Bacterial Endosymbiont Diversity among Bemisia tabaci (Hemiptera: Aleyrodidae) Populations in Florida
Abstract
1. Introduction
2. Material and Methods
2.1. Field Collection
2.2. Whitefly Identification
2.3. Facultative Endosymbionts Identification
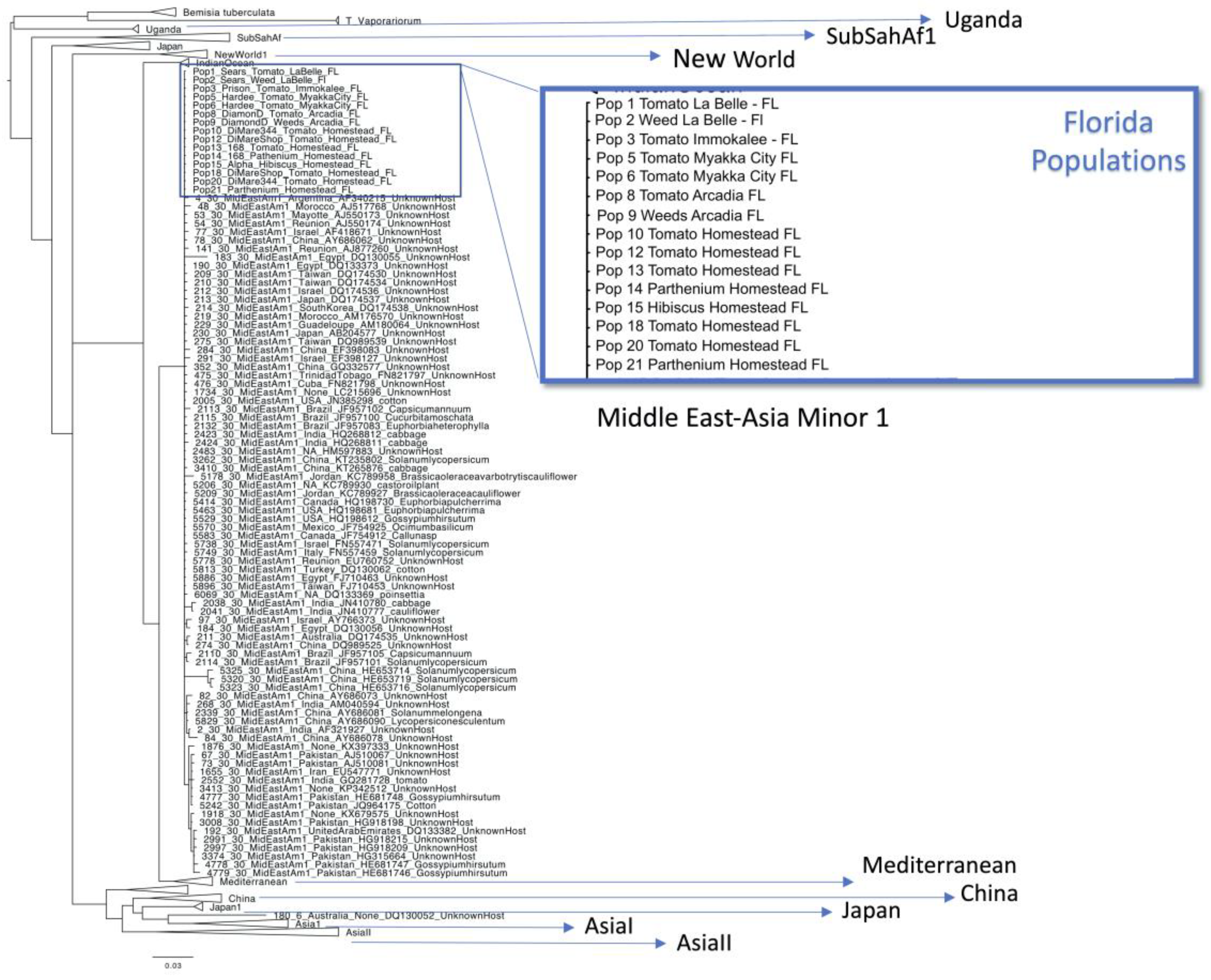
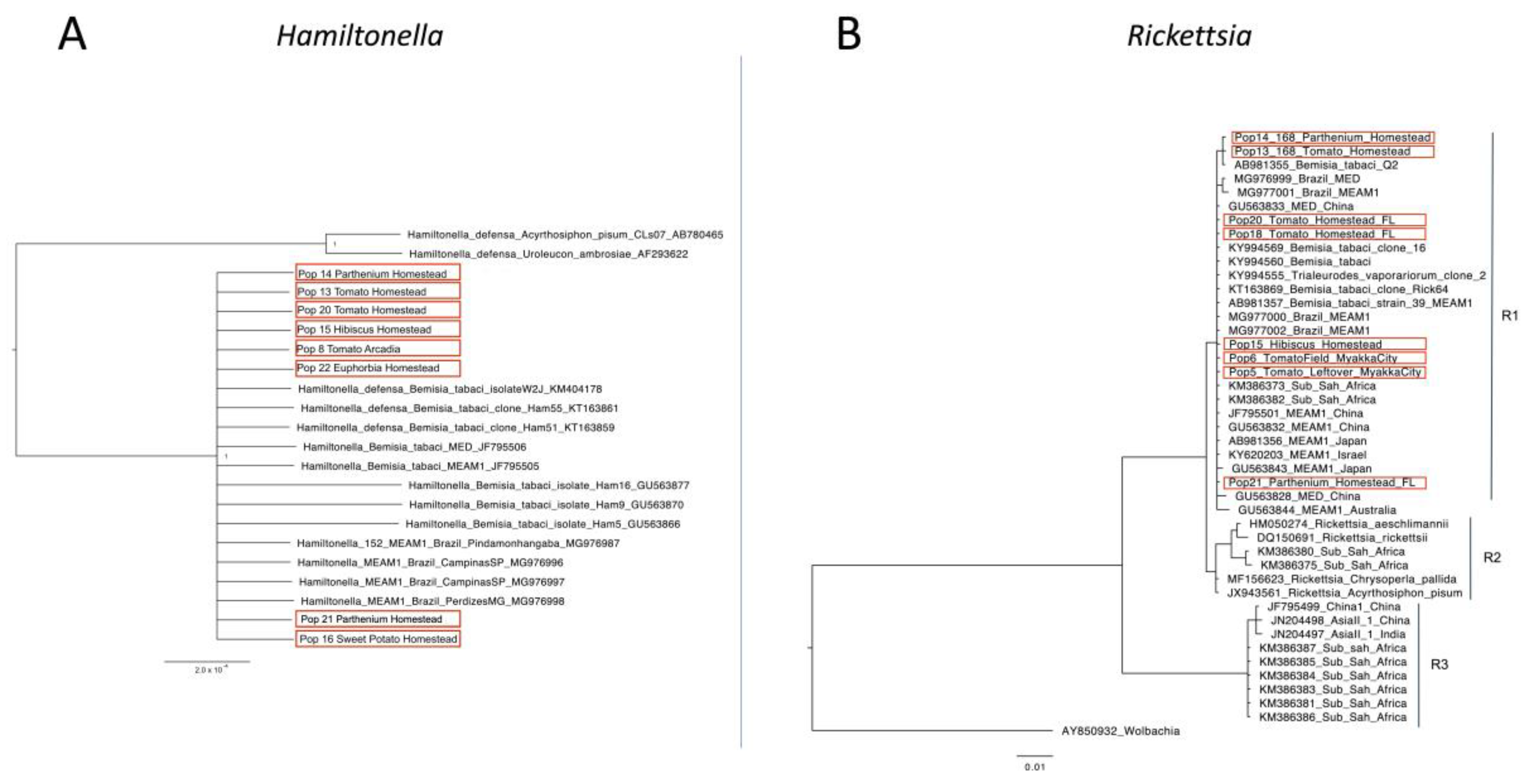
2.4. Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging Virus Diseases Transmitted by Whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef]
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The Sweetpotato or Silverleaf Whiteflies: Biotypes of Bemisia tabaci or a Species Complex? Annu. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- Perring, T.M. The Bemisia tabaci species complex. Crop Prot. 2001, 20, 725–737. [Google Scholar] [CrossRef]
- Rossitto De Marchi, B.; Massaharu Marubayashi, J.; Madoglio Favara, G.; Atsushi Yuki, V.; Watanabe, L.F.L.F.M.; Barbosa, L.F.L.F.; Pavan, M.A.M.A.; Krause-Sakate, R. Comparative transmission of five viruses by Bemisia tabaci NW2 and MEAM1. Trop. Plant Pathol. 2017, 1. [Google Scholar]
- Polston, J.E.; De Barro, P.; Boykin, L.M. Transmission specificities of plant viruses with the newly identified species of the Bemisia tabaci species complex. Pest Manag. Sci. 2014, 70, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.R.; Kontsedalov, S.; Khasdan, V.; Ishaaya, I. Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch. Insect Biochem. 2005, 58, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Kanakala, S.; Ghanim, M. Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 2019, 14, e0213946. [Google Scholar] [CrossRef] [PubMed]
