Population Genetic Structure of Codling Moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), in Different Localities and Host Plants in Chile
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Material
2.2. Microsatellite Analysis
2.3. Data Analysis
3. Results
3.1. Basic Statistics, HWE, and Linkage Disequilibrium
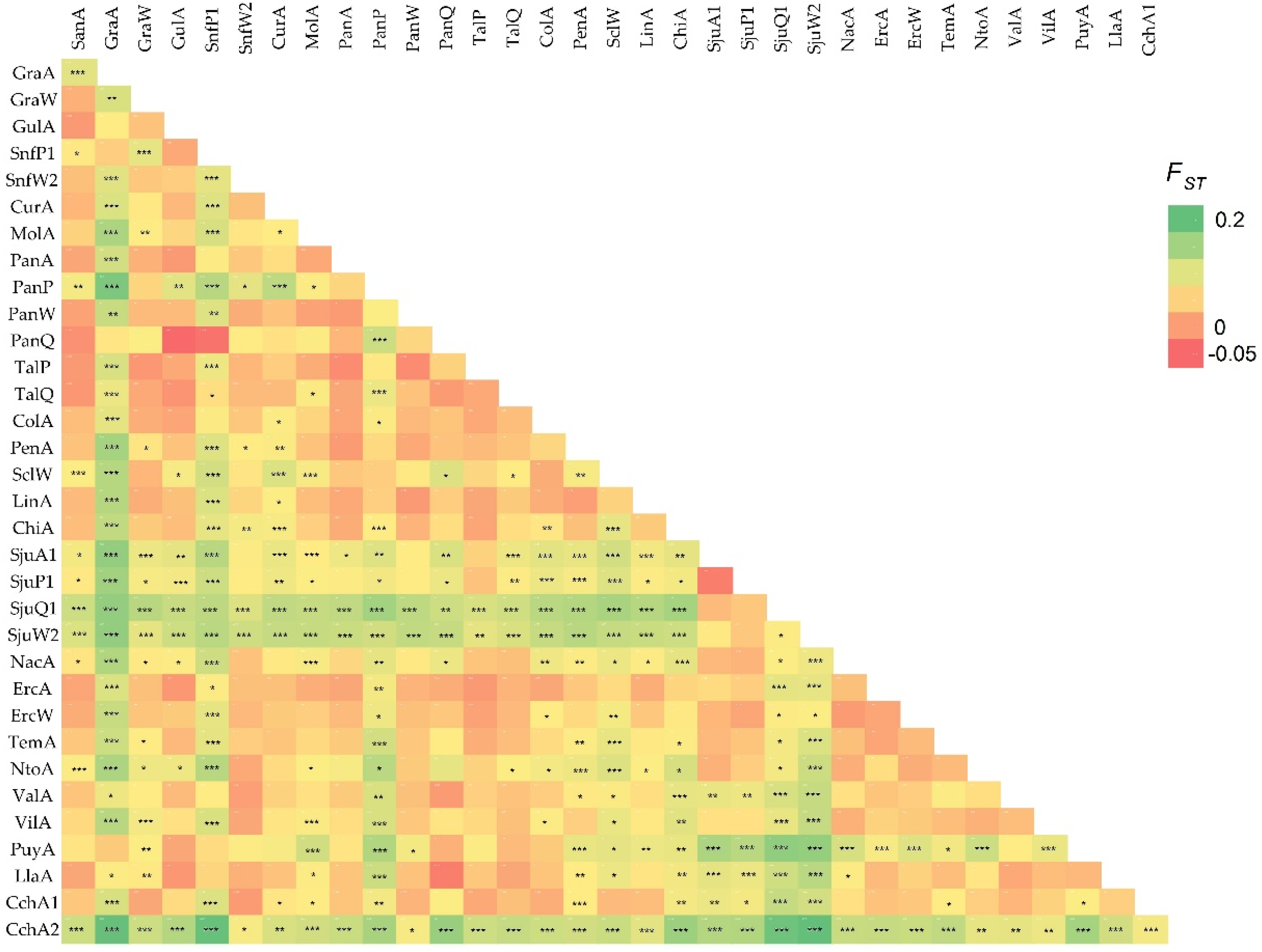
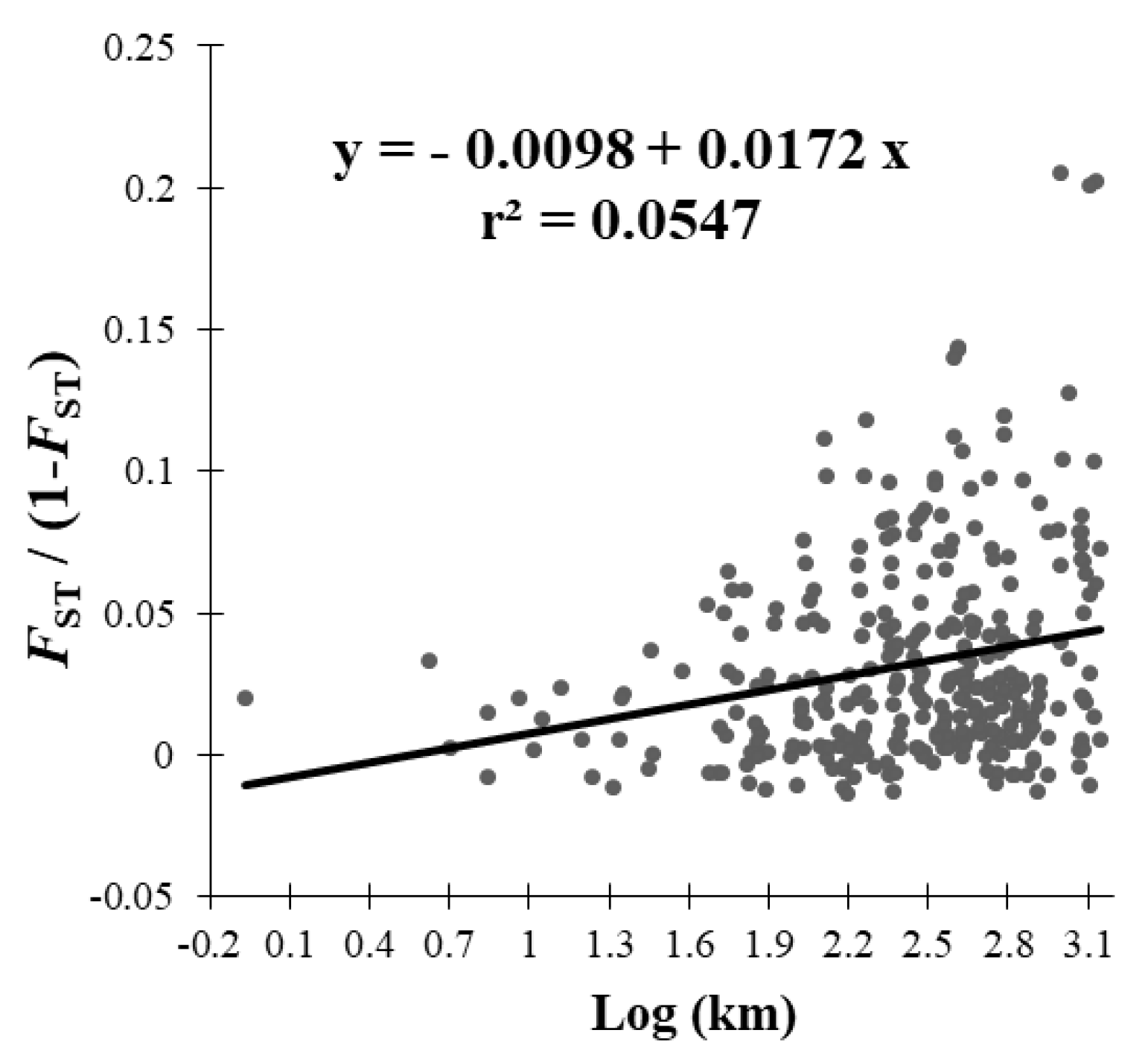
3.2. AMOVA and FST Analysis
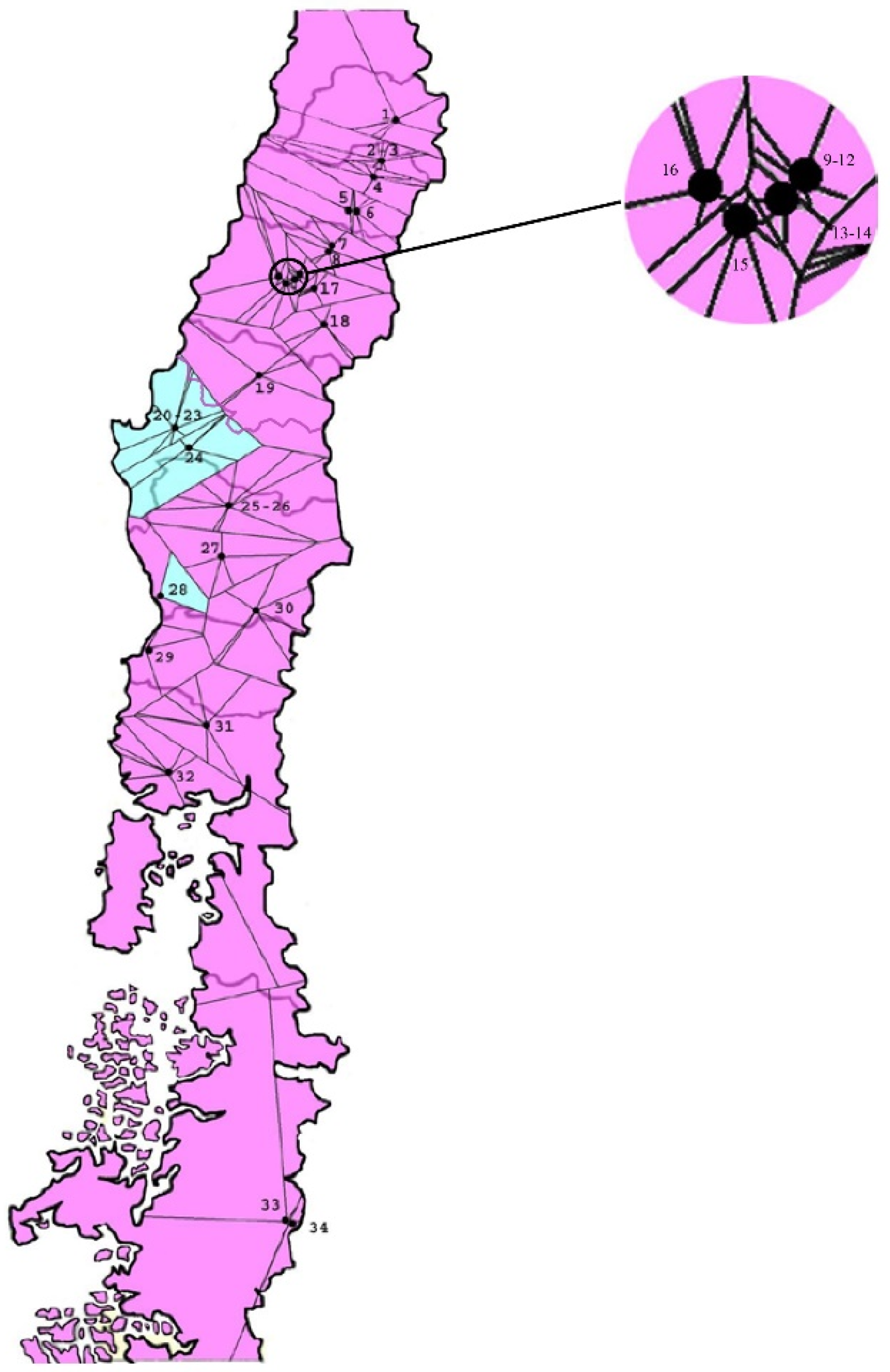
3.3. Bayesian Cluster Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, M.; Duan, X.; Li, Y.; Men, Q. Codling moth Cydia pomonella (L.). In Biological Invasions and Its Management in China; Wan, F., Jiang, M., Zhan, A., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 285–298. [Google Scholar]
- Knight, A.; Judd, G.J.R.; Gillian, T.; Fuentes-Contreras, E.; Walker, W.B. Integrated management of tortricid pests of tree fruits. In Integrated Management of Diseases and Insect Pests of Tree Fruit; Xu, X., Fountain, M., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2019; pp. 1–47. [Google Scholar]
- Willet, M.J.; Neven, L.; Miller, C.E. The occurrence of codling moth in low latitude countries: Validation of pest distribution reports. Horttechnology 2009, 19, 633–637. [Google Scholar] [CrossRef]
- Franck, P.; Timm, A.E. Population genetic structure of Cydia pomonella: A review and case study comparing spatiotemporal variation. J. Appl. Entomol. 2010, 134, 191–200. [Google Scholar] [CrossRef]
- Knight, A.L. Codling moth areawide integrated pest management. In Areawide Pest Management: Theory and Implementation; Koul, O., Cuperus, G.W., Elliott, N., Eds.; CAB International: Wallingford, UK, 2008; pp. 159–190. [Google Scholar]
- Kovaleski, A.; Mumford, J. Pulling out the evil by the root: The codling moth Cydia pomonella eradication programme in Brazil. In Area-Wide Control of Insect Pests. From Research to Field Implementation; Vreysen, M.J.B., Robinson, A.S., Hendrichs, J., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 581–590. [Google Scholar]
- Bloem, S.; McCluskey, A.; Fugger, R.; Arthurs, S.; Wood, S.; Carpenter, J. Suppression of the codling moth Cydia pomonella in British Columbia, Canada using an area-wide integrated approach with an SIT component. In Area-Wide Control of Insect Pests. From Research to Field Implementation; Vreysen, M.J.B., Robinson, A.S., Hendrichs, J., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 591–601. [Google Scholar]
