The ABCB Multidrug Resistance Proteins Do Not Contribute to Ivermectin Detoxification in the Colorado Potato Beetle, Leptinotarsa decemlineata (Say)
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Chemicals
2.3. Identification of MDR Genes in L. decemlineata
2.4. Tissue Expression of MDR Genes in L. decemlineata
2.5. Double-Stranded RNA Synthesis
2.6. Feeding and Survival Assays
2.6.1. RNAi Silencing Assays
2.6.2. dsRNA + Ivermectin Survival Assays
2.6.3. Verapamil + Ivermectin Survival Assays
2.7. cDNA Synthesis
2.8. qRT-PCR
3. Results
3.1. MDR Genes Identified in L. decemlineata
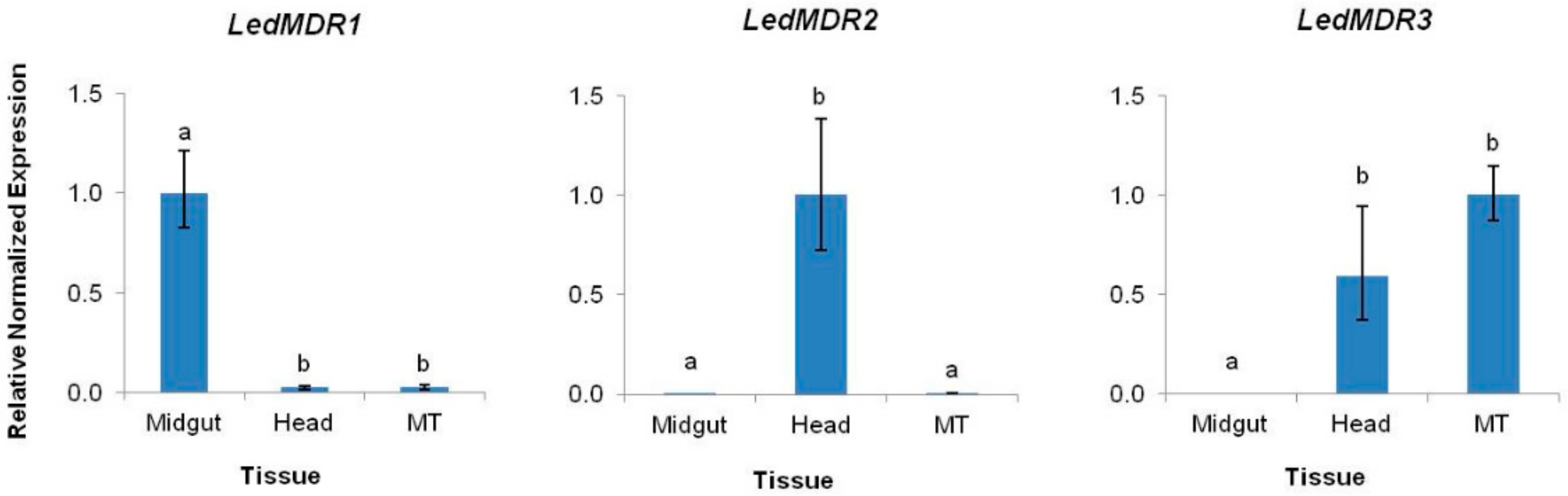
3.2. LedMDR Genes Show Variable Tissue Expression
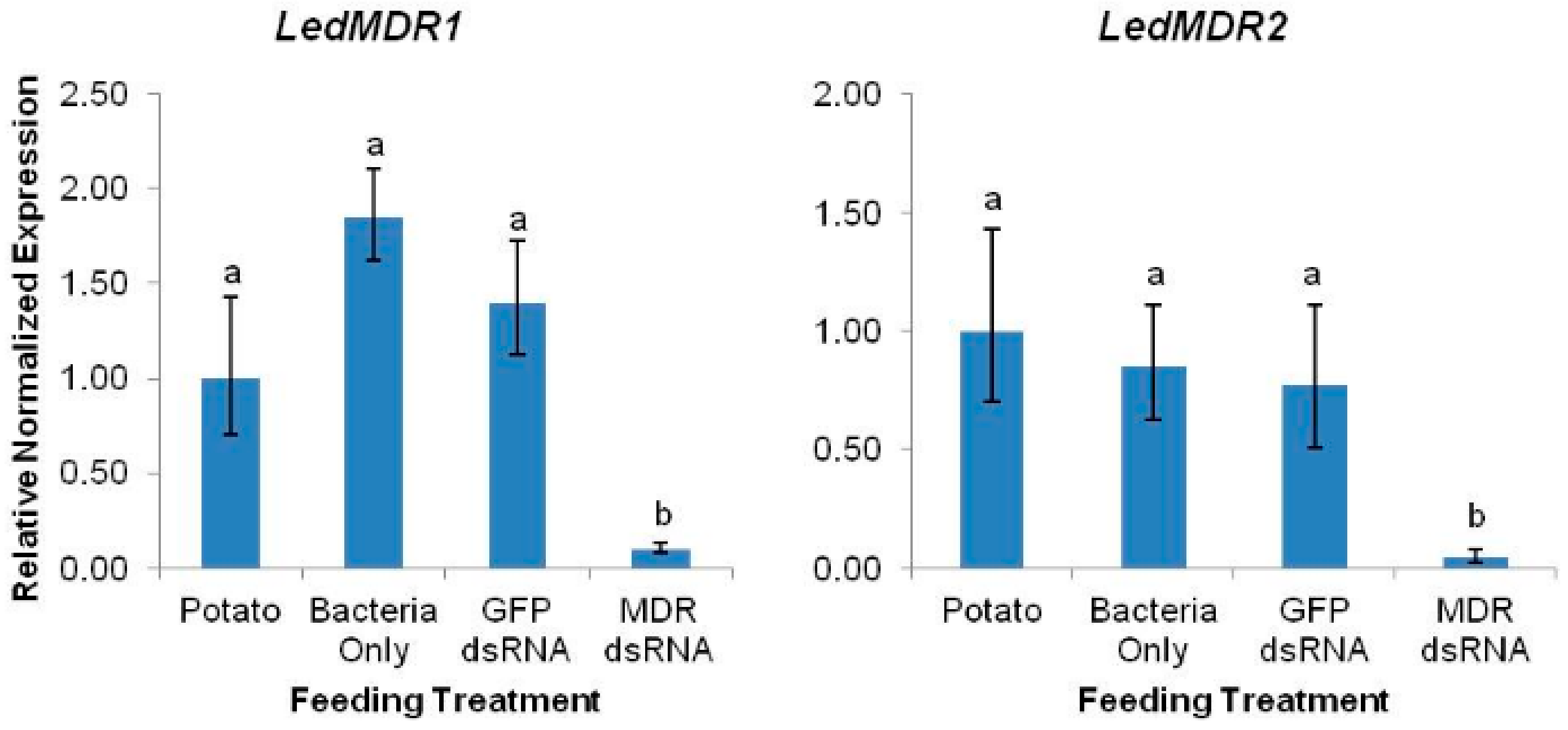
3.3. LedMDR Genes Were Effectively Silenced by Ingested dsRNA
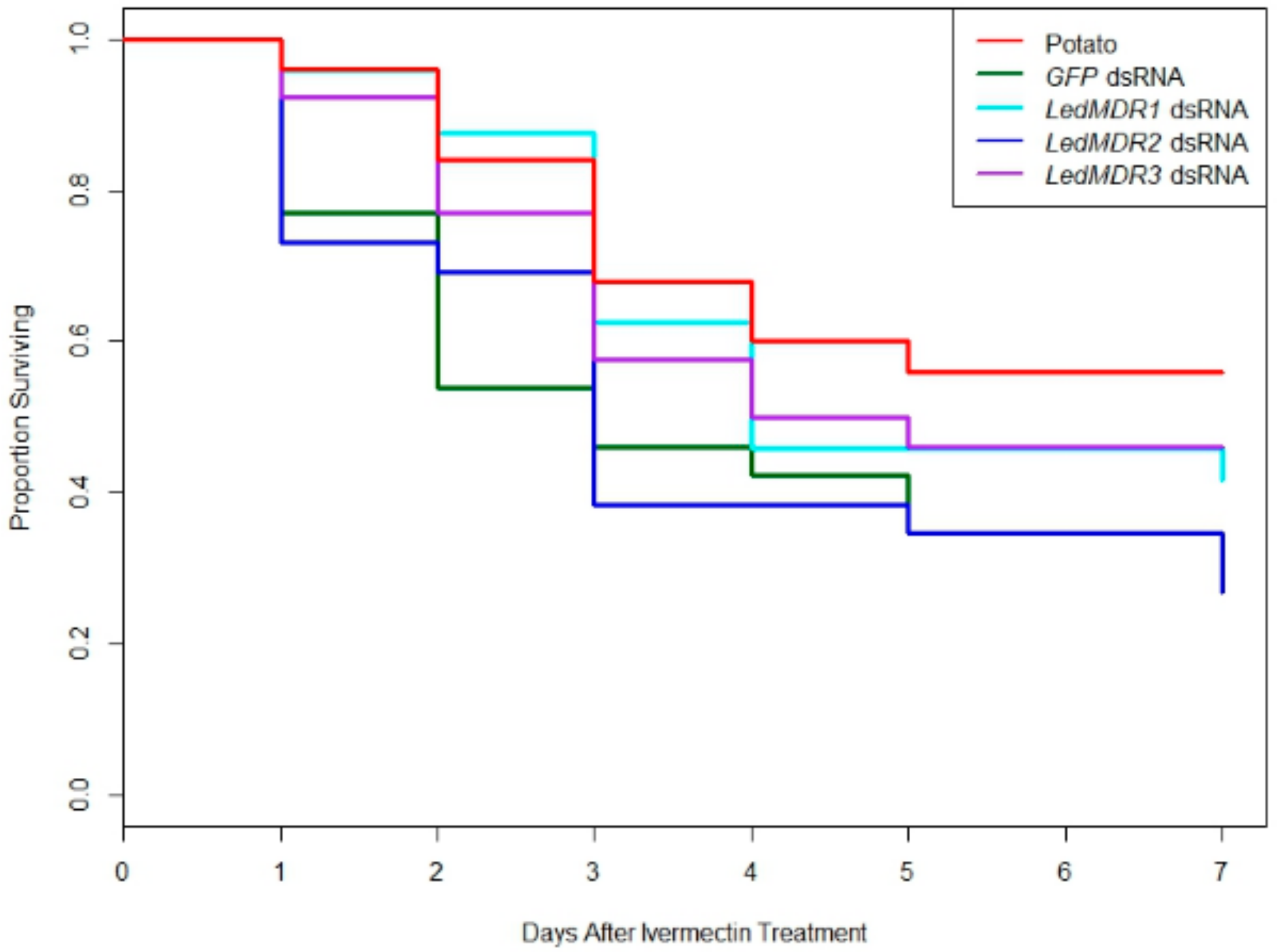
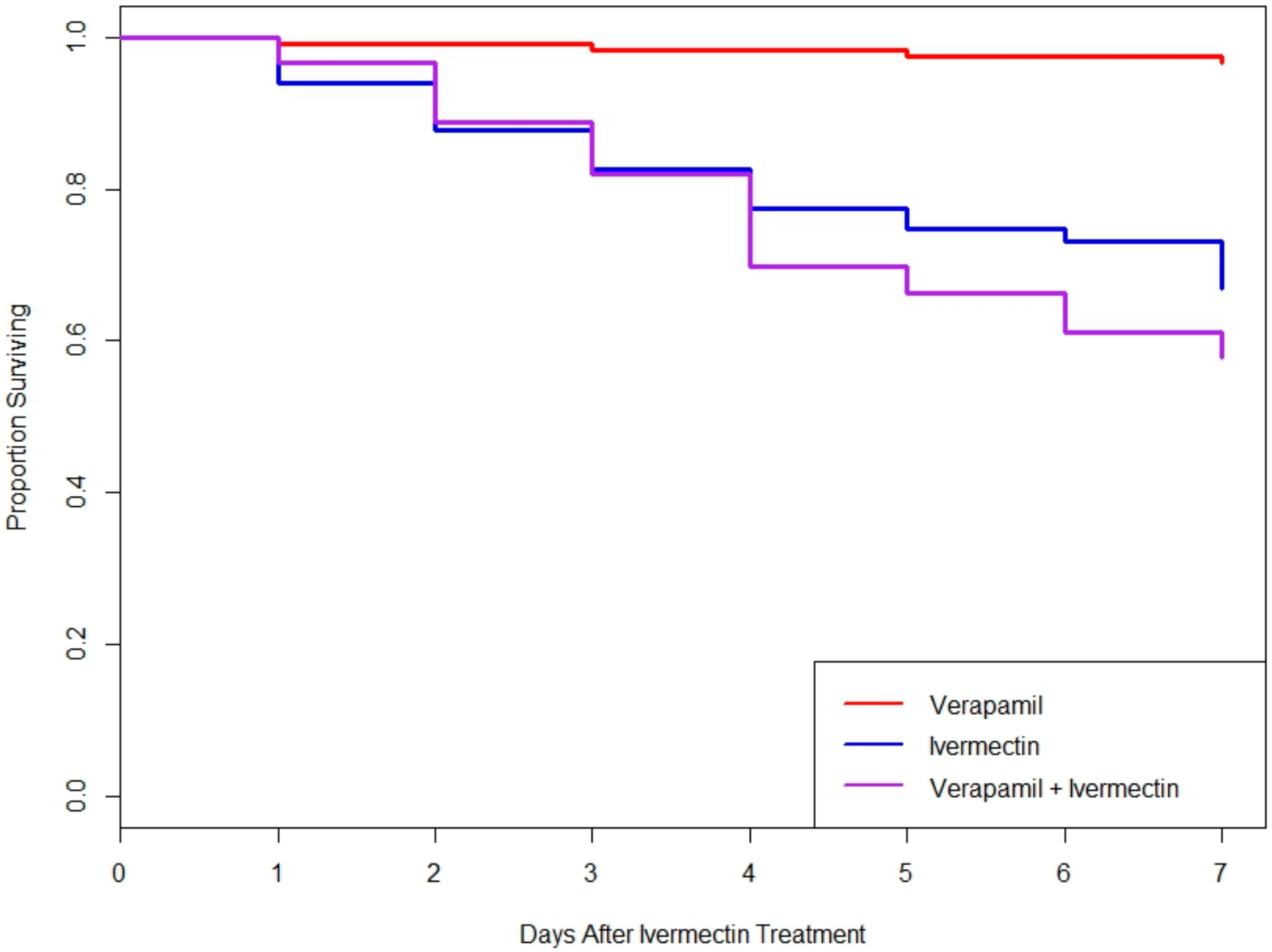
3.4. Neither Silencing of MDR Genes nor MDR Protein Inhibition Increased Toxicity of Ivermectin in L. decemlineata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alyokhin, A. Colorado potato beetle management on potatoes: Current challenges and future prospects. Fruitvegetable Cereal Sci. Biotechnol. 2009, 3, 10–19. [Google Scholar]
- Alyokhin, A.; Baker, M.; Mota-Sanchez, D.; Dively, G.; Grafius, E. Colorado potato beetle resistance to insecticides. Am. J. Potato Res. 2008, 85, 395–413. [Google Scholar] [CrossRef]
- Whalon, M.E.; Mota-Sanchez, D.; Hollingworth, R.M. The MSU Arthropod Pesticide Resistance Database. Available online: https://www.pesticideresistance.org/ (accessed on 28 September 2017).