- Mckenzie, C.L.; Hodges, G.; Osborne, L.S.; Byrne, F.J.; Shatters, R.G. Distribution of Bemisia tabaci (Hemiptera: Aleyrodidae) Biotypes in Florida-Investigating the Q Invasion. J. Econ. Entomol. 2009, 102, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Vegetables Annual Summary. Available online: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1183files/97/viewDocumentInfo.html (accessed on 19 February 2020).
- Bellows, T.S.; Perring, T.M.; Gill, R.J.; Headrick, D.H. Description of a Species of Bemisia (Homoptera: Aleyrodidae). Ann. Entomol. Soc. Am. 1994, 87, 195–206. [Google Scholar] [CrossRef]
- Sloan, D.B.; Moran, N.A. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 2012, 8, 986–989. [Google Scholar] [CrossRef] [PubMed]
- Kontsedalov, S.; Zchori-Fein, E.; Chiel, E.; Gottlieb, Y.; Inbar, M.; Ghanim, M. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag. Sci. 2008, 64, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Morin, S.; Ghanim, M.; Sobol, I.; Czosnek, H. The GroEL Protein of the Whitefly Bemisia tabaci Interacts with the Coat Protein of Transmissible and Nontransmissible Begomoviruses in the Yeast Two-Hybrid System. Virology 2000, 276, 404–416. [Google Scholar] [CrossRef]
- Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Kontsedalov, S.; Skaljac, M.; Brumin, M.; Sobol, I.; Czosnek, H.; Vavre, F.; Fleury, F.; et al. The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 2010, 84, 9310–9317. [Google Scholar] [CrossRef]
- Brumin, M.; Kontsedalov, S.; Ghanim, M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011, 18, 57–66. [Google Scholar] [CrossRef]