- Duan, X.L.; Li, Y.T.; Men, Q.L.; Zhang, M.; Qiao, X.F.; Harari, A.; Chen, M.H. Limited gene flow among Cydia pomonella (Lepidoptera: Tortricidae) populations in two isolated regions in China: Implications for utilization of the SIT. Fla. Entomol. 2016, 99, 23–29. [Google Scholar] [CrossRef][Green Version]
- Franck, P.; Ricci, B.; Klein, E.K.; Olivares, J.; Simon, S.; Cornuet, J.M.; Lavigne, C. Genetic inferences about the population dynamics of codling moth females at a local scale. Genetica 2011, 139, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Dorn, S. Microsatellites reveal genetic differentiation among populations in an insect species with high genetic variability in dispersal, the codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae). Bull. Entomol. Res. 2010, 100, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Men, Q.L.; Chen, M.H.; Zhang, Y.L.; Feng, J.N. Genetic structure and diversity of a newly invasive species, the codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae) in China. Biol. Invasions 2013, 15, 447–458. [Google Scholar] [CrossRef]
- Meraner, A.; Brandstatter, A.; Thaler, R.; Aray, B.; Unterlechner, M.; Niederstatter, H.; Parson, W.; Zelger, R.; Dalla Via, J.; Dallinger, R. Molecular phylogeny and population structure of the codling moth (Cydia pomonella) in Central Europe: I. Ancient clade splitting revealed by mitochondrial haplotype markers. Mol. Phylogenet. Evol. 2008, 48, 825–837. [Google Scholar] [CrossRef]
- Li, Y.; Duan, X.; Qiao, X.; Li, X.; Wang, K.; Men, Q.; Chen, M. Mitochondrial DNA revealed the extent of genetic diversity and invasion origin of populations from two separate invaded areas of a newly invasive pest, Cydia pomonella (L.) (Lepidoptera: Tortricidae) in China. Bull. Entomol. Res. 2015, 105, 485–496. [Google Scholar] [CrossRef]
- Fuentes-Contreras, E.; Espinoza, J.L.; Lavandero, B.; Ramirez, C.C. Population genetic structure of codling moth (Lepidoptera: Tortricidae) from apple orchards in central Chile. J. Econ. Entomol. 2008, 101, 190–198. [Google Scholar] [CrossRef]
- Margaritopoulos, J.T.; Voudouris, C.C.; Olivares, J.; Sauphanor, B.; Mamuris, Z.; Tsitsipis, J.A.; Franck, P. Dispersal ability in codling moth: Mark-release-recapture experiments and kinship analysis. Agric. For. Entomol. 2012, 14, 399–407. [Google Scholar] [CrossRef]