- Panini, M.; Manicardi, G.C.; Moores, G.D.; Mazzoni, E. An overview of the main pathways of metabolic resistance in insects. Invertebr. Surviv. J. 2016, 13, 326–335. [Google Scholar]
- Albers, S.-V.; Koning, S.M.; Konings, W.N.; Driessen, A.J.M. Insights into ABC transport in Archaea. J. Bioenerg. Biomembr. 2004, 36, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Chen, J. ATP-binding cassette transporters in bacteria. In Annual Review of Biochemistry; Annual Reviews Inc.: Palo Alto, CA, USA, 2004; Volume 73, pp. 241–268. [Google Scholar]
- Dassa, E.; Bouige, P. The ABC of ABCs: A phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001, 152, 211–229. [Google Scholar] [CrossRef]
- Croop, J.M.; Raymond, M.; Haber, D.; Devault, A.; Arceci, R.J.; Gros, P.; Housman, D.E. The three mouse multidrug resistance (MDR) genes are expressed in a tissue-specific manner in normal mouse tissues. Mol. Cell. Biol. 1989, 9, 1346–1350. [Google Scholar] [CrossRef]
- Fojo, A.T.; Ueda, K.; Slamon, D.J.; Poplack, D.G.; Gottesman, M.M.; Pastan, I. Expression of a multidrug-resistance gene in human tumors and tissues. Proc. Natl. Acad. Sci. USA 1987, 84, 265–269. [Google Scholar] [CrossRef]
- Fromm, M.F. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol. Sci. 2004, 25, 423–429. [Google Scholar] [CrossRef]
- Huai-Yun, H.; Secrest, D.T.; Mark, K.S.; Carney, D.; Brandquist, C.; Elmquist, W.F.; Miller, D.W. Expression of multidrug resistance-associated protein (MRP) in brain microvessel endothelial cells. Biochem. Biophys. Res. Commun. 1998, 243, 816–820. [Google Scholar] [CrossRef]
- Schinkel, A.H. P-glycoprotein, a gatekeeper in the blood-brain barrier. Adv. Drug Deliv. Rev. 1999, 36, 179–194. [Google Scholar] [CrossRef]
- Simmons, J.; D’Souza, O.; Rheault, M.; Donly, C. Multidrug resistance protein gene expression in Trichoplusia ni caterpillars. Insect Mol. Biol. 2013, 22, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Tapadia, M.G.; Lakhotia, S.C. Expression of mdr49 and mdr65 multidrug resistance genes in larval tissues of Drosophila melanogaster under normal and stress conditions. Cell Stress Chaperones 2005, 10, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, A. P-glycoprotein-mediated efflux transport of anticancer drugs at the blood-brain barrier. Ther. Drug Monit. 1998, 20, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef]
- Figueira-Mansur, J.; Ferreira-Pereira, A.; Mansur, J.F.; Franco, T.A.; Alvarenga, E.S.L.; Sorgine, M.H.F.; Neves, B.C.; Melo, A.C.A.; Leal, W.S.; Masuda, H.; et al. Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegypti larvae. Insect Mol. Biol. 2013, 22, 648–658. [Google Scholar] [CrossRef]
- Hawthorne, D.J.; Dively, G.P. Killing them with kindness? In-hive medications may inhibit xenobiotic efflux transporters and endanger honey bees. PLoS ONE 2011, 6, e26796. [Google Scholar] [CrossRef]
- Podsiadlowski, L.; Matha, V.; Vilcinskas, A. Detection of a P-glycoprotein related pump in Chironomus larvae and its inhibition by verapamil and cyclosporin A. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998, 121, 443–450. [Google Scholar] [CrossRef]
- Porretta, D.; Epis, S.; Mastrantonio, V.; Ferrari, M.; Bellini, R.; Favia, G.; Urbanelli, S. How heterogeneous is the involvement of ABC transporters against insecticides? Acta Trop. 2016, 157, 131–135. [Google Scholar] [CrossRef]
- Porretta, D.; Gargani, M.; Bellini, R.; Medici, A.; Punelli, F.; Urbanelli, S. Defence mechanisms against insecticides temephos and diflubenzuron in the mosquito Aedes caspius: The P-glycoprotein efflux pumps. Med Vet. Entomol. 2008, 22, 48–54. [Google Scholar] [CrossRef]
- Sun, H.; Buchon, N.; Scott, J.G. Mdr65 decreases toxicity of multiple insecticides in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2017, 89, 11–16. [Google Scholar] [CrossRef]
