder in Canada: Do we have a problem? HiveLights 2007, 20, 14–16. [Google Scholar]
- CAPA, Canadian Association of Professional Apiculturists. Statement on Honey bee Wintering Losses in Canada (2016). Available online: http://www.capabees.com/shared/2015/07/2016-CAPA-Statement-on-Colony-Losses-July-19.pdf (accessed on 15 May 2020).
- Bixby, M.A. Canadian beekeeping perspective on colony health and growing our local queen supply. HiveLights 2017, 30, 13–15. [Google Scholar]
- Meixner, M.D.; Büchler, R.; Costa, C.; Roy, M.F.; Hatjina, F.; Kryger, P.; Uzunov, A.; Carreck, N.L. Honey bee genotypes and the environment. J. Apic. Res. 2014, 53, 183–187. [Google Scholar] [CrossRef]
- Seeley, T. Darwinian beekeeping: An evolutionary approach to apiculture. Am. Bee J. 2017, 157, 277–282. [Google Scholar]
- Pourjardieu, B.; Mallard, J. Les bases de la génétique quantitative: Les méthodes d’estimation de l’héritabilité et des corrélations génétiques. In Productions Animales—Génétique Quantitative, special ed.; INRA Production Animales: Paris, France, 1992; Volume 1, pp. 87–92. [Google Scholar]
- Minvielle, F. Que Sais-Je ? La Selection Animale, 1st ed.; Les presses universitaires de France: Paris, France, 1998; pp. 16–24. [Google Scholar]
- Kruuk, L.E.B. Estimating genetic parameters in natural populations using the ‘animal model’. Philos. Trans. R. Soc. B 2004, 359, 873–890. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman, P., Ed.; Longman: Burnt Mill, UK, 1996; pp. 135–165. [Google Scholar]
- Néron, F.; Guéguen, R. Petit Précis d’Elevage, 1st ed.; Agricole, F., Ed.; Agriproduction: Paris, France, 2018; pp. 310–443. [Google Scholar]
- Chevalet, C.; Cornuet, J.M. Evolution de la consanguinite dans une population d’abeilles. Apidologie 1982, 13, 157–168. [Google Scholar] [CrossRef]
- Büchler, R.; Andonov, S.; Bienefeld, K.; Costa, C.; Kezic, N.; Kryger, P.; Spivak, M.; Uzunov, A.; Wilde, J.; Hatjina, F. Standard methods for rearing and selection of apis mellifera queens. J. Apic. Res. 2013, 52, 1–29. [Google Scholar] [CrossRef]
- Laidlaw, H.H.; Page, R.E. Queen Rearing and Bee Breeding, 1st ed.; Wicwas Press: Cheshire, CT, USA, 1997; pp. 135–145. [Google Scholar]
- Hellmich, R.L.; Waller, G.D. Preparing for africanized honey bees: Evaluating control in mating apiaries. Am. Bee J. 1990, 130, 537–542. [Google Scholar]
- Guzman-Novoa, E. Elemental Genetics and Breeding for the Honeybee, 1st ed.; Association, O.B., Ed.; Guelph University Press: Bayfield, ON, Canada, 2007; pp. 47–49. [Google Scholar]
- Sorel, A.; Martin, G.; Houle, E.; Giovenazzo, P. Finding DCA’s. Available online: https://www.beeculture.com/finding-dcas/ (accessed on 15 May 2020).
- Ambrose, J.T. Management for honey production. In Hive and the Honey bee, 3rd ed.; Graham, J., Ed.; Dadant & Sons: Hamilton, IL, USA, 2008; Volume 1, pp. 602–654. [Google Scholar]
- FAQ, Fédération des Apiculteurs du Québec. Plan Stratégique 2010–2015 du Secteur Apicole du Québec—Rapport Final. Available online: https://www.apiculteursduquebec.com/documents/Plan_strategique.pdf (accessed on 15 May 2020).
