RNAi-Based Functional Genomics in Hemiptera
Abstract
Simple Summary
Abstract
1. Introduction
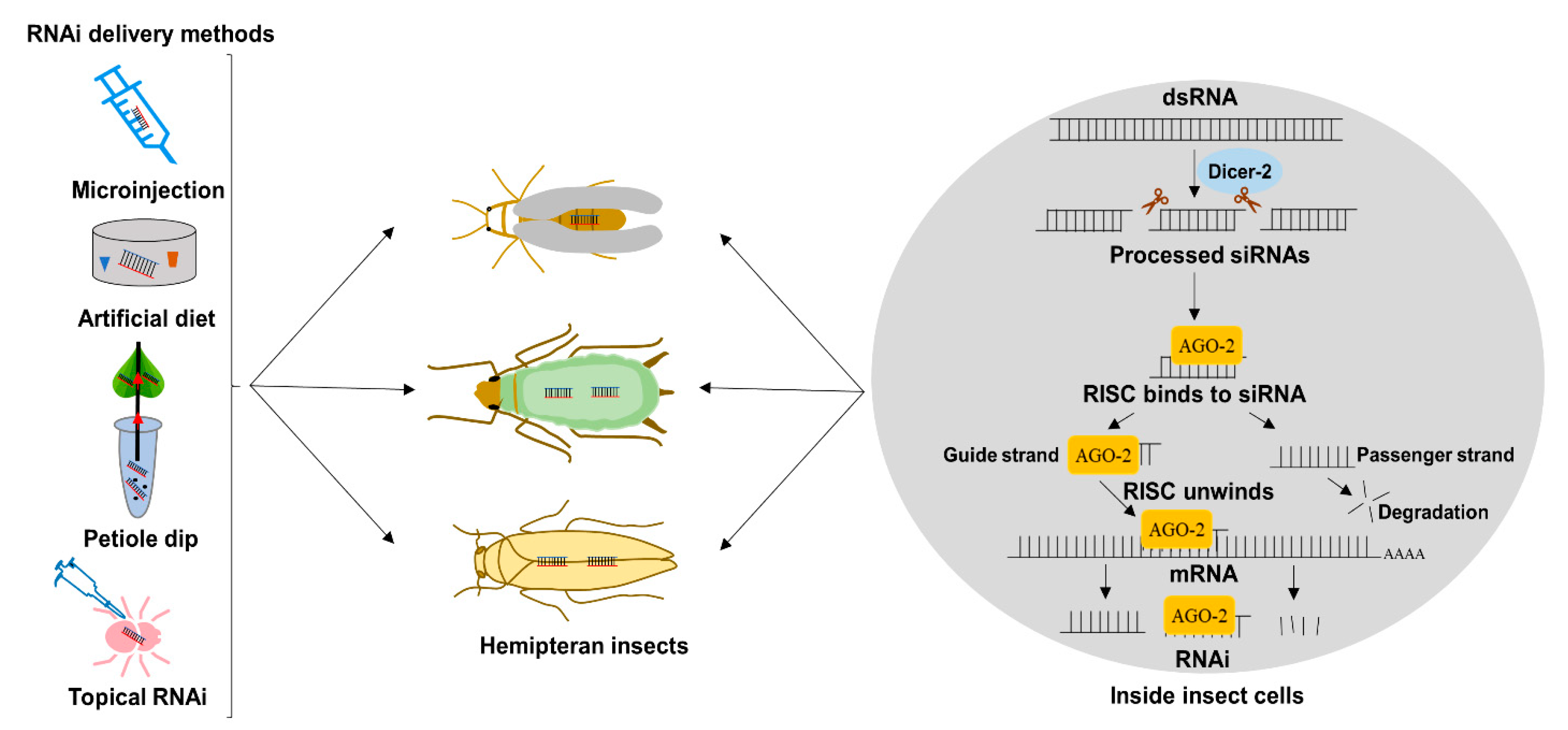
2. RNAi Delivery Strategies for Functional Genomics Studies in Hemiptera
2.1. Delivery by Microinjection
2.2. Delivery by Artificial Diet
2.3. Delivery of dsRNA by Topical Application to Insect and Plants
3. Variables Affecting RNAi Phenotypes in Functional Genomics Studies in Hemipteran Insects
3.1. Life-Stage of Insect
3.2. Target Gene Selection
3.3. Nucleases
3.4. Core RNAi Machinery
4. Applications of RNAi in Functional Genomic Studies of Hemiptera Insects
4.1. Parental RNAi
4.2. Embryonic RNAi
4.3. Post-Embryonic RNAi
4.4. Life Stage-Specific RNAi
4.5. RNAi in Studying Reproduction-Related Genes
4.6. RNAi in Behavioural Biology
4.7. RNAi in Exploring Biosynthetic Pathways
4.8. RNAi in Immunity-Related Genes
4.9. RNAi Studies to Uncover the Mechanism of Resistance Against Chemicals
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Panfilio, K.A.; Angelini, D.R. By land, air, and sea: Hemipteran diversity through the genomic lens. Curr. Opin. Insect Sci. 2018, 25, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Panfilio, K.A.; Jentzsch, I.M.V.; Benoit, J.B.; Erezyilmaz, D.; Suzuki, Y.; Colella, S.; Robertson, H.M.; Poelchau, M.F.; Waterhouse, R.M.; Ioannidis, P.; et al. Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Boil. 2019, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Bellés, X. Beyond Drosophila: RNAi In Vivo and Functional Genomics in Insects. Annu. Rev. Èntomol. 2010, 55, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Legeai, F.; Shigenobu, S.; Gauthier, J.-P.; Colbourne, J.K.; Rispe, C.; Collin, O.; Richards, S.; Wilson, A.C.C.; Murphy, T.; Tagu, D. AphidBase: A centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Boil. 2010, 19, 5–12. [Google Scholar] [CrossRef]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Boil. 2016, 14, 1–15. [Google Scholar] [CrossRef]
- Lin, S.-C.; Chang, Y.-Y.; Chan, C.-C. Strategies for gene disruption in Drosophila. Cell Biosci. 2014, 4, 63. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.-W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9. [Google Scholar] [CrossRef]
- Lim, Z.X.; Robinson, K.E.; Jain, R.; Chandra, G.S.; Asokan, R.; Asgari, S.; Mitter, N. Diet-delivered RNAi in Helicoverpa armigera—Progresses and challenges. J. Insect Physiol. 2016, 85, 86–93. [Google Scholar] [CrossRef]
- Bally, J.; Fishilevich, E.; Bowling, A.J.; Pence, H.E.; Narva, K.E.; Waterhouse, P.M. Improved insect-proofing: Expressing double-stranded RNA in chloroplasts. Pest Manag. Sci. 2018, 74, 1751–1758. [Google Scholar] [CrossRef]
- Yu, X.-D.; Liu, Z.; Huang, S.-L.; Chen, Z.-Q.; Sun, Y.; Duan, P.-F.; Ma, Y.-Z.; Xia, L.-Q. RNAi-mediated plant protection against aphids. Pest Manag. Sci. 2016, 72, 1090–1098. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Dyson, P.J. Gene silencing in non-model insects: Overcoming hurdles using symbiotic bacteria for trauma-free sustainable delivery of RNA interference. Bioessays 2017, 39, 1600247. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.-W.; Zhuo, J.-C.; Zhang, C.-X.; Wang, D. Characterization of NlHox3, an essential gene for embryonic development in Nilaparvata lugens. Arch. Insect Biochem. Physiol. 2018, 98, e21448. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-L.; An, Z.-F.; Liu, X.-D. Wingless gene cloning and its role in manipulating the wing dimorphism in the white-backed planthopper, Sogatella furcifera. BMC Mol. Boil. 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Amiri, A.; Bandani, A.R.; Alizadeh, H. Molecular identification of cysteine and trypsin protease, effect of different hosts on protease expression, and RNAi mediated silencing of cysteine protease gene in the sunn pest. Arch. Insect Biochem. Physiol. 2015, 91, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.D.; Wouters, R.H.M.; Mugford, S.T.; Hogenhout, S.A. Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 2014, 66, 541–548. [Google Scholar] [CrossRef]
- Shang, F.; Niu, J.-Z.; Ding, B.-Y.; Zhang, Q.; Ye, C.; Zhang, W.; Smagghe, G.; Wang, J.-J. Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Boil. 2017, 27, 221–233. [Google Scholar] [CrossRef]
- Yu, N.; Christiaens, O.; Liu, J.; Niu, J.; Cappelle, K.; Caccia, S.; Huvenne, H.; Smagghe, G. Delivery of dsRNA for RNAi in insects: An overview and future directions. Insect Sci. 2012, 20, 4–14. [Google Scholar] [CrossRef]