- Franck, P.; Reyes, M.; Olivares, J.; Sauphanor, B. Genetic architecture in codling moth populations: Comparison between microsatellite and insecticide resistance markers. Mol. Ecol. 2007, 16, 3554–3564. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.M. Codling moth occurrence, host race formation, and damage. In Tortricid Pests. Their Biology, Natural Enemies and Control; van der Geest, L.P.S., Evenhuis, H.H., Eds.; Elsevier: Amsterdam, The Netherlands, 1991; pp. 313–328. [Google Scholar]
- Cisneros, F.H.; Barnes, M.M. Contribution to the biological and ecological characterization of apple and walnut host races of codling moth, Laspeyresia pomonella (L.): Moth longevity and oviposition capacity. Environ. Entomol. 1974, 3, 402–406. [Google Scholar] [CrossRef]
- Phillips, P.H.; Barnes, M.M. Host race formation among sympatric apple, walnut and plum populations of the codling moth, Laspeyresia pomonella. Ann. Entomol. Soc. Am. 1975, 68, 1053–1060. [Google Scholar] [CrossRef]
- Ter-Hovhannesyan, A.; Azizyan, A. Interactions between plants and codling moth (Cydia pomonella L.). IOBC/WPRS Bull. 2003, 26, 91–96. [Google Scholar]
- Timm, A.E.; Geertsema, H.; Warnich, L. Population genetic structure of economically important Tortricidae (Lepidoptera) in South Africa: A comparative analysis. Bull. Entomol. Res. 2010, 100, 421–431. [Google Scholar] [CrossRef]
- Timm, A.E.; Geertsema, H.; Warnich, L. Gene flow among Cydia pomonella (Lepidoptera: Tortricidae) geographic and host populations in South Africa. J. Econ. Entomol. 2006, 99, 341–348. [Google Scholar] [CrossRef]
- Thaler, R.; Brandstatter, A.; Meraner, A.; Chabicovski, M.; Parson, W.; Zelger, R.; Dalla Via, J.; Dallinger, R. Molecular phylogeny and population structure of the codling moth (Cydia pomonella) in Central Europe: II. AFLP analysis reflects human-aided local adaptation of a global pest species. Mol. Phylogenet. Evol. 2008, 48, 838–849. [Google Scholar] [CrossRef]
- Cheney, S.; Hadapad, A.B.; Zebitz, C.P.W. AFLP analysis of genetic differentiation in CpGV resistant and susceptible Cydia pomonella (L.) populations. Mitt. Dtsch. Ges. Allg. Angew. Entomol. 2008, 16, 117–120. [Google Scholar]
- Khaghaninia, S.; Mohammadi, S.A.; Sarafrazi, A.L.; Nejad, K.H.I. Population variation of codling moth Cydia pomonella (Lep.; Tortricidae) based on molecular data from northwestern Iran. Turk. J. Zool. 2011, 35, 571–578. [Google Scholar]
- Zada, H.; Salijoqui, A.-U.-R.; Ali, I.; Ahmad, B.; Khan, A.W.; Ahmad, S. Molecular characterization of codling moth Cydia pomonella (Linnaeus) (Lepidoptera: Tortricidae) in Swat Valley Pakistan using random amplified polymorphic DNA (RAPD) polymerase chain reaction. Pak. J. Zool. 2019, 51, 1547–1554. [Google Scholar] [CrossRef]