- Giovenazzo, P.; Martin, G. Available online: http://crsad.qc.ca/uploads/tx_centrerecherche/195_Rapport_final.pdf (accessed on 15 May 2020).
- Bixby, M.; Mcafee, A. Queens rules! Growing Canada’s queen breeding industry: Results from the 2016–2017 queen breeder survey. Am. Bee J. 2017, 157, 893–895. [Google Scholar]
- Spivak, M.; Downey, D.L. Field assays for hygienic behavior in honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 1998, 91, 64–70. [Google Scholar] [CrossRef]
- Spivak, M.; Reuter, G.S. Honey bee hygienic behavior. Am. Bee J. 1998, 138, 283–286. [Google Scholar]
- Imdorf, A.; Charrière, J.D.; Kilchenmann, V.; Bogdanov, S.; Fluri, P. Alternative strategy in central europe for the control of varroa destructor in honey bee colonies. Apiacta 2003, 38, 258–278. [Google Scholar]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D.L.; Locke, B.; Delaplane, K.S.; Wauquiez, Q.; Tannahill, C.; Frey, E.; Ziegelmann, B.; et al. Standard methods for varroa research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Giovenazzo, P.; Dubreuil, P. Evaluation of spring organic treatments against varroa destructor (Acari: Varroidae) in honey bee apis mellifera (Hymenoptera: Apidae) colonies in Eastern Canada. Exp. Appl. Acarol. 2011, 55, 65–76. [Google Scholar] [CrossRef]
- Delaplane, K.S.; van der Steen, J.; Guzman-Novoa, E. Standard methods for estimating strength parameters of Apis mellifera colonies. J. Apic. Res. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Klei, B. Approximative Variance for Heritability estimates. Available online: http://nce.ads.uga.edu/html/projects/AI_SE_revised.pdf (accessed on 13 July 2020).