- Will, T.; Vilcinskas, A. The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Boil. 2015, 57, 34–40. [Google Scholar] [CrossRef]
- Ghanim, M.; Kontsedalov, S.; Czosnek, H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem. Mol. Boil. 2007, 37, 732–738. [Google Scholar] [CrossRef]
- Rosa, C.; Kamita, S.G.; Falk, B.W. RNA interference is induced in the glassy winged sharpshooter Homalodisca vitripennis by actin dsRNA. Pest Manag. Sci. 2012, 68, 995–1002. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Katoch, R.; Sethi, A.; Thakur, N.; Murdock, L.L. RNAi for Insect Control: Current Perspective and Future Challenges. Appl. Biochem. Biotechnol. 2013, 171, 847–873. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, H.-C.; Miao, X.-X. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2012, 20, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, D.; Yao, Q.; Zhang, J.; Dong, X.; Tian, H.; Zhang, W. Feeding-based RNA interference of atrehalose phosphate synthasegene in the brown planthopper, Nilaparvata lugens. Insect Mol. Boil. 2010, 19, 777–786. [Google Scholar] [CrossRef]
- Price, D.R.; Gatehouse, J.A. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008, 26, 393–400. [Google Scholar] [CrossRef]
- Guo, L.; Xie, W.; Liu, Y.; Yang, Z.; Yang, X.; Xia, J.; Wang, S.; Wu, Q.; Zhang, Y. Identification and characterization of doublesex in Bemisia tabaci. Insect Mol. Boil. 2018, 27, 620–632. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Y.; Francis, F.; Cheng, D.; Sun, J.; Chen, J.-L. Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem. Mol. Boil. 2015, 64, 16–24. [Google Scholar] [CrossRef]
- Vyas, M.; Raza, A.; Ali, M.Y.; Ashraf, M.A.; Mansoor, S.; Shahid, A.A.; Brown, J.K. Knock down of Whitefly Gut Gene Expression and Mortality by Orally Delivered Gut Gene-Specific dsRNAs. PLoS ONE 2017, 12, e0168921. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; Singh, P.K.; Tuli, R. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef]
- Tariq, K.; Ali, A.; Davies, T.G.E.; Naz, E.; Naz, L.; Sohail, S.; Hou, M.; Ullah, F. RNA interference-mediated knockdown of voltage-gated sodium channel (MpNav) gene causes mortality in peach-potato aphid, Myzus persicae. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Araujo, R.; Santos, A.; Pinto, F.; Gontijo, N.F.; Lehane, M.; Pereira, M.H. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Boil. 2006, 36, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.W.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2018, 75, 18–28. [Google Scholar] [CrossRef]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 2018, 75, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Boil. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Niu, J.; Yang, W.; Tian, Y.; Fan, J.; Ye, C.; Shang, F.; Ding, B.; Zhang, J.; An, X.; Yang, L.; et al. Topical dsRNA delivery induces gene silencing and mortality in the pea aphid. Pest Manag. Sci. 2019, 75, 2873–2881. [Google Scholar] [CrossRef]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-Stranded RNA Uptake through Topical Application, Mediates Silencing of Five CYP4 Genes and Suppresses Insecticide Resistance in Diaphorina citri. PLoS ONE 2014, 9, e110536. [Google Scholar] [CrossRef]
- Zheng, Y.; Hu, Y.; Yan, S.; Zhou, H.; Song, D.; Yin, M.; Shen, J.; Songm, D. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag. Sci. 2019, 75, 1993–1999. [Google Scholar] [CrossRef]
- Thairu, M.W.; Skidmore, I.H.; Bansal, R.; Nováková, E.; Hansen, T.E.; Li-Byarlay, H.; Wickline, S.A.; Hansen, A.K. Efficacy of RNA interference knockdown using aerosolized short interfering RNAs bound to nanoparticles in three diverse aphid species. Insect Mol. Boil. 2017, 14, 356–368. [Google Scholar] [CrossRef]
- Hunter, W.B.; Glick, E.; Paldi, N.; Bextine, B.R. Advances in RNA interference: dsRNA Treatment in Trees and Grapevines for Insect Pest Suppression. Southwest Èntomol. 2012, 37, 85–87. [Google Scholar] [CrossRef]
- Ghosh, S.K.B.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double strand RNA delivery system for plant-sap-feeding insects. PLoS ONE 2017, 12, e0171861. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Figueira, A. Management of insect pest by RNAi—A new tool for crop protection. In RNA Interference; Abdurakhmonov, I.Y., Ed.; InTechOpen: Rijeka, Croatia, 2016; pp. 371–390. [Google Scholar]