- Franck, P.; Guerin, B.; Loiseau, A.; Sauphanor, B. Isolation and characterization of microsatellite loci in the codling moth Cydia pomonella L. (Lepidoptera, Tortricidae). Mol. Ecol. Notes 2005, 5, 99–102. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Gu, H.N.; Dorn, S. Isolation of microsatellite loci in the codling moth, Cydia pomonella (Lepidoptera: Tortricidae). Mol. Ecol. Notes 2005, 5, 226–227. [Google Scholar] [CrossRef]
- Pajač, I.; Barić, B.; Šimon, S.; Mikac, K.M.; Pejić, I. An initial examination of the population genetic structure of Cydia pomonella (Lepidoptera: Tortricidae) in Croatian apple orchards. J. Food Agric. Environ. 2011, 9, 459–464. [Google Scholar]
- Voudouris, C.C.; Franck, P.; Olivares, J.; Sauphanor, B.; Mamuris, Z.; Tsitsipis, J.A.; Margaritopoulos, J.T. Comparing the genetic structure of codling moth Cydia pomonella (L.) from Greece and France: Long distance gene-flow in a sedentary pest species. Bull. Entomol. Res. 2012, 102, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Fuentes-Contreras, E.; Barros, W.; Ramirez, C.C. Utilización de microsatélites para la determinación de la variabilidad genética de la polilla de la manzana Cydia pomonella L. (Lepidoptera: Tortricidae) en Chile Central. Agric. Téc. (Chile) 2007, 67, 244–252. [Google Scholar]
- Fuentes-Contreras, E.; Basoalto, E.; Franck, P.; Lavandero, B.; Knight, A.L.; Ramírez, C.C. Measuring local genetic variability in populations of codling moth (Lepidoptera: Tortricidae) across an unmanaged and commercial orchard interface. Environ. Entomol. 2014, 43, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Chappell, T.M.; Kennedy, G.G.; Walgenbach, J.F. Predicting codling moth (Cydia pomonella) phenology in North Carolina on the basis of temperature and improved generation turnover estimates. Pest Manag. Sci. 2015, 71, 1425–1432. [Google Scholar] [CrossRef]
- Boivin, T.; Chadoeuf, J.; Bouvier, J.C.; Beslay, D.; Sauphanor, B. Modelling the interactions between phenology and insecticide resistance genes in the codling moth Cydia pomonella. Pest Manag. Sci. 2005, 61, 53–67. [Google Scholar] [CrossRef]
- FIA. Agenda de Innovación Agraria Territorial: Región de Aysén del General Carlos Ibañez del Campo; Gráfica Barclau: Santiago, Chile, 2009; p. 80. [Google Scholar]