- Pirchner, F. Population Genetics in Animal Breeding, 2nd ed.; Frape, D.L., Ed.; Plenium press: New York, NY, USA, 1983; pp. 30–45. [Google Scholar]
- Pirchner, F.; Ruttner, F.; Ruttner, H. Erbliche Unterschiede zwischen ertragseigenschaften von Bienen. Proc. Inter. Cong. Entomol. 1962, 11, 510–516. [Google Scholar]
- Rinderer, T. Bee Genetic and Breeding, 1st ed.; Rinderer, T.E., Ed.; Academic press: Orlando, FL, USA, 1986; pp. 57–68. [Google Scholar]
- Brascamp, E.W.; Willam, A.; Boigenzahn, C.; Bijma, P.; Veerkamp, R.F. Heritabilities and genetic correlations for honey yield, gentleness, calmness and swarming behaviour in austrian honey bees. Apidologie 2016, 47, 739–748. [Google Scholar] [CrossRef]
- Brascamp, E.W.; Willam, A.; Boigenzahn, C.; Bijma, P.; Veerkamp, R.F. Correction to: Heritabilities and genetic correlations for honey yield, gentleness, calmness and swarming behaviour in austrian honey. Apidologie 2018, 49, 462–463. [Google Scholar] [CrossRef]
- Andonov, S.; Costa, C.; Uzunov, A.; Bergomi, P.; Lourenco, D.; Misztral, I. Modeling honey yield, defensive and swarming behaviors of Italian honey bees (Apis mellifera ligustica) using linear-threshold approaches. BMC Genet. 2019, 20, 78. [Google Scholar] [CrossRef]
- Guichard, M.; Neuditschko, M.; Soland, G.; Fried, P.; Grandjean, M.; Gerster, S.; Dainat, B.; Bijma, P.; Brascamp, E.W. Estimates of genetic parameters for production, behaviour, and health traits in two Swiss honey bee populations. Apidologie 2020. [Google Scholar] [CrossRef]