- Powell, M.; Pyati, P.; Cao, M.; Bell, H.; Gatehouse, J.A.; Fitches, E.C. Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Riga, M.; Denecke, S.; Livadaras, I.; Geibel, S.; Nauen, R.; Vontas, J. Development of efficient RNAi in Nezara viridula for use in insecticide target discovery. Arch. Insect Biochem. Physiol. 2019, 103, e21650. [Google Scholar] [CrossRef] [PubMed]
- Vélez, A.M.; Fishilevich, E.; Matz, N.; Storer, N.P.; Narva, K.; Siegfried, B.D. Parameters for Successful Parental RNAi as An Insect Pest Management Tool in Western Corn Rootworm, Diabrotica virgifera virgifera. Genes 2016, 8, 7. [Google Scholar] [CrossRef]
- Pinzón, N.; Bertrand, S.; Subirana, L.; Busseau, I.; Escriva, H.; Seitz, H. Functional lability of RNA-dependent RNA polymerases in animals. PLoS Genet. 2019, 15, e1007915. [Google Scholar] [CrossRef]
- Zha, W.; Peng, X.; Chen, R.; Du, B.; Zhu, L.; He, G. Knockdown of Midgut Genes by dsRNA-Transgenic Plant-Mediated RNA Interference in the Hemipteran Insect Nilaparvata lugens. PLoS ONE 2011, 6, e20504. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef]
- Jaubert-Possamai, S.; Le Trionnaire, G.; Bonhomme, J.; Christophides, G.K.; Rispe, C.; Tagu, D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007, 7, 63. [Google Scholar] [CrossRef]
- Allen, M.L.; Walker, W.B., III. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef]
- Ghodke, A.B.; Good, R.T.; Golz, J.F.; Russell, D.A.; Edwards, O.; Robin, C. Extracellular endonucleases in the midgut of Myzus persicae may limit the efficacy of orally delivered RNAi. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Van Eck, J.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Boil. 2017, 88, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Peters, R.S.; Mayer, C.; Liu, S.; Zhou, X.; et al. Phylogenetic Origin and Diversification of RNAi Pathway Genes in Insects. Genome Boil. Evol. 2016, 8, 3784–3793. [Google Scholar] [CrossRef] [PubMed]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Boil. 2008, 9, R10. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; An, X.; Jiang, Y.-D.; Ding, B.-Y.; Shang, F.; Christiaens, O.; Taning, C.N.T.; Smagghe, G.; Niu, J.; Wang, J.-J. Induction of RNAi Core Machinery’s Gene Expression by Exogenous dsRNA and the Effects of Pre-exposure to dsRNA on the Gene Silencing Efficiency in the Pea Aphid (Acyrthosiphon pisum). Front. Physiol. 2019, 9, 1906. [Google Scholar] [CrossRef]
- Bansal, R.; Michel, A.P. Core RNAi Machinery and Sid1, a Component for Systemic RNAi, in the Hemipteran Insect, Aphis glycines. Int. J. Mol. Sci. 2013, 14, 3786–3801. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Dixit, S.; Sharma, S.; Singh, H.; Kumar, J.; Verma, P.C.; Chandrashekar, K. siRNA Machinery in Whitefly (Bemisia tabaci). PLoS ONE 2013, 8, e83692. [Google Scholar] [CrossRef]
- Nicholson, S.J.; Nickerson, M.L.; Dean, M.; Song, Y.; Hoyt, P.; Rhee, H.; Kim, C.; Puterka, G.J. The genome of Diuraphis noxia, a global aphid pest of small grains. BMC Genomics 2015, 16, 429. [Google Scholar] [CrossRef]
- Taning, C.N.T.; De Andrade, E.C.; Hunter, W.B.; Christiaens, O.; Smagghe, G. Asian Citrus Psyllid RNAi Pathway—RNAi evidence. Sci. Rep. 2016, 6, 38082. [Google Scholar] [CrossRef]
- Sparks, M.E.; Shelby, K.S.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the Invasive Brown Marmorated Stink Bug, Halyomorpha halys (Stål) (Heteroptera: Pentatomidae). PLoS ONE 2014, 9, e111646. [Google Scholar] [CrossRef]
- Davis-Vogel, C.; Van Allen, B.; Van Hemert, J.L.; Sethi, A.; Nelson, M.E.; Sashital, D.G. Identification and comparison of key RNA interference machinery from western corn rootworm, fall armyworm, and southern green stink bug. PLoS ONE 2018, 13, e0203160. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Zhan, S.; Huang, Y.P. Genome editing in insects: Current status and challenges. Natl. Sci. Rev. 2019, 6, 399–401. [Google Scholar] [CrossRef]
- Swevers, L.; Smagghe, G. Use of RNAi for Control of Insect Crop Pests; Springer Science and Business Media LLC: Berlin, Germany, 2012; pp. 177–197. [Google Scholar]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.-C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef]