- Artigas, J.N. Entomología Económica; Universidad de Concepción: Concepción, Chile, 1994. [Google Scholar]
- González, R.H. Las Polillas de la Fruta en Chile (Lepidoptera: Tortricidae, Pyralidae); Universidad de Chile: Santiago, Chile, 2003; Volume 9, p. 188. [Google Scholar]
- Lacoste, P.; Yuri, J.A.; Aranda, M.; Castro, A.; Quinteros, K.; Solar, M.; Soto, N.; Chávez, C.; Gaete, J.; Rivas, J. Geography of the fruit growing in Chile and Cuyo (1700–1850). Estud. Ibero-Am. 2011, 37, 62–85. [Google Scholar]
- Basoalto, E.; Miranda, M.; Knight, A.L.; Fuentes-Contreras, E. Landscape analysis of adult codling moth (Lepidoptera: Tortricidae) distribution and dispersal within typical agroecosystems dominated by apple production in central Chile. Environ. Entomol. 2010, 39, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Sunnucks, P.; Hales, D.F. Numerous transposed sequences of mitochondrial cytochrome oxidase I–II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 1996, 13, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT: A Program to Estimate and Test Gene Diversities and Fixation Indices; Version 2.9.3; Lausanne University: Lausanne, Switzerland, 2001. [Google Scholar]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Schneider, S.; Roessli, D.; Excoffier, L. ARLEQUIN: A Software for Population Genetics Data Analysis; Version 2.0; University of Geneva: Geneva, Switzerland, 2000. [Google Scholar]
- Addinsoft. XLSTAT-PRO; Version 7.5.2; Addinsoft: New York, NY, USA, 2006. [Google Scholar]
- Chen, C.; Durand, E.; Forbes, F.; Francois, O. Bayesian clustering algorithms ascertaining spatial population structure: A new computer program and a comparison study. Mol. Ecol. Notes 2007, 7, 747–756. [Google Scholar] [CrossRef]
- Wan, F.H.; Yin, C.L.; Tang, R.; Chen, M.H.; Wu, Q.; Huang, C.; Qian, W.Q.; Rota-Stabelli, O.; Yang, N.W.; Wang, S.P.; et al. A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 2019, 10, 4237. [Google Scholar] [CrossRef]
- Muñoz, J.G. Cultivos frutales en 1579, orgullo de los santiaguinos. Estud. Av. 2001, 16, 103–115. [Google Scholar]
- Young, G.F.W. Bernardo Philippi, initiator of German colonization in Chile. Hisp. Am. Hist. Rev. 1971, 51, 478–496. [Google Scholar] [CrossRef]
- Monaghan, J. Chile, Perú, and the California Gold Rush of 1849; University of California Press: Berkeley, CA, USA, 1973. [Google Scholar]
- Simpson, C.B. The Codling Moth; United States Department of Agriculture: Washington, DC, USA, 1903; Volume 41.