- Soller, M.; Bar-Cohen, R. Some observations on the heritability and genetic correlation between honey production and brood area in the honeybee. J. Apic. Res. 1967, 6, 37–43. [Google Scholar] [CrossRef]
- Facchini, E.; Bijma, P.; Pagnacco, G.; Rizzi, R.; Brascamp, E.W. Hygienic behaviour in honeybees: A comparison of two recording methods and estimation of genetic parameters. Apidologie 2019, 50, 163–172. [Google Scholar] [CrossRef]
- Guarna, M.M.; Hoover, S.E.; Huxter, E.; Higo, H.; Moon, K.M.; Domanski, D.; Bixby, M.E.; Melathopoulos, A.; Ibrahim, A.; Peirson, M.; et al. Peptide biomarkers used for the selective breeding of a complex polygenic trait in honey bees. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Harbo, J.R.; Harris, J.W. Selecting honey bees for resistance to Varroa jacobsoni. Apidologie 1999, 30, 183–196. [Google Scholar] [CrossRef]
- Tedeschi, L.O. Assessment of the adequacy of mathematical models. Agric. Syst. 2006, 89, 225–247. [Google Scholar] [CrossRef]
- Gabka, J. Correlations between the strength, amount of brood, and honey production of the honey bee colony. Med. Weter. 2014, 70, 754–756. [Google Scholar]
- Noel, A.; Le Conte, Y.; Mondet, F. Varroa destructor: How does it harm Apis mellifera honey bees and what can be done about it? Emerg. Top. Life Sci. 2020, 4, 45–57. [Google Scholar]
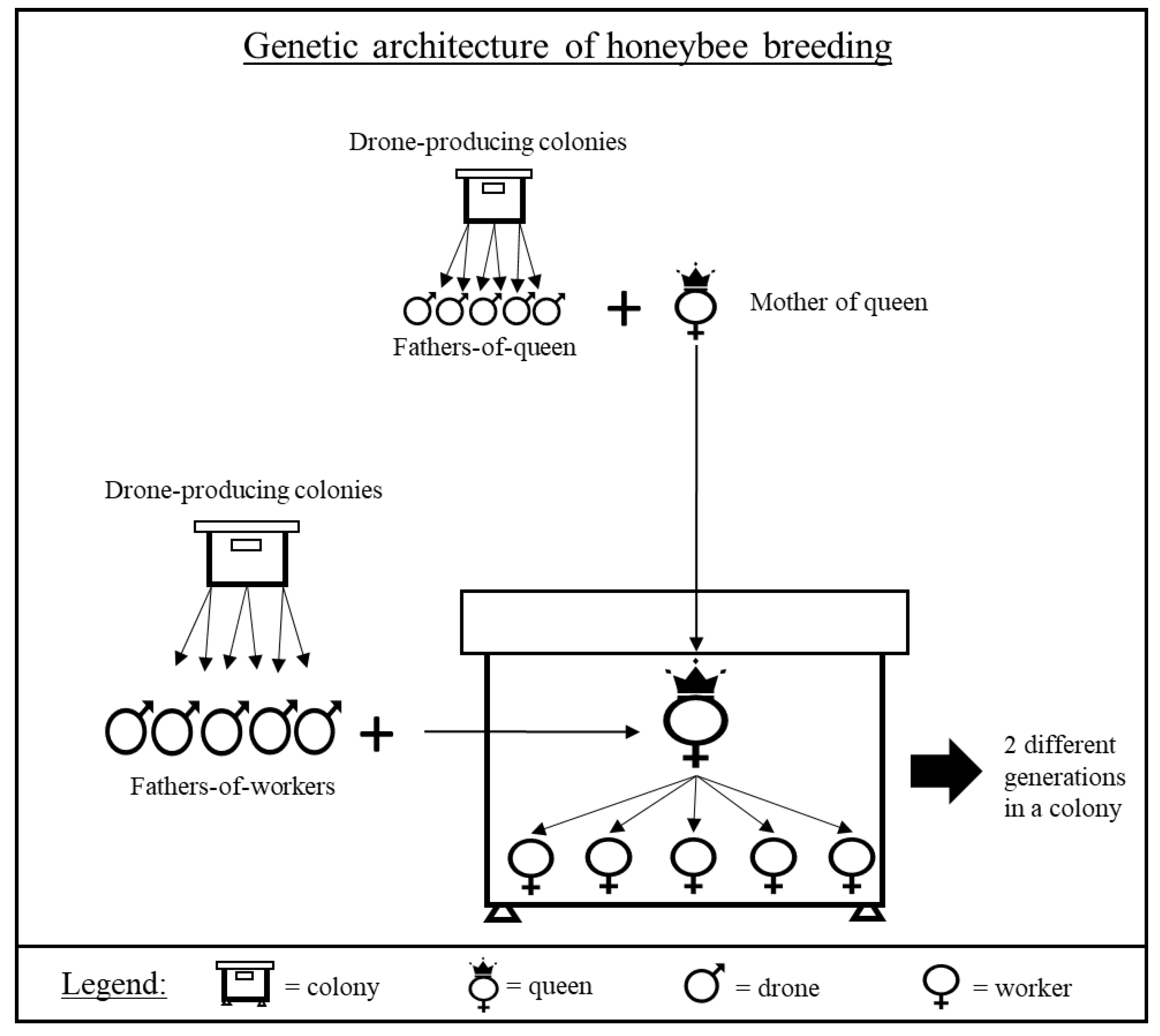



| Generation | Number of Colonies | Number of Queen-Producing Colonies for Breeding of Next Generation |
|---|---|---|
| 2010 | 26 | 7 |
| 2011 | 60 | 11 |
| 2012 | 38 | 12 |
| 2013 | 45 | 13 |
| 2014 | 109 | 11 |
| 2015 | 144 | 14 |
| 2016 | 97 | 9 |