- Lozano, J.; Gómez-Orte, E.; Lee, H.-J.; Bellés, X.; Lozano-Fernandez, J. Super-induction of Dicer-2 expression by alien double-stranded RNAs: An evolutionary ancient response to viral infection? Dev. Genes Evol. 2012, 222, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Sparks, C.; Jones, H.; Riley, M.; Francis, F.; Du, W.; Xia, L. Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants. Plant Biotechnol. J. 2019, 17, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Hattori, M. Gene silencing by parental RNA interference in the green rice leafhopper, Nephotettix cincticeps (hemiptera: Cicadellidae). Arch. Insect Biochem. Physiol. 2016, 91, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, E.; Vélez, A.M.; Khajuria, C.; Frey, M.L.; Hamm, R.L.; Wang, H.; Schulenberg, G.A.; Bowling, A.J.; Pence, H.E.; Gandra, P.; et al. Use of chromatin remodeling ATPases as RNAi targets for parental control of western corn rootworm (Diabrotica virgifera virgifera) and Neotropical brown stink bug (Euschistus heros). Insect Biochem. Mol. Boil. 2016, 71, 58–71. [Google Scholar] [CrossRef]
- Brito, T.; Julio, A.; Berni, M.; Poncio, L.D.C.; Bernardes, E.S.; Araújo, H.M.M.; Sammeth, M.; Pane, A. Transcriptomic and functional analyses of the piRNA pathway in the Chagas disease vector Rhodnius prolixus. PLoS Negl. Trop. Dis. 2018, 12, e0006760. [Google Scholar] [CrossRef]
- Will, T.; Schmidtberg, H.; Skaljac, M.; Vilcinskas, A. Heat shock protein 83 plays pleiotropic roles in embryogenesis, longevity, and fecundity of the pea aphid Acyrthosiphon pisum. Dev. Genes Evol. 2016, 227, 1–9. [Google Scholar] [CrossRef]
- Simonet, P.; Gaget, K.; Parisot, N.; Duport, G.; Rey, M.; Febvay, G.; Charles, H.; Callaerts, P.; Colella, S.; Calevro, F. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Bewick, A.J.; Sanchez, Z.; McKinney, E.C.; Moore, A.J.; Moore, P.J.; Schmitz, R.J. Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus. Epigenet. Chromatin 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Panfilio, K.A. Late extraembryonic morphogenesis and its zenRNAi-induced failure in the milkweed bug Oncopeltus fasciatus. Dev. Boil. 2009, 333, 297–311. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Walter-Nuno, A.B.; Oliveira, M.P.; Oliveira, M.F.; Gonçalves, R.L.; Ramos, I.B.; Koerich, L.B.; Oliveira, P.L.; Paiva-Silva, G.O. Silencing of Maternal Heme-binding Protein Causes Embryonic Mitochondrial Dysfunction and Impairs Embryogenesis in the Blood Sucking Insect Rhodnius prolixus. J. Boil. Chem. 2013, 288, 29323–29332. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Kikuchi, Y.; Miura, T.; Fukatsu, T. Ultrabithorax is essential for bacteriocyte development. Proc. Natl. Acad. Sci. USA 2015, 112, 9376–9381. [Google Scholar] [CrossRef]
- Chesebro, J.; Hrycaj, S.; Mahfooz, N.; Popadic, A. Diverging functions of Scr between embryonic and post-embryonic development in a hemimetabolous insect, Oncopeltus fasciatus. Dev. Biol. 2009, 329, 142–151. [Google Scholar] [CrossRef]
- Yan, T.; Chen, H.; Sun, Y.; Yu, X.-D.; Xia, L. RNA Interference of the Ecdysone Receptor Genes EcR and USP in Grain Aphid (Sitobion avenae) Affects Its Survival and Fecundity upon Feeding on Wheat Plants. Int. J. Mol. Sci. 2016, 17, 2098. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, M.; Reding, K.; Pick, L. Establishment of molecular genetic approaches to study gene expression and function in an invasive hemipteran, Halyomorpha halys. Evodevo 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Li, X.; Liu, F.; Wu, C.; Zhao, J.; Cai, W.; Hua, H. Decapentaplegic function in wing vein development and wing morph transformation in brown planthopper, Nilaparvata lugens. Dev. Boil. 2019, 449, 143–150. [Google Scholar] [CrossRef]
- Lin, X.; Yao, Y.; Jin, M.; Li, Q. Characterization of the Distal-less gene homologue, NlDll, in the brown planthopper, Nilaparvata lugens (Stål). Gene 2014, 535, 112–118. [Google Scholar] [CrossRef]
- Wu, W.-J.; Wang, Y.; Huang, H.-J.; Bao, Y.-Y.; Zhang, C. Ecdysone receptor controls wing morphogenesis and melanization during rice planthopper metamorphosis. J. Insect Physiol. 2012, 58, 420–426. [Google Scholar] [CrossRef]
- El-Shesheny, I.; Hajeri, S.; El-Hawary, I.; Gowda, S.; Killiny, N. Silencing Abnormal Wing Disc Gene of the Asian Citrus Psyllid, Diaphorina citri Disrupts Adult Wing Development and Increases Nymph Mortality. PLoS ONE 2013, 8, e65392. [Google Scholar] [CrossRef]