- Nahuelhual, L.; Carmona, A.; Lara, A.; Echeverría, C.; González, M.E. Land-cover change to forest plantations: Proximate causes and implications for the landscape in south-central Chile. Landsc. Urban Plan. 2012, 107, 12–20. [Google Scholar] [CrossRef]
- Bùes, R.; Toubon, J.F.; Poitout, H.S. Variabilité écophysiologique et enzymatique de Cydia pomonella L en fonction del’origine géographique et de la plante hôte. Agronomie 1995, 15, 221–231. [Google Scholar] [CrossRef]
- Torres, F.; Rodriguez, M.A.; Lavandero, B.; Fuentes-Contreras, E. Body mass and wing geometric morphology of the codling moth (Lepidoptera: Tortricidae) according to sex, location and host plant in the region of Maule, Chile. Cienc. Investig. Agrar. 2015, 42, 397–406. [Google Scholar] [CrossRef]
- Gu, H.N.; Hughes, J.; Dorn, S. Trade-off between mobility and fitness in Cydia pomonella L. (Lepidoptera: Tortricidae). Ecol. Entomol. 2006, 31, 68–74. [Google Scholar] [CrossRef]
- Keil, S.; Gu, H.N.; Dorn, S. Response of Cydia pomonella to selection on mobility: Laboratory evaluation and field verification. Ecol. Entomol. 2001, 26, 495–501. [Google Scholar] [CrossRef]
- Schumacher, P.; Weber, D.C.; Hagger, C.; Dorn, S. Heritability of flight distance for Cydia pomonella. Entomol. Exp. Appl. 1997, 85, 169–175. [Google Scholar] [CrossRef]
- Schumacher, P.; Weyeneth, A.; Weber, D.C.; Dorn, S. Long flights in Cydia pomonella L. (Lepidoptera: Tortricidae) measured by a flight mill: Influence of sex, mated status and age. Physiol. Entomol. 1997, 22, 149–160. [Google Scholar] [CrossRef]
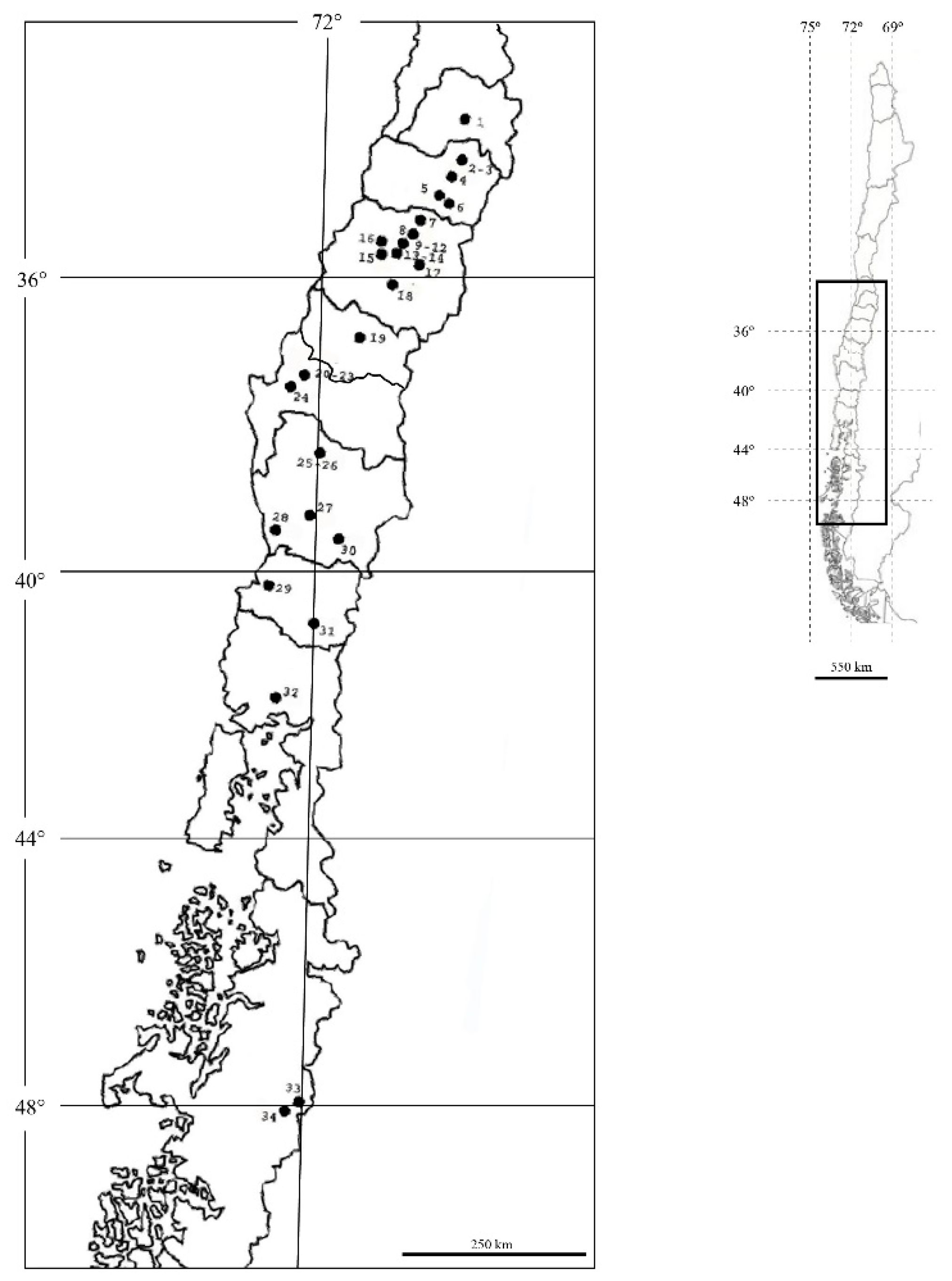
| Code | Location | Region/Country Zone | Host | Latitude | Longitude | |
|---|---|---|---|---|---|---|
| (1) | SanA | Santiago | Metropolitan C a | Apple | 33°31′57.5″ S | 70°32′40.1″ W |
| (2) | GraA | Graneros | O’Higgins C | Apple | 34°0.4′3.8″ S | 70°42′43.7″ W |
| (3) | GraW | Graneros | O’Higgins C | Walnut | 34°0.4′3.8″ S | 70°42′43.7″ W |
| (4) | GulA | Gultro | O’Higgins C | Apple | 34°11′51.9″ S | 70°46′31.5″ W |
| (5) | SnfP1 | San Fernando 1 | O’Higgins C | Pear | 34°36′8.5″ S | 71°2′9.7″ W |
| (6) | SnfW2 | San Fernando 2 | O’Higgins C | Walnut | 34°36′18″ S | 70°58′43″ W |
| (7) | CurA | Curicó | Maule C | Apple | 35°1′12.2″ S | 71°14′26.2″ W |
| (8) | MolA | Molina | Maule C | Apple | 35°5′52″ S | 71°16′26.27″ W |
| (9) | PanA | Panguilemo | Maule C | Apple | 35°22′13.4″ S | 71°35′50.3″ W |
| (10) | PanP | Panguilemo | Maule C | Pear | 35°22′13.4″ S | 71°35′50.3″ W |
| (11) | PanW | Panguilemo | Maule C | Walnut | 35°22′13.4″ S | 71°35′50,3″ W |
| (12) | PanQ | Panguilemo | Maule C | Quince | 35°22′13.4″ S | 71°35′50,3″ W |
| (13) | TalP | Talca | Maule C | Pear | 35°24′48.6″ S | 71°38′24.2″ W |
| (14) | TalQ | Talca | Maule C | Quince | 35°24′47.8″ S | 71°38′24.1″ W |
| (15) | ColA | Colín | Maule C | Apple | 35°27′56.5″ S | 71°44′4.5″ W |
| (16) | PenA | Pencahue | Maule C | Apple | 35°23′9.8″ S | 71°48′38.4″ W |
| (17) | SclW | San Clemente | Maule C | Walnut | 35°31′24.7″ S | 71°26′0.3″ W |