- Zchori-Fein, E.; Brown, J.K. Diversity of Prokaryotes Associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 2002, 95, 711–718. [Google Scholar] [CrossRef]
- Weeks, A.R.; Velten, R.; Stouthamer, R. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proceedings. Biol. Sci. 2003, 270, 1857–1865. [Google Scholar] [CrossRef]
- Everett, K.D.E.; Thao, M.; Horn, M.; Dyszynski, G.E.; Baumann, P. Novel chlamydiae in whiteflies and scale insects: Endosymbionts “Candidatus Fritschea bemisiae” strain Falk and “Candidatus Fritschea eriococci” strain Elm. Int. J. Syst. Evol. Microbiol. 2005, 55, 1581–1587. [Google Scholar] [CrossRef]
- Gottlieb, Y.; Ghanim, M.; Chiel, E.; Gerling, D.; Portnoy, V.; Steinberg, S.; Tzuri, G.; Horowitz, A.R.; Belausov, E.; Mozes-Daube, N.; et al. Identification and Localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 2006, 72, 3646–3652. [Google Scholar] [CrossRef]
- Pan, H.; Li, X.; Ge, D.; Wang, S.; Wu, Q.; Xie, W.; Jiao, X.; Chu, D.; Liu, B.; Xu, B.; et al. Factors Affecting Population Dynamics of Maternally Transmitted Endosymbionts in Bemisia tabaci. PLoS ONE 2012, 7, e30760. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 1991, 10, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Kontsedalov, S.; Abu-Moch, F.; Lebedev, G.; Czosnek, H.; Horowitz, A.R.; Ghanim, M. Bemisia tabaci Biotype Dynamics and Resistance to Insecticides in Israel During the Years 2008-2010. J. Integr. Agric. 2012, 11, 312–320. [Google Scholar] [CrossRef]
- De Barro, P.J.; Scott, K.D.; Graham, G.C.; Lange, C.L.; Schutze, M.K. Isolation and characterization of microsatellite loci in Bemisia tabaci. Mol. Ecol. Notes 2002, 3, 40–43. [Google Scholar] [CrossRef]