- Yoon, K.S.; Strycharz, J.P.; Baek, J.H.; Sun, W.; Kim, J.H.; Kang, J.S.; Pittendrigh, B.R.; Lee, S.H.; Clark, J.M. Brief exposures of human body lice to sublethal amounts of ivermectin over-transcribes detoxification genes involved in tolerance. Insect Mol. Biol. 2011, 20, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Buss, D.S.; McCaffery, A.R.; Callaghan, A. Evidence for P-glycoprotein modification of insecticide toxicity in mosquitoes of the Culex pipiens complex. Med Vet. Entomol. 2002, 16, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Van Leeuwen, T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 2014, 45, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Jiang, C.; Zhou, X.; Qian, K.; Wang, L.; Shen, Y.; Zhao, Y. Increased expression of P-glycoprotein is associated with chlorpyrifos resistance in the german cockroach (blattodea: Blattellidae). J. Econ. Entomol. 2016, 109, 2500–2505. [Google Scholar] [CrossRef]
- Kang, X.L.; Zhang, M.; Wang, K.; Qiao, X.F.; Chen, M.H. Molecular cloning, expression pattern of multidrug resistance associated protein 1 (MRP1, ABCC1) gene, and the synergistic effects of verapamil on toxicity of two insecticides in the bird cherry-oat aphid. Arch. Insect Biochem. Physiol. 2016, 92, 65–84. [Google Scholar] [CrossRef]
- Le Gall, V.L.; Klafke, G.M.; Torres, T.T. Detoxification mechanisms involved in ivermectin resistance in the cattle tick, Rhipicephalus (boophilus) microplus. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Luo, L.; Sun, Y.J.; Wu, Y.J. Abamectin resistance in Drosophila is related to increased expression of P-glycoprotein via the dEGFR and dAkt pathways. Insect Biochem. Mol. Biol. 2013, 43, 627–634. [Google Scholar] [CrossRef]
- Pohl, P.C.; Klafke, G.M.; Carvalho, D.D.; Martins, J.R.; Daffre, S.; Da Silva Vaz, I.; Masuda, A. ABC transporter efflux pumps: A defense mechanism against ivermectin in Rhipicephalus (boophilus) microplus. Int. J. Parasitol. 2011, 41, 1323–1333. [Google Scholar] [CrossRef]
- Dermauw, W.; Wybouw, N.; Rombauts, S.; Menten, B.; Vontas, J.; Grbić, M.; Clark, R.M.; Feyereisen, R.; Van Leeuwen, T. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. USA 2013, 110, E113–E122. [Google Scholar] [CrossRef]
- Lanning, C.L.; Fine, R.L.; Corcoran, J.J.; Ayad, H.M.; Rose, R.L.; Abou-Donia, M.B. Tobacco budworm P-glycoprotein: Biochemical characterization and its involvement in pesticide resistance. Biochim. Et Biophys. Acta Gen. Subj. 1996, 1291, 155–162. [Google Scholar] [CrossRef]
- Riga, M.; Tsakireli, D.; Ilias, A.; Morou, E.; Myridakis, A.; Stephanou, E.G.; Nauen, R.; Dermauw, W.; Van Leeuwen, T.; Paine, M.; et al. Abamectin is metabolized by CYP392A16, a cytochrome P450 associated with high levels of acaricide resistance in Tetranychus urticae. Insect Biochem. Mol. Biol. 2014, 46, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Pu, J.; Chen, F.; Wang, J.; Han, Z. Multiple ATP-binding cassette transporters are involved in insecticide resistance in the small brown planthopper, Laodelphax striatellus. Insect Mol. Biol. 2017, 26, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Pauchet, Y.; Bretschneider, A.; Augustin, S.; Heckel, D.G. A P-glycoprotein is linked to resistance to the Bacillus thuringiensis Cry3Aa toxin in a leaf beetle. Toxins 2016, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Roulet, A.; Puel, O.; Gesta, S.; Lepage, J.F.; Drag, M.; Soll, M.; Alvinerie, M.; Pineau, T. MDR1-deficient genotype in Collie dogs hypersensitive to the P-glycoprotein substrate ivermectin. Eur. J. Pharmacol. 2003, 460, 85–91. [Google Scholar] [CrossRef]
- Xu, M.; Molento, M.; Blackhall, W.; Ribeiro, P.; Beech, R.; Prichard, R. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol. Biochem. Parasitol. 1998, 91, 327–335. [Google Scholar] [CrossRef]
- Schoville, S.D.; Chen, Y.H.; Andersson, M.N.; Benoit, J.B.; Bhandari, A.; Bowsher, J.H.; Brevik, K.; Cappelle, K.; Chen, M.J.M.; Childers, A.K.; et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef]