- Xu, N.; Chen, H.; Xue, W.; Yuan, X.; Xia, P.; Xu, H.-J. The MTase15 regulates reproduction in the wing-dimorphic planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Boil. 2019, 28, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Huang, B.; Jiang, Y.-P.; Gu, H.-T.; Xia, T.; Yang, G.-Q.; Liu, F.; Wu, J.-C. Carboxylesterase Precursor (EST-1) Mediated the Fungicide Jinggangmycin-Suppressed Reproduction of Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Èntomol. 2017, 110, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Blariza, M.J.; Grosso, C.G.; García, B.A. Silencing of Two Vitellogenin Genes Inhibits Oviposition in the Chagas Disease Vector Triatoma infestans (Hemiptera: Reduviidae). Am. J. Trop. Med. Hyg. 2017, 97, 477–480. [Google Scholar] [CrossRef]
- Basnet, S.; Kamble, S.T. Knockdown of the Chromatin Remodeling Gene Brahma by RNA Interference Reduces Reproductive Fitness and Lifespan in Common Bed Bug (Hemiptera: Cimicidae). J. Med Èntomol. 2017, 55, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Smykal, V.; Bajgar, A.; Provazník, J.; Fexova, S.; Buricova, M.; Takaki, K.; Hodkova, M.; Jindra, M.; Dolezel, D. Juvenile hormone signaling during reproduction and development of the linden bug, Pyrrhocoris apterus. Insect Biochem. Mol. Boil. 2014, 45, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-D.; Yuan, X.; Bai, Y.-L.; Wang, G.-Y.; Zhou, W.-W.; Zhu, Z.-R. Knockdown of timeless Disrupts the Circadian Behavioral Rhythms in Laodelphax striatellus (Hemiptera: Delphacidae). Environ. Èntomol. 2018, 47, 1216–1225. [Google Scholar] [CrossRef]
- Lan, H.; Hong, X.; Huang, R.; Lin, X.; Li, Q.; Li, K.; Zhou, T. RNA interference-mediated knockdown and virus-induced suppression of Troponin C gene adversely affect the behavior or fitness of the green rice leafhopper, Nephotettix cincticeps. Arch. Insect Biochem. Physiol. 2017, 97, e21438. [Google Scholar] [CrossRef]
- Zhang, X.; Pengsakul, T.; Tukayo, M.; Yu, L.; Fang, W.; Luo, D. Host-location behavior of the tea green leafhopper Empoasca vitis Göthe (Hemiptera: Cicadellidae): Olfactory and visual effects on their orientation. Bull. Èntomol. Res. 2017, 108, 423–433. [Google Scholar] [CrossRef]
- Li, X.; Qu, M.-J.; Zhang, Y.; Li, J.-W.; Liu, T. Expression of Neuropeptide F Gene and Its Regulation of Feeding Behavior in the Pea Aphid, Acyrthosiphon pisum. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Paim, R.M.M.; Araujo, R.; Lehane, M.J.; Gontijo, N.F.; Pereira, M.H. Long-term effects and parental RNAi in the blood feeder Rhodnius prolixus (Hemiptera; Reduviidae). Insect Biochem. Mol. Boil. 2013, 43, 1015–1020. [Google Scholar] [CrossRef]
- Ikeno, T.; Ishikawa, K.; Numata, H.; Goto, S.G. Circadian clock gene Clockis involved in the photoperiodic response of the bean bugRiptortus pedestris. Physiol. Èntomol. 2013, 38, 157–162. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Du, J.; Zhang, J.; Shen, J.; Cai, W. Three Melanin Pathway Genes, TH, yellow, and aaNAT, Regulate Pigmentation in the Twin-Spotted Assassin Bug, Platymeris biguttatus (Linnaeus). Int. J. Mol. Sci. 2019, 20, 2728. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-J.; Li, Z.-X. The terpenoid backbone biosynthesis pathway directly affects the biosynthesis of alarm pheromone in the aphid. Insect Mol. Boil. 2018, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, M.M.; Parks, M.C.; Tibbetts, A.E.; Swart, J.S.; Richards, E.; Vanegas, J.C.; Cenzer, M.L.; Crowley, L.; Simmons, W.R.; Hou, W.S.; et al. Manipulation of insulin signaling phenocopies evolution of a host-associated polyphenism. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.-J.; Dong, X.-L.; Kang, K.; Chen, H.; Dai, X.-Y.; Wu, G.-A.; Zheng, L.; Yu, Y.; Zhai, Y.-F. FoxO Transcription Factor Regulate Hormone Mediated Signaling on Nymphal Diapause. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, X.; Gu, S.-H.; Zhou, Y.; Wang, S.; Guo, Y.-Y. Silencing of odorant binding protein gene AlinOBP4 by RNAi induces declining electrophysiological responses of Adelphocoris lineolatus to six semiochemicals. Insect Sci. 2016, 24, 789–797. [Google Scholar] [CrossRef]
- Walker, W.B.; Allen, M.L. Expression and RNA Interference of Salivary Polygalacturonase Genes in the Tarnished Plant Bug, Lygus lineolaris. J. Insect Sci. 2010, 10, 1–13. [Google Scholar] [CrossRef]
- Brent, C.S.; Hull, J.J. RNA interference-mediated knockdown of eye coloration genes in the western tarnished plant bug (Lygus hesperus Knight). Arch. Insect Biochem. Physiol. 2018, 100, e21527. [Google Scholar] [CrossRef]