- Frohlich, D.R.; Torres-Jerez, I.; Bedford, I.D.; Markham, P.G.; Brown, J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999, 8, 1683–1691. [Google Scholar] [CrossRef]
- Heddi, A.; Grenier, A.M.; Khatchadourian, C.; Charles, H.; Nardon, P. Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 1999, 96, 6814–6819. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 2004, 70, 3401–3406. [Google Scholar] [CrossRef]
- Boykin, L.M.; De Barro, P.J. A practical guide to identifying members of the Bemisia tabaci species complex: And other morphologically identical species. Front. Ecol. Evol. 2014, 2, 45. [Google Scholar] [CrossRef]
- Fonsah, E.G.; Chen, Y.; Diffie, S.; Srinivansan, R.; Riley, D. Economic Productivity and Profitability Analysis for Whiteflies and Tomato Yellow Leaf Curl Virus (TYLCV) Management Options. J. Agric. Environ. Sci. 2018, 7, 1–9. [Google Scholar]
- Bockoven, A.A.; Bondy, E.C.; Flores, M.J.; Kelly, S.E.; Ravenscraft, A.M.; Hunter, M.S. What Goes Up Might Come Down: The Spectacular Spread of an Endosymbiont Is Followed by Its Decline a Decade Later. Microb. Ecol. 2019, 1–13. [Google Scholar] [CrossRef]
- de Moraes, L.A.; Muller, C.; de Freitas Bueno, R.C.O.; Santos, A.; Bello, V.H.; De Marchi, B.R.; Watanabe, L.F.M.; Marubayashi, J.M.; Santos, B.R.; Yuki, V.A.; et al. Distribution and phylogenetics of whiteflies and their endosymbiont relationships after the Mediterranean species invasion in Brazil. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E.; et al. Rapid Spread of a Bacterial Symbiont in an Invasive Whitefly Is Driven by Fitness Benefits and Female Bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef] [PubMed]