- Zhao, J.Z.; Bishop, B.A.; Grafius, E.J. Inheritance and synergism of resistance to imidacloprid in the Colorado potato beetle (Coleoptera: Chrysomelidae). J. Econ. Entomol. 2000, 93, 1508–1514. [Google Scholar] [CrossRef]
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Kumar, A.; Congiu, L.; Lindström, L.; Piiroinen, S.; Vidotto, M.; Grapputo, A. Sequencing, de novo assembly and annotation of the Colorado potato beetle, Leptinotarsa decemlineata, transcriptome. PLoS ONE 2014, 9, e86012. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to rt-qPCR-publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time rt-PCR. Nucleic Acids Res. 2001, 29, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Broehan, G.; Kroeger, T.; Lorenzen, M.; Merzendorfer, H. Functional analysis of the ATP-binding cassette (ATP) transporter gene family of Tribolium castaneum. BMC Genom. 2013, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Sharom, F.J. The P-glycoprotein multidrug transporter. Essays Biochem. 2011, 50, 161–178. [Google Scholar] [CrossRef]
- Merzendorfer, H. ABC transporters and their role in protecting insects from pesticides and their metabolites. In Advances in Insect Physiology; Elsevier Ltd.: Oxford, UK, 2014; Volume 46, pp. 1–72. [Google Scholar]
- Strauss, A.S.; Wang, D.; Stock, M.; Gretscher, R.R.; Groth, M.; Boland, W.; Burse, A. Tissue-specific transcript profiling for ABC transporters in the sequestering larvae of the phytophagous leaf beetle Chrysomela populi. PLoS ONE 2014, 9, e98637. [Google Scholar] [CrossRef]
- Pignatelli, P.; Ingham, V.A.; Balabanidou, V.; Vontas, J.; Lycett, G.; Ranson, H. The Anopheles gambiae ATP-binding cassette transporter family: Phylogenetic analysis and tissue localization provide clues on function and role in insecticide resistance. Insect Mol. Biol. 2018, 27, 110–122. [Google Scholar] [CrossRef]
- Saremba, B.M.; Murch, S.J.; Tymm, F.J.M.; Rheault, M.R. The metabolic fate of dietary nicotine in the cabbage looper, Trichoplusia ni (hübner). J. Insect Physiol. 2018, 109, 1–10. [Google Scholar] [CrossRef]
- Self, L.S.; Guthrie, F.E.; Hodgson, E. Metabolism of nicotine by tobacco-feeding insects. Nature 1964, 204, 300–301. [Google Scholar] [CrossRef]
- Sajitha, T.P.; Manjunatha, B.L.; Siva, R.; Gogna, N.; Dorai, K.; Ravikanth, G.; Uma Shaanker, R. Mechanism of resistance to camptothecin, a cytotoxic plant secondary metabolite, by Lymantria sp. larvae. J. Chem. Ecol. 2018, 44, 611–620. [Google Scholar] [CrossRef]
- Gómez, N.E.; Witte, L.; Hartmann, T. Chemical defense in larval tortoise beetles: Essential oil composition of fecal shields of Eurypedus nigrosignata and foliage of its host plant, Cordia curassavica. J. Chem. Ecol. 1999, 25, 1007–1027. [Google Scholar] [CrossRef]
- Strauss, A.S.; Peters, S.; Boland, W.; Burse, A. ABC transporter functions as a pacemaker for sequestration of plant glucosides in leaf beetles. eLife 2013, 2013, 1–16. [Google Scholar] [CrossRef]
- Mayer, F.; Mayer, N.; Chinn, L.; Pinsonneault, R.L.; Kroetz, D.; Bainton, R.J. Evolutionary conservation of vertebrate blood-brain barrier chemoprotective mechanisms in Drosophila. J. Neurosci. 2009, 29, 3538–3550. [Google Scholar] [CrossRef]
- Murray, C.L.; Quaglia, M.; Arnason, J.T.; Morris, C.E. A putative nicotine pump at the metabolic blood–brain barrier of the tobacco hornworm. J. Neurobiol. 1994, 25, 23–34. [Google Scholar] [CrossRef]
- Schinkel, A.H.; Wagenaar, E.; Mol, C.A.A.M.; Van Deemter, L. P-glycoprotein in the blood-brain barrier of mice influences the brain penetration and pharmacological activity of many drugs. J. Clin. Investig. 1996, 97, 2517–2524. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, A.H.; Wagenaar, E.; Van Deemter, L.; Mol, C.A.A.M.; Borst, P. Absence of the MDR1a P-glycoprotein in mice affects tissue distribution and pharmacokinetics of dexamethasone, digoxin, and cyclosporin A. J. Clin. Investig. 1995, 96, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, A.; Tamai, I.; Sakata, A.; Tenda, Y.; Terasaki, T. Restricted transport of cyclosporin A across the blood-brain barrier by a multidrug transporter, P-glycoprotein. Biochem. Pharmacol. 1993, 46, 1096–1099. [Google Scholar] [CrossRef]