- Luo, J.; Li, Z.; Ma, C.; Zhang, Z.; Hull, J.J.; Lei, C.; Jin, S.; Chen, L. Knockdown of a metathoracic scent gland desaturase enhances the production of (E)-4-oxo-2-hexenal and suppresses female sexual attractiveness in the plant bug Adelphocoris suturalis. Insect Mol. Boil. 2017, 26, 642–653. [Google Scholar] [CrossRef]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. Proc. R. Soc. B 2019, 286, 20182207. [Google Scholar] [CrossRef]
- Wang, Z.-Z.; Bing, X.-L.; Liu, S.-S.; Chen, X.-X. RNA interference of an antimicrobial peptide, Btdef, reduces Tomato yellow leaf curl China virus accumulation in the whitefly Bemisia tabaci. Pest Manag. Sci. 2016, 73, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Eliautout, R.; Dubrana, M.-P.; Vincent-Monégat, C.; Vallier, A.; Braquart-Varnier, C.; Poirié, M.; Saillard, C.; Heddi, A.; Arricau-Bouvery, N. Immune response and survival of Circulifer haematoceps to Spiroplasma citri infection requires expression of the gene hexamerin. Dev. Comp. Immunol. 2016, 54, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Redinbaugh, M.G.; Michel, A.P. Molecular interactions and immune responses between Maize fine streak virus and the leafhopper vector Graminella nigrifrons through differential expression and RNA interference. Insect Mol. Boil. 2015, 24, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zheng, L.; Mao, Q.; Liu, J.; Wang, H.; Jia, D.; Chen, H.; Wu, W.; Wei, T. Fibrillar structures induced by a plant reovirus target mitochondria to activate typical apoptotic response and promote viral infection in insect vectors. PLoS Pathog. 2019, 15, e1007510. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, X.; Ali, E.; Liao, X.; Jin, R.; Ren, Z.; Wan, H.; Li, J. Characterization of nitenpyram resistance in Nilaparvata lugens (Stål). Pestic. Biochem. Physiol. 2019, 157, 26–32. [Google Scholar] [CrossRef]
- Tian, F.; Li, C.; Wang, Z.; Liu, J.; Zeng, X. Identification of detoxification genes in imidacloprid-resistant Asian citrus psyllid (Hemiptera: Lividae) and their expression patterns under stress of eight insecticides. Pest Manag. Sci. 2018, 75, 1400–1410. [Google Scholar] [CrossRef]
- Chen, X.; Xia, J.; Shang, Q.; Song, D.; Gao, X. UDP-glucosyltransferases potentially contribute to imidacloprid resistance in Aphis gossypii glover based on transcriptomic and proteomic analyses. Pestic. Biochem. Physiol. 2019, 159, 98–106. [Google Scholar] [CrossRef]
- Gong, Y.-H.; Yu, X.-R.; Shang, Q.-L.; Shi, X.-Y.; Gao, X.-W. Oral Delivery Mediated RNA Interference of a Carboxylesterase Gene Results in Reduced Resistance to Organophosphorus Insecticides in the Cotton Aphid, Aphis gossypii Glover. PLoS ONE 2014, 9, e102823. [Google Scholar] [CrossRef]
- Xiao, D.; Lu, Y.-H.; Shang, Q.; Song, D.-L.; Gao, X. Gene silencing of two acetylcholinesterases reveals their cholinergic and non-cholinergic functions in Rhopalosiphum padi and Sitobion avenae. Pest Manag. Sci. 2014, 71, 523–530. [Google Scholar] [CrossRef]
- Thakur, N.; Mundey, J.K.; Upadhyay, S.K. RNAi—Implications in Entomological Research and Pest Control. RNA Interference; Abdurakhmonov, I.Y., Ed.; InTech: Rijeka, Croatia, 2016; pp. 341–369. [Google Scholar]
- Abdellatef, E.; Will, T.; Koch, A.M.; Imani, J.; Vilcinskas, A.; Kogel, K.-H. Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 2015, 13, 849–857. [Google Scholar] [CrossRef]
- Pick, L. Hox genes, evo-devo, and the case of the ftz gene. Chromosoma 2015, 125, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Angelini, D.R.; Liu, P.Z.; Hughes, C.L.; Kaufman, T.C. Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera). Dev. Boil. 2005, 287, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Z.; Kaufman, T.C. hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 2004, 131, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.-L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Sapountzis, P.; Duport, G.; Balmand, S.; Gaget, K.; Jaubert-Possamai, S.; Febvay, G.; Charles, H.; Rahbé, Y.; Colella, S.; Calevro, F. New insight into the RNA interference response against cathepsin-L gene in the pea aphid, Acyrthosiphon pisum: Molting or gut phenotypes specifically induced by injection or feeding treatments. Insect Biochem. Mol. Boil. 2014, 51, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Jiang, Y.-D.; An, X.; Yang, L.; Shang, F.; Niu, J.; Wang, J.-J. Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Perrimon, N.; Ni, J.-Q.; Perkins, L. In vivo RNAi: Today and Tomorrow. Cold Spring Harb. Perspect. Boil. 2010, 2, a003640. [Google Scholar] [CrossRef]