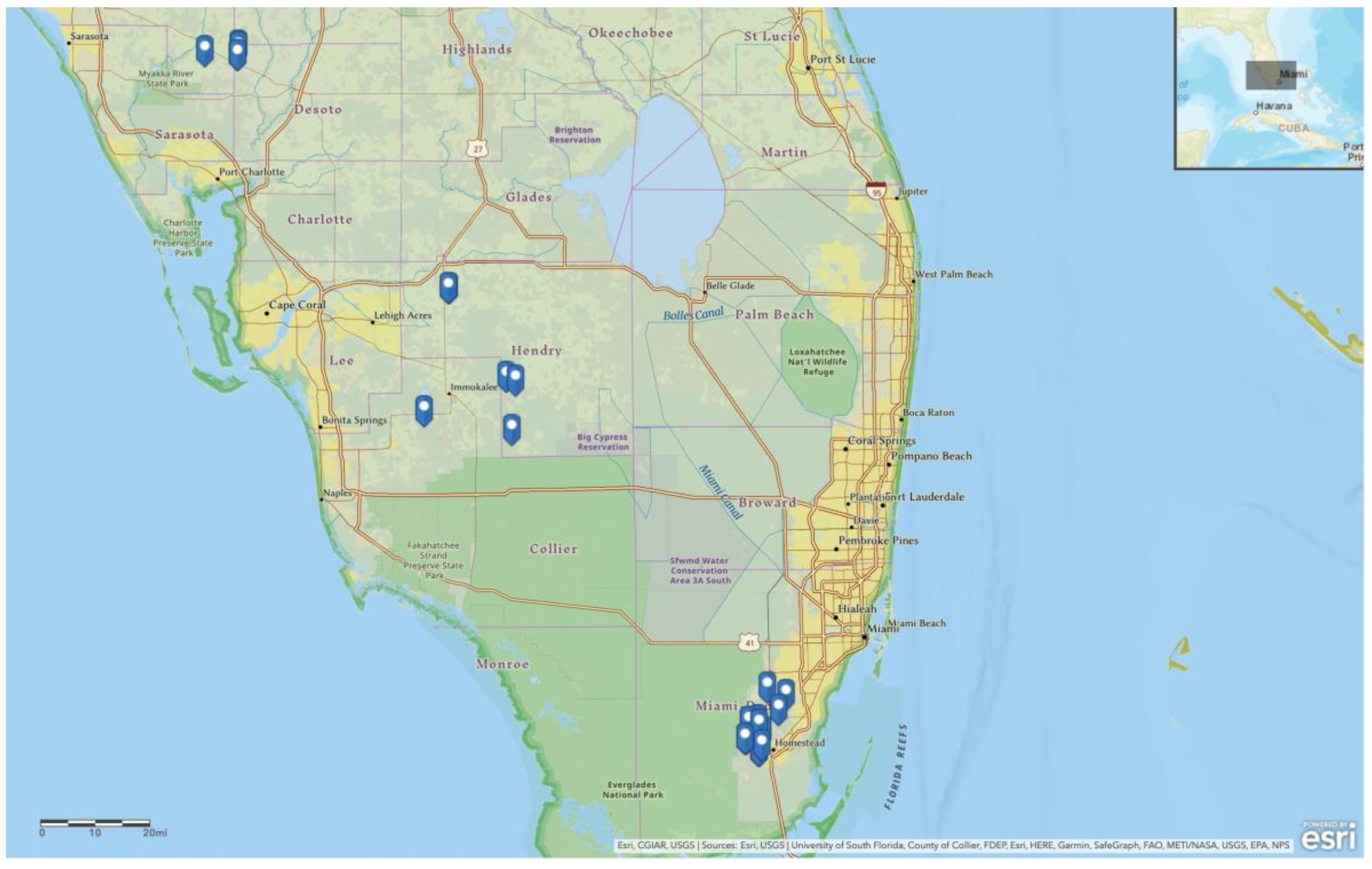
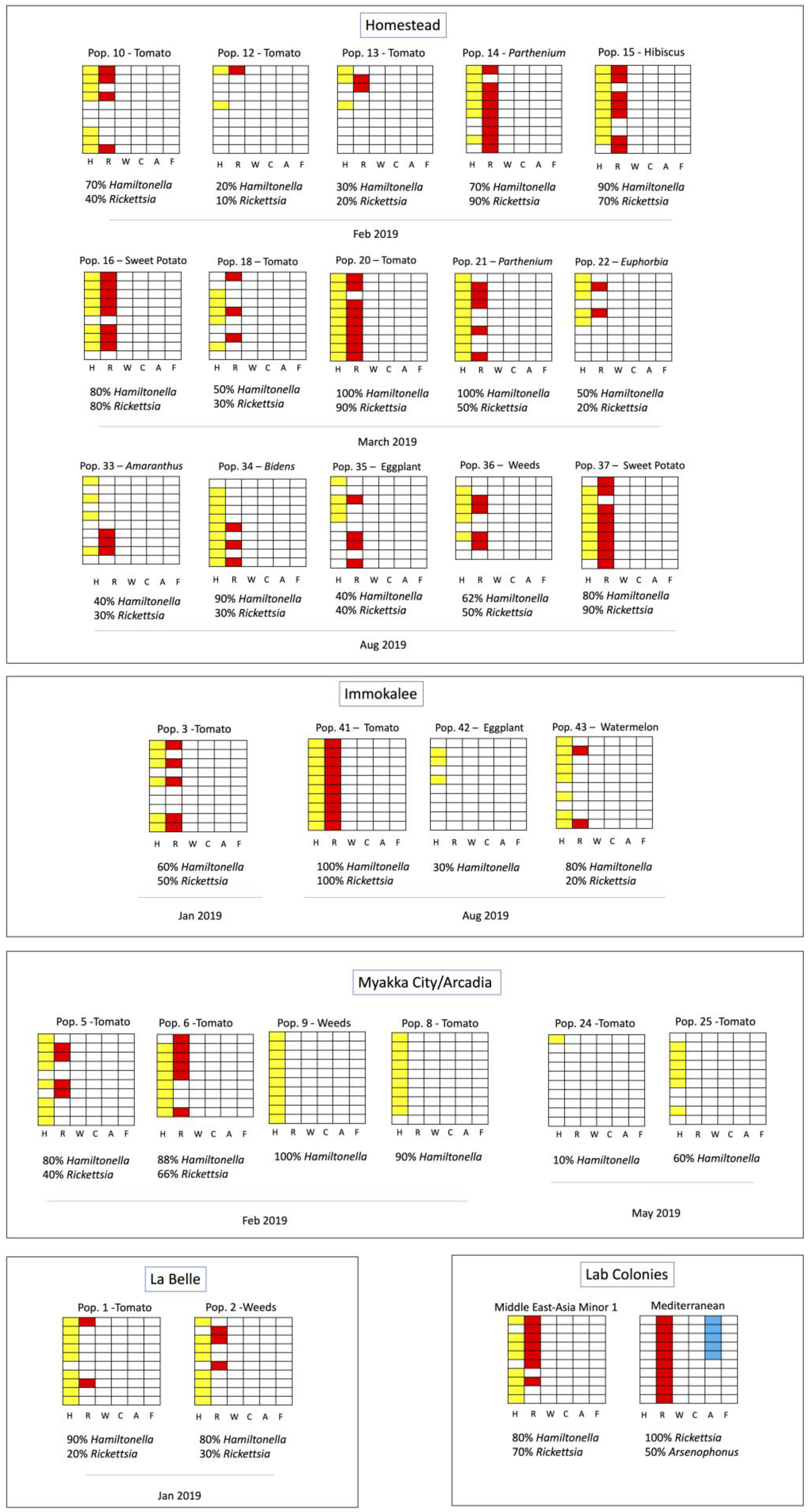
| Pop. ID | Collection Date | Town | Host Crop | Species | Hamiltonella GenBank Accession | Rickettsia GenBank Accession | mtCOI GenBank Accession | Endosymbionts | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | R | W | C | A | F | ||||||||
| B Colony | 1990s | Bradenton | Gossypium hirsutum | MEAM1 | - | - | - | 8/10 | 7/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Q Colony | Jul 2017 | Palm Beach | G. hirsutum * | MED | - | - | - | 0/10 | 10/10 | 0/10 | 0/10 | 5/10 | 0/10 |
| 1 | Jan 2019 | LaBelle | Solanum lycopersicum | MEAM1 | - | - | MN972494 | 9/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 2 | Jan 2019 | LaBelle | Weeds | MEAM1 | - | - | MN972495 | 8/10 | 3/10 | 0/6 | 0/10 | 0/10 | 0/10 |
| 3 | Jan 2019 | Immokalee | S. lycopersicum | MEAM1 | - | - | MN972496 | 6/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 5 | Jan 2019 | Myakka City | S. lycopersicum | MEAM1 | - | MN973880 | MN972497 | 8/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 6 | Feb 2019 | Myakka City | S. lycopersicum | MEAM1 | - | MN973881 | MN972498 | 8/9 | 6/8 | 0/9 | 0/10 | 0/10 | 0/10 |
| 8 | Feb 2019 | Arcadia | S. lycopersicum | MEAM1 | MN973892 | - | MN972508 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 9 | Feb 2019 | Arcadia | Weeds | MEAM1 | - | - | MN972499 | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 10 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | - | - | MN972500 | 7/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 12 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | - | - | MN972501 | 2/7 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 13 | Feb 2019 | Homestead | S. lycopersicum | MEAM1 | MN973889 | MN973886 | MN972502 | 3/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 14 | Feb 2019 | Homestead | Parthenium sp. | MEAM1 | MN973888 | MN973887 | MN972503 | 7/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 15 | Feb 2019 | Homestead | Hibiscus sp. | MEAM1 | MN973891 | MN973882 | MN972504 | 9/10 | 7/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 16 | Mar 2019 | Homestead | Ipomoea batatas | MEAM1 | MN973895 | - | - | 8/10 | 8/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 18 | Mar 2019 | Homestead | S. lycopersicum | MEAM1 | - | MN973883 | MN972505 | 5/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 20 | Mar 2019 | Homestead | S. lycopersicum | MEAM1 | MN973890 | MN973884 | MN972506 | 10/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 21 | Mar 2019 | Homestead | Parthenium sp. | MEAM1 | MN973894 | MN973885 | MN972507 | 10/10 | 5/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 22 | Mar 2019 | Homestead | Euphorbia SP. | MEAM1 | MN973893 | - | - | 5/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 24 | May 2019 | Myakka City | S. lycopersicum | MEAM1 | - | - | - | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 25 | May 2019 | Myakka City | S. lycopersicum | MEAM1 | - | - | - | 6/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 33 | Aug 2019 | Homestead | Amaranthus sp. | MEAM1 | - | - | - | 4/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 34 | Aug 2019 | Homestead | Bidens sp. | MEAM1 | - | - | - | 9/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 