- Groen, S.C.; LaPlante, E.R.; Alexandre, N.M.; Agrawal, A.A.; Dobler, S.; Whiteman, N.K. Multidrug transporters and organic anion transporting polypeptides protect insects against the toxic effects of cardenolides. Insect Biochem. Mol. Biol. 2017, 81, 51–61. [Google Scholar] [CrossRef]
- Shukle, R.H.; Yoshiyama, M.; Morton, P.K.; Johnson, A.J.; Schemerhorn, B.J. Tissue and developmental expression of a gene from hessian fly encoding an ABC-active-transporter protein: Implications for Malpighian tubule function during interactions with wheat. J. Insect Physiol. 2008, 54, 146–154. [Google Scholar] [CrossRef]
- Gaertner, L.S.; Murray, C.L.; Morris, C.E. Transepithelial transport of nicotine and vinblastine in isolated Malpighian tubules of the tobacco hornworm (Manduca sexta) suggests a P-glycoprotein-like mechanism. J. Exp. Biol. 1998, 201, 2637–2645. [Google Scholar]
- Rheault, M.R.; Plaumann, J.S.; O’Donnell, M.J. Tetraethylammonium and nicotine transport by the Malpighian tubules of insects. J. Insect Physiol. 2006, 52, 487–498. [Google Scholar] [CrossRef]
- Tian, L.; Yang, J.; Hou, W.; Xu, B.; Xie, W.; Wang, S.; Zhang, Y.; Zhou, X.; Qingjun, W.U. Molecular cloning and characterization of a P-glycoprotein from the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Mol. Sci. 2013, 14, 22891–22905. [Google Scholar] [CrossRef] [PubMed]
- James, C.E.; Davey, M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009, 39, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Blackhall, W.J.; Liu, H.Y.; Xu, M.; Prichard, R.K.; Beech, R.N. Selection at a P-glycoprotein gene in ivermectin- and moxidectin-selected strains of Haemonchus contortus. Mol. Biochem. Parasitol. 1998, 95, 193–201. [Google Scholar] [CrossRef]
- Srinivas, R.; Udikeri, S.S.; Jayalakshmi, S.K.; Sreeramulu, K. Identification of factors responsible for insecticide resistance in Helicoverpa armigera. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004, 137, 261–269. [Google Scholar] [CrossRef]
- Zuo, Y.Y.; Huang, J.L.; Wang, J.; Feng, Y.; Han, T.T.; Wu, Y.D.; Yang, Y.H. Knockout of a P-glycoprotein gene increases susceptibility to abamectin and emamectin benzoate in Spodoptera exigua. Insect Mol. Biol. 2018, 27, 36–45. [Google Scholar] [CrossRef]
- Yoon, K.S.; Nelson, J.O.; Marshall Clark, J. Selective induction of abamectin metabolism by dexamethasone, 3-methylcholanthrene, and phenobarbital in Colorado potato beetle, Leptinotarsa decemlineata (say). Pestic. Biochem. Physiol. 2002, 73, 74–86. [Google Scholar] [CrossRef]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Lu, X.B.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015, 22, 203–219. [Google Scholar] [CrossRef]
- Qian, L.; Cao, G.; Song, J.; Yin, Q.; Han, Z. Biochemical mechanisms conferring cross-resistance between tebufenozide and abamectin in Plutella xylostella. Pestic. Biochem. Physiol. 2008, 91, 175–179. [Google Scholar] [CrossRef]
- Mounsey, K.E.; Pasay, C.J.; Arlian, L.G.; Morgan, M.S.; Holt, D.C.; Currie, B.J.; Walton, S.F.; McCarthy, J.S. Increased transcription of Glutathione S-transferases in acaricide exposed scabies mites. Parasites Vectors 2010, 3, 43. [Google Scholar] [CrossRef]
- Wei, Q.B.; Lei, Z.R.; Nauen, R.; Cai, D.C.; Gao, Y.L. Abamectin resistance in strains of vegetable leafminer, Liriomyza sativae (diptera: Agromyzidae) is linked to elevated Glutathione S-transferase activity. Insect Sci. 2015, 22, 243–250. [Google Scholar] [CrossRef]