- Yang, Y.; Wan, P.-J.; Hu, X.-X.; Li, G.-Q. RNAi mediated knockdown of the ryanodine receptor gene decreases chlorantraniliprole susceptibility in Sogatella furcifera. Pestic. Biochem. Physiol. 2014, 108, 58–65. [Google Scholar] [CrossRef]
- Scott, J.G. Investigating Mechanisms of Insecticide Resistance: Methods, Strategies, and Pitfalls. In Pesticide Resistance in Arthropods; Springer Science and Business Media LLC: Berlin, Germany, 1990; pp. 39–57. [Google Scholar]
- Ding, B.-Y.; Shang, F.; Zhang, Q.; Xiong, Y.; Yang, Q.; Niu, J.; Smagghe, G.; Wang, J.-J. Silencing of Two Insulin Receptor Genes Disrupts Nymph-Adult Transition of Alate Brown Citrus Aphid. Int. J. Mol. Sci. 2017, 18, 357. [Google Scholar] [CrossRef]
- Moriyama, M.; Hosokawa, T.; Tanahashi, M.; Nikoh, N.; Fukatsu, T. Suppression of Bedbug’s Reproduction by RNA Interference of Vitellogenin. PLoS ONE 2016, 11, e0153984. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.W.; Jin, X.; Xu, X.L.; Zeng, B.P. Characterization of the akirin gene and its role in the NF-kappaB signaling pathway of Sogatella furcifera. Front. Physiol. 2018, 9, 1411. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Pan, Y.; Yang, C.; Gao, X.; Xi, J.; Wu, Y.; Huang, X.; Zhu, E.; Xin, X.; Zhan, C.; et al. Over-expression of CYP6A2 is associated with spirotetramat resistance and cross-resistance in the resistant strain of Aphis gossypii Glover. Pestic. Biochem. Physiol. 2016, 126, 64–69. [Google Scholar] [CrossRef] [PubMed]

| Insect Species | No. of Core RNAi Genes | Reference | ||
|---|---|---|---|---|
| Dcr2 | R2D2 | Ago2 | ||
| Acyrthosiphon pisum | 1 | 1 | 1 | [55] |
| Aphis glycines | 1 | 1 | 1 | [56] |
| Bemisia tabaci | 1 | 1 | 1 | [57] |
| Diuraphis noxia | 1 | 1 | 2 | [58] |
| Diaphorina citri | 1 | 0 | 1 | [59] |
| Halyomorpha halys | 1 | 0 | 1 | [60] |
| Myzus persicae | 1 | 1 | 2 | [51] |
| Nezara viridula | 1 | 2 | 2 | [61] |
| Nilaparvata lugens | 1 | 1 | 1 | [62] |
| RNAi Studies | Family | Insect Species | Target Gene and Studied Functions | Ref |
|---|---|---|---|---|
| Parental RNAi | Aphididae | Sitobion avenae | Zinc finger protein SaZFP | [66] |
| Myzus persicae | Salivary proteins MpC002, MpPInto2 | [15] | ||
| Cicadellidae | Nephotettix cincticeps | Role of Laccase-2 in the cuticle pigmentation | [67] | |
| Pentatomidae | Euschistus heros | Chromatin-remodeling ATPases, brahma, mi-2, iswi, parental gene silencing | [68] | |
| Reduviidae | Rhodnius prolixus | piwi orthologs, role in oogenesis | [69] | |
| Embryonic RNAi | Delphacidae | Nilaparvata lugens | Hox3-like gene involved in embryonic development | [12] |
| Aphididae | Acyrthosiphon pisum | Heat shock protein 83 (HSP83) crucial in fecundity and embryogenesis | [70] | |
| Phenylalanine hydroxylase important for embryonic development | [71] | |||
| Lygaeidae | Oncopeltus fasciatus | DNA methyltransferase 1 (Dnmt1) essential for egg production and embryo viability | [72] | |
| Zen gene, role in mid embryogenesis | [73] | |||
| Reduviidae | Rhodnius prolixus | Heme-binding protein, role in embryogenesis | [74] | |
| Post-embryonic RNAi | Lygaeidae | Nysius plebeius | Ultrabithorax require for bacteriocyte development | [75] |
| Oncopeltus fasciatus | Sex combs reduced (Scr), role in wing suppression and wing program | [76] | ||
| Aphididae | Sitobion avenae | Ecdysone receptor (EcR) and ultraspiracle (USP) essential in growth and development | [77] | |
| Scutelleridae | Eurygaster integriceps | Cysteine gene involved in growth and development | [14] | |
| Pentatomidae | Halyomorpha halys | Sex combs reduced (Scr) involve in development | [78] | |
| Regeneration-dependant RNAi | Delphacidae | Nilaparvata lugens | Decapentaplegic gene, role in wing vein development and wing morph transformation | [79] |
| Distal-less gene homologue, NlDll, role in leg development and wing structure | [80] | |||
| Sogatella furcifera | Role of Wingless gene (Wg) in the development and growth of wings | [13] | ||
| Laodelphax striatellus | Ecdysone receptor involved in wing morphogenesis and melanisation | [81] | ||
| Liviidae | Diaphorini citri | Abnormal wing disc (awd) gene involved in wing development and metamorphosis | [82] | |
| RNAi in studying reproduction-related genes | Delphacidae | Nilaparvata lugens | S-Adenosyl-l-methionine-dependent methyltransferases (SAMMTases), regulates reproduction | [83] |
| Sogatella furcifera | Carboxylesterase precursor (EST-1), role in fungicide suppressed reproduction | [84] | ||