| 2017 | 85 | 11 |
| Total | 604 |
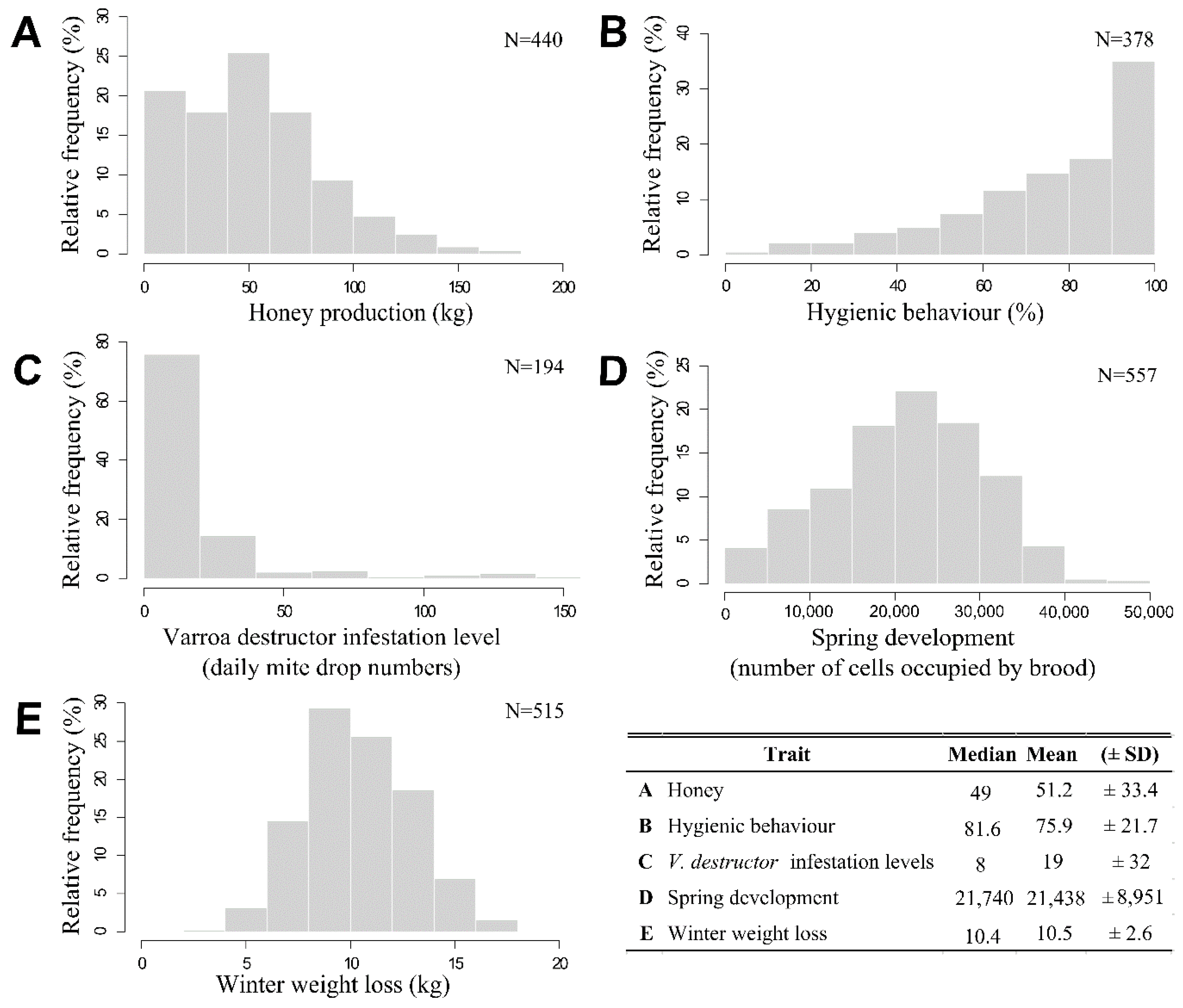
| Trait | n | VA (±SE) | VR (±SE) | h2 (±SE) |
|---|---|---|---|---|
| Production trait | ||||
| Honey production (kg) | 440 | 74.6 (±50.3) | 300.2 (±47.6) | 0.20 (±0.13) |
| Health traits | ||||
| Hygienic behavior (%) | 378 | 0.32 (±0.24) | 1.45 (±0.23) | 0.18 (±0.13) |
| Varroa destructor infestation levels (daily mite drop numbers) | 194 | 0.35 (±0.47) | 0.43 (±0.41) | 0.44 (±0.56) |
| Hardiness traits | ||||
| Spring development (number of cells occupied by brood) | 557 | 11,098,058 (±5,514,355) | 25,989,661 (±4,731,354) | 0.30 (±0.14) |
| Winter weight loss (kg) | 515 | 0.40 (±0.37) | 3.35 (±0.41) | 0.11 (±0.09) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maucourt, S.; Fortin, F.; Robert, C.; Giovenazzo, P. Genetic Parameters of Honey Bee Colonies Traits in a Canadian Selection Program. Insects 2020, 11, 587. https://doi.org/10.3390/insects11090587
Maucourt S, Fortin F, Robert C, Giovenazzo P. Genetic Parameters of Honey Bee Colonies Traits in a Canadian Selection Program. Insects. 2020; 11(9):587. https://doi.org/10.3390/insects11090587
Chicago/Turabian StyleMaucourt, Ségolène, Frédéric Fortin, Claude Robert, and Pierre Giovenazzo. 2020. "Genetic Parameters of Honey Bee Colonies Traits in a Canadian Selection Program" Insects 11, no. 9: 587. https://doi.org/10.3390/insects11090587
APA StyleMaucourt, S., Fortin, F., Robert, C., & Giovenazzo, P. (2020). Genetic Parameters of Honey Bee Colonies Traits in a Canadian Selection Program. Insects, 11(9), 587. https://doi.org/10.3390/insects11090587