| (18) | LinA | Linares | Maule C | Apple | 35°57′10.7″ S | 71°19′29.1″ W |
| (19) | ChiA | Chillán | Ñuble S | Apple | 36°32′50.5″ S | 72°1′27.6″ W |
| (20) | SjuA1 | Santa Juana 1 | Bío Bío S | Apple | 37°10′11.1″ S | 72°56′25.3″ W |
| (21) | SjuP1 | Santa Juana 1 | Bío Bío S | Pear | 37°10′10.6″ S | 72°56′25.4″ W |
| (22) | SjuQ1 | Santa Juana 1 | Bío Bío S | Quince | 37°10′11.7″ S | 72°56′25.9″ W |
| (23) | SjuW2 | Santa Juana 2 | Bío Bío S | Walnut | 37°10′37″ S | 72°56′13″ W |
| (24) | NacA | Nacimiento | Bío Bío S | Apple | 37°24′21.1″ S | 72°47′38.8″ W |
| (25) | ErcA | Ercilla | Araucanía S | Apple | 38°5′29.5″ S | 72°21′5.4″ W |
| (26) | ErcW | Ercilla | Araucanía S | Walnut | 38°5′29.5″ S | 72°21′5.4″ W |
| (27) | TemA | Temuco | Araucanía S | Apple | 38°41′8.6″ S | 72°25′37.5″ W |
| (28) | NtoA | Nueva Toltén | Araucanía S | Apple | 39°9′35,7″ S | 73°6′2.8″ W |
| (29) | ValA | Valdivia | Los Ríos S | Apple | 39°46′29.7″ S | 73°14′52.8″ W |
| (30) | VilA | Villarrica | Los Lagos S | Apple | 39°78′86″ S | 72°3′32″ W |
| (31) | PuyA | Puyehue | Los Lagos S | Apple | 40°41′10″ S | 72°35′45″ W |
| (32) | LlaA | Llanquihue | Los Lagos S | Apple | 41°15′12″ S | 73°0′12″ W |
| (33) | CchA1 | Chile Chico 1 | Aysén S | Apple | 46°32′29.8″ S | 71°43′21.7″ W |
| (34) | CchA2 | Chile Chico 2 | Aysén S | Apple | 46°33′38″ S | 71°40′25″ W |
| (35) | VleA | Valence | Avignon F | Apple | 44°58′43″ N | 4°55′45″ E |
| (36) | VleP | Valence | Avignon F | Pear | 44°58′32″ N | 4°55′53″ E |
| (37) | VleW | Valence | Avignon F | Walnut | 44°58′31″ N | 4°56′01″ E |
| Sample | N | NA | a | Na | HE | FIS |
|---|---|---|---|---|---|---|
| SanA | 19 | 5.2 | 3.7 | 0.022 | 0.605 | −0.184 |
| GraA | 20 | 3.6 | 2.8 | 0.000 | 0.496 | −0.028 |
| GraW | 20 | 4.4 | 3.5 | 0.027 | 0.575 | −0.147 |
| GulA | 17 | 4.2 | 3.5 | 0.002 | 0.608 | −0.025 * |
| SnfP1 | 19 | 4.6 | 3.4 | 0.027 | 0.578 | −0.049 |
| SnfW2 | 20 | 4.4 | 3.5 | 0.006 | 0.565 | 0.140 |
| CurA | 19 | 4.8 | 3.5 | 0.000 | 0.565 | 0.020 |
| MolA | 19 | 5.0 | 3.8 | 0.027 | 0.644 | −0.143 |
| PanA | 17 | 4.2 | 3.3 | 0.004 | 0.602 | 0.101 |
| PanP | 15 | 4.4 | 3.5 | 0.000 | 0.556 | 0.001 |
| PanW | 17 | 4.4 | 3.5 | 0.024 | 0.592 | 0.046 |
| PanQ | 7 | 3.0 | 3.0 | 0.006 | 0.588 | −0.215 |
| TalP | 20 | 4.8 | 3.4 | 0.027 | 0.592 | 0.043 |
| TalQ | 19 | 5.2 | 3.6 | 0.018 | 0.568 | −0.017 |
| ColA | 20 | 5.0 | 3.8 | 0.030 | 0.635 | −0.087 * |
| PenA | 20 | 4.2 | 3.3 | 0.009 | 0.582 | −0.203 * |
| SclW | 19 | 5.0 | 3.6 | 0.015 | 0.585 | 0.046 |
| LinA | 19 | 4.6 | 3.6 | 0.001 | 0.603 | −0.082 |
| ChiA | 19 | 4.0 | 3.2 | 0.000 | 0.587 | −0.087 |
| SjuA1 | 15 | 3.8 | 3.2 | 0.009 | 0.558 | −0.004 |
| SjuP1 | 20 | 5.0 | 3.4 | 0.000 | 0.575 | −0.061 |
| SjuQ1 | 18 | 4.4 | 3.4 | 0.000 | 0.525 | −0.078 |
| SjuW2 | 19 | 4.4 | 3.2 | 0.006 | 0.535 | −0.102 |
| NacA | 17 | 3.8 | 2.9 | 0.000 | 0.516 | −0.048 |
| ErcA | 20 | 5.0 | 3.9 | 0.001 | 0.645 | −0.117 |
| ErcW | 20 | 5.0 | 3.7 | 0.000 | 0.589 | −0.031 |