- Çağatay, N.S.; Menault, P.; Riga, M.; Vontas, J.; Ay, R. Identification and characterization of abamectin resistance in Tetranychus urticae Koch populations from greenhouses in Turkey. Crop Prot. 2018, 112, 112–117. [Google Scholar] [CrossRef]
- Stumpf, N.; Nauen, R. Biochemical markers linked to abamectin resistance in Tetranychus urticae (Acari: Tetranychidae). Pestic. Biochem. Physiol. 2002, 72, 111–121. [Google Scholar] [CrossRef]
- Clements, J.; Schoville, S.; Peterson, N.; Huseth, A.S.; Lan, Q.; Groves, R.L. RNA interference of three up-regulated transcripts associated with insecticide resistance in an imidacloprid resistant population of Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2017, 135, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, M.; Palli, S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 2017, 83, 1–12. [Google Scholar] [CrossRef]
- Gaddelapati, S.C.; Kalsi, M.; Roy, A.; Palli, S.R. Cap ‘n’ collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Biochem. Mol. Biol. 2018, 99, 54–62. [Google Scholar] [CrossRef]
- Clements, J.; Schoville, S.; Clements, N.; Chapman, S.; Groves, R.L. Temporal patterns of imidacloprid resistance throughout a growing season in Leptinotarsa decemlineata populations. Pest Manag. Sci. 2017, 73, 641–650. [Google Scholar] [CrossRef]
- Clements, J.; Schoville, S.; Peterson, N.; Lan, Q.; Groves, R.L. Characterizing molecular mechanisms of imidacloprid resistance in select populations of Leptinotarsa decemlineata in the Central Sands region of Wisconsin. PLoS ONE 2016, 11, e0147844. [Google Scholar] [CrossRef]
| Primer | Sequence |
|---|---|
| Primers for Fragment + L4440 Construction | |
| LedMDR1 For | [GAGCGGCCGC]TGTTCATGATTTATTCTAGT * |
| LedMDR1 Rev | [GCAGGTCGAC]ATCTGGTCTGACGGATAG * |
| LedMDR2 For | [GAGCGGCCGC]AAGTTAGCCGTGGAAGCCAT * |
| LedMDR2 Rev | [GCAGGTCGAC]TCCCACGCACAATTCACTGG * |
| LedMDR3 For | [TAGCGGCCGC]AGTGGGAAGACGCCATCAGT * |
| LedMDR3 Rev | [GCAGGTCGAC]TGCCATACCACCAACATAACGA * |
| qPCR Primers | |
| qLedL8e For | GGTAACCATCAACACATTGG |
| qLedL8e Rev | TCTTGGCATCCACTTTACC |
| qLedMDR1 For | TAGACCTCACATGGTTCAGG |
| qLedMDR1 Rev | TTAGACTTCCGTTGACTTCTTC |
| qLedMDR2 For | TAGTTTCCCAGGAGCCGAAC |
| qLedMDR2 Rev | TTCGCACTCTTTGCAGCTTTC |
| qLedMDR3 For | TCGTTGGTATCTGCTCTCTTCG |
| qLedMDR3 Rev | TGAGGTGCCATTATTCGATCTG |
| Name | Protein Type | Accession | Genome Transcript(S) * |
|---|---|---|---|
| LedABCB 1 | Full | BK010703 | LDEC021233/014301 |
| LedABCB 2 | Full | BK010704 | LDEC008907/008908/008909/008910 |
| LedABCB 3 | Full | BK010705 | LDEC008906 |
| LedABCB member 6, mitochondrial-like | half | XM_023161180 | LDEC007355/022912 |
| LedABCB member 7, mitochondrial-like | half | XM_023169211 | LDEC002144/024539 |
| LedABCB member 8, mitochondrial-like | half | XM_023160888 | LDEC019485 |
| LedABCB member 10, mitochondrial-like | half | XM_023173355 | LDEC024415/010427 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Favell, G.; McNeil, J.N.; Donly, C. The ABCB Multidrug Resistance Proteins Do Not Contribute to Ivermectin Detoxification in the Colorado Potato Beetle, Leptinotarsa decemlineata (Say). Insects 2020, 11, 135. https://doi.org/10.3390/insects11020135
Favell G, McNeil JN, Donly C. The ABCB Multidrug Resistance Proteins Do Not Contribute to Ivermectin Detoxification in the Colorado Potato Beetle, Leptinotarsa decemlineata (Say). Insects. 2020; 11(2):135. https://doi.org/10.3390/insects11020135
Chicago/Turabian StyleFavell, Grant, Jeremy N. McNeil, and Cam Donly. 2020. "The ABCB Multidrug Resistance Proteins Do Not Contribute to Ivermectin Detoxification in the Colorado Potato Beetle, Leptinotarsa decemlineata (Say)" Insects 11, no. 2: 135. https://doi.org/10.3390/insects11020135
APA StyleFavell, G., McNeil, J. N., & Donly, C. (2020). The ABCB Multidrug Resistance Proteins Do Not Contribute to Ivermectin Detoxification in the Colorado Potato Beetle, Leptinotarsa decemlineata (Say). Insects, 11(2), 135. https://doi.org/10.3390/insects11020135