| Reduviidae | Triatoma infestans | Vitellogenin Gene, role in ovipostion | [85] | |
| Aphididae | Aphis citricidus | Vitellogenin (Vg) and Vg receptor, Role in development and reproduction | [16] | |
| Aleyrodidae | Bemisia tabaci | Role of doublesex gene in sex determination | [26] | |
| Cimicidae | Cimex lectularius | Chromatin remodelling gene Brahma function in reproduction and survival | [86] | |
| Pyrrhocoridae | Pyrrhocoris apterus | Methoprene-tolerant (Met) involve in reproduction and development | [87] | |
| RNAi in behavioural biology | Delphacidae | Laodelphax striatellus | Odorant-binding proteins, role in host-seeking behaviour | [33] |
| Timeless gene crucial for circadian rhythms | [88] | |||
| Cicadellidae | Nephotettix cincticeps | Troponin C involve in behaviour and fitness | [89] | |
| Empoasca vitis Göthe | Opsin genes are critical for host orientation behaviour | [90] | ||
| Aphididae | Acyrthosiphon pisum | Neuropeptide F regulates feeding behaviour | [91] | |
| Sitobion avenae | SaveOrco gene, role in aphid’s response to pheromones | [27] | ||
| Reduviidae | Rhodnius prolixus | Nitrophorins, reduced anticoagulant activity and poor feeding behaviours | [92] | |
| Alydidae | Riptortus pedestris | Role of Clock gene in the circadian rhythm | [93] | |
| RNAi in exploring biosynthetic pathways | Reduviidae | Platymeris biguttatus | Tyrosine hydroxylase (TH), yellow, arylalkylamine-N-acetyltransferase (aaNAT), role in pigmentation | [94] |
| Aphididae | Aphis gossypii | Farnesyl diphosphate synthase (FPPS), role in the biosynthesis of alarm pheromone | [95] | |
| Rhopalidae | Jadera haematoloma | Forkhead-box O (FoxO) critical for the evolution of polyphenism | [96] | |
| Delphacidae | Laodelphax striatellus | Forkhead-box O (FoxO) control hormone-mediated signalling pathway in nymphal diapause | [97] | |
| Miridae | Adelphocoris lineolatus | Odorant binding proteins play a role in the identification of volatile compounds and sex pheromones | [98] | |
| Lygus lineolaris | Polygalacturonase gene mainly express in salivary gland demonstrating that PGs are salivary enzyme | [99] | ||
| Lygus hesperus Knight | Cardinal gene is essential for eye colouration | [100] | ||
| Adelphocoris suturalis | Desaturase-like genes, role in sex pheromones biosynthetic pathway | [101] | ||
| RNAi in immunity-related genes | Pentatomidae | Plautia stali | Immune deficiency (IMD) pathway genes, role in controlling arrays of microbes | [102] |
| Aleyrodidae | Bemisia tabaci | A defensin-like antimicrobial peptide is involved in begomovirus infection | [103] | |
| Reduviidae | Rhodnius prolixus | Immune-deficiency pathway (IMD) regulates the activity of the antimicrobial peptide | ||
| Cicadellidae | Circulifer haematoceps | Hexamerin gene is required for immune response and survival against the bacterium | [104] | |
| Graminella nigrifrons | Peptidoglycan recognition proteins involved in immune response and pathogen transmission | [105] | ||
| Recilia dorsalis | Caspase is involved in promoting virus infection | [106] | ||
| RNAi studies to uncover the mechanism of resistance against chemicals | Delphacidae | Nilaparvata lugens | Cytochrome P450 (CYP) genes involve in nitenpyram resistance | [107] |
| Liviidae | Diaphorini citri | CYP knockdown increased susceptibility to imidacloprid | [108] | |
| Aphididae | Aphis gossypii | UDP-glycosyltransferases (UGTs), role in imidacloprid resistance | [109] | |
| Carboxylesterase silencing led to increased sensitivity to organophosphorus | [110] | |||
| Rhopalosiphum padi | Acetylcholinesterase gene is responsible for resistance to pirimicarb and malathion | [111] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, R.G.; Robinson, K.E.; Fletcher, S.J.; Mitter, N. RNAi-Based Functional Genomics in Hemiptera. Insects 2020, 11, 557. https://doi.org/10.3390/insects11090557
Jain RG, Robinson KE, Fletcher SJ, Mitter N. RNAi-Based Functional Genomics in Hemiptera. Insects. 2020; 11(9):557. https://doi.org/10.3390/insects11090557
Chicago/Turabian StyleJain, Ritesh G., Karl E. Robinson, Stephen J. Fletcher, and Neena Mitter. 2020. "RNAi-Based Functional Genomics in Hemiptera" Insects 11, no. 9: 557. https://doi.org/10.3390/insects11090557
APA StyleJain, R. G., Robinson, K. E., Fletcher, S. J., & Mitter, N. (2020). RNAi-Based Functional Genomics in Hemiptera. Insects, 11(9), 557. https://doi.org/10.3390/insects11090557