| TemA | 17 | 4.2 | 3.3 | 0.006 | 0.583 | −0.050 |
| NtoA | 8 | 3.8 | 3.7 | 0.032 | 0.566 | −0.148 |
| ValA | 17 | 4.2 | 3.3 | 0.025 | 0.582 | 0.049 |
| VilA | 17 | 3.8 | 3.2 | 0.006 | 0.535 | −0.078 |
| PuyA | 20 | 4.0 | 3.1 | 0.007 | 0.525 | 0.038 |
| LlaA | 19 | 4.8 | 3.6 | 0.000 | 0.591 | 0.056 |
| CchA1 | 20 | 4.6 | 3.6 | 0.026 | 0.582 | −0.095 |
| CchA2 | 17 | 4.0 | 2.9 | 0.037 | 0.436 | −0.149 |
| VleA | 29 | 7.6 | 4.7 | 0.000 | 0.696 | −0.060 |
| VleP | 24 | 6.2 | 4.3 | 0.045 | 0.660 | 0.117 * |
| VleW | 56 | 9.0 | 4.4 | 0.023 | 0.675 | 0.064 |
| Variation Source | df | Sum of Squares | Variance Components | Percentage of Variation | Fixation Index a,b |
|---|---|---|---|---|---|
| Among groups (zone) | 1 | 7.485 | 0.00652 | 0.44 | FCT = 0.00445 * |
| Among locations within groups (location) | 24 | 74.320 | 0.03646 | 2.49 | FSC = 0.02499 *** |
| Within locations | 1192 | 1695.813 | 1.42266 | 97.07 | FST = 0.02932 *** |
| Total | 1217 | 1777.618 | 1.46564 | 100 | |
| Among groups (host plant) | 3 | 9.252 | 0.00068 | 0.05 | FCT = 0.00046 |
| Among locations within groups (location) | 30 | 87.336 | 0.04172 | 2.85 | FSC = 0.02854 *** |
| Within locations | 1184 | 1681.030 | 1.41979 | 97.10 | FST 0.02899 *** |
| Total | 1217 | 1777.618 | 1.46218 | 100 |
| GraA | PanA | PanP | PanW | SjuA1 | SjuP1 | SjuQ1 | ErcA | |
|---|---|---|---|---|---|---|---|---|
| GraW | 0.057 a ** | |||||||
| PanP | 0.011 | - | ||||||
| PanW | −0.011 | 0.023 | - | |||||
| PanQ | −0.001 | 0.068 ** | 0.012 | |||||
| SjuP1 | −0.023 | - | ||||||
| SjuQ1 | 0.001 | 0.006 | - | |||||
| SjuW2 | 0.020 | 0.006 | 0.025 * | |||||
| ErcW | −0.008 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basoalto, A.; Ramírez, C.C.; Lavandero, B.; Devotto, L.; Curkovic, T.; Franck, P.; Fuentes-Contreras, E. Population Genetic Structure of Codling Moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), in Different Localities and Host Plants in Chile. Insects 2020, 11, 285. https://doi.org/10.3390/insects11050285
Basoalto A, Ramírez CC, Lavandero B, Devotto L, Curkovic T, Franck P, Fuentes-Contreras E. Population Genetic Structure of Codling Moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), in Different Localities and Host Plants in Chile. Insects. 2020; 11(5):285. https://doi.org/10.3390/insects11050285
Chicago/Turabian StyleBasoalto, Alejandra, Claudio C. Ramírez, Blas Lavandero, Luis Devotto, Tomislav Curkovic, Pierre Franck, and Eduardo Fuentes-Contreras. 2020. "Population Genetic Structure of Codling Moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), in Different Localities and Host Plants in Chile" Insects 11, no. 5: 285. https://doi.org/10.3390/insects11050285
APA StyleBasoalto, A., Ramírez, C. C., Lavandero, B., Devotto, L., Curkovic, T., Franck, P., & Fuentes-Contreras, E. (2020). Population Genetic Structure of Codling Moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), in Different Localities and Host Plants in Chile. Insects, 11(5), 285. https://doi.org/10.3390/insects11050285