35 | Aug 2019 | Homestead | Solanum melongena | MEAM1 | - | - | - | 4/10 | 4/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 36 | Aug 2019 | Homestead | Weeds | MEAM1 | - | - | - | 5/8 | 4/8 | 0/10 | 0/10 | 0/10 | 0/10 |
| 37 | Aug 2019 | Homestead | I. batatas | MEAM1 | - | - | - | 8/10 | 9/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 41 | Aug 2019 | Immokalee | S. lycopersicum | MEAM1 | - | - | - | 10/10 | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 42 | Aug 2019 | Immokalee | S. melongena | MEAM1 | - | - | - | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| 43 | Aug 2019 | Immokalee | Citrullus lanatus | MEAM1 | - | - | - | 8/10 | 2/10 | 0/10 | 0/10 | 0/10 | 0/10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossitto De Marchi, B.; Smith, H.A. Bacterial Endosymbiont Diversity among Bemisia tabaci (Hemiptera: Aleyrodidae) Populations in Florida. Insects 2020, 11, 179. https://doi.org/10.3390/insects11030179
Rossitto De Marchi B, Smith HA. Bacterial Endosymbiont Diversity among Bemisia tabaci (Hemiptera: Aleyrodidae) Populations in Florida. Insects. 2020; 11(3):179. https://doi.org/10.3390/insects11030179
Chicago/Turabian StyleRossitto De Marchi, Bruno, and Hugh A. Smith. 2020. "Bacterial Endosymbiont Diversity among Bemisia tabaci (Hemiptera: Aleyrodidae) Populations in Florida" Insects 11, no. 3: 179. https://doi.org/10.3390/insects11030179
APA StyleRossitto De Marchi, B., & Smith, H. A. (2020). Bacterial Endosymbiont Diversity among Bemisia tabaci (Hemiptera: Aleyrodidae) Populations in Florida. Insects, 11(3), 179. https://doi.org/10.3390/insects11030179






