Infection Patterns and Fitness Effects of Rickettsia and Sodalis Symbionts in the Green Lacewing Chrysoperla carnea
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Endosymbiont Screening in Natural and Laboratory Ch. carnea Populations
2.2. Molecular Characterization of Endosymbionts
2.3. Endosymbiont–Host Interaction
2.3.1. Insect Rearing
2.3.2. Vertical Transmission Rate of Endosymbionts in Ch. carnea
2.3.3. Effect of Endosymbionts on Host Reproductive Success
2.3.4. Endosymbiont Titer in Single and Co-Infected Ch. carnea
3. Results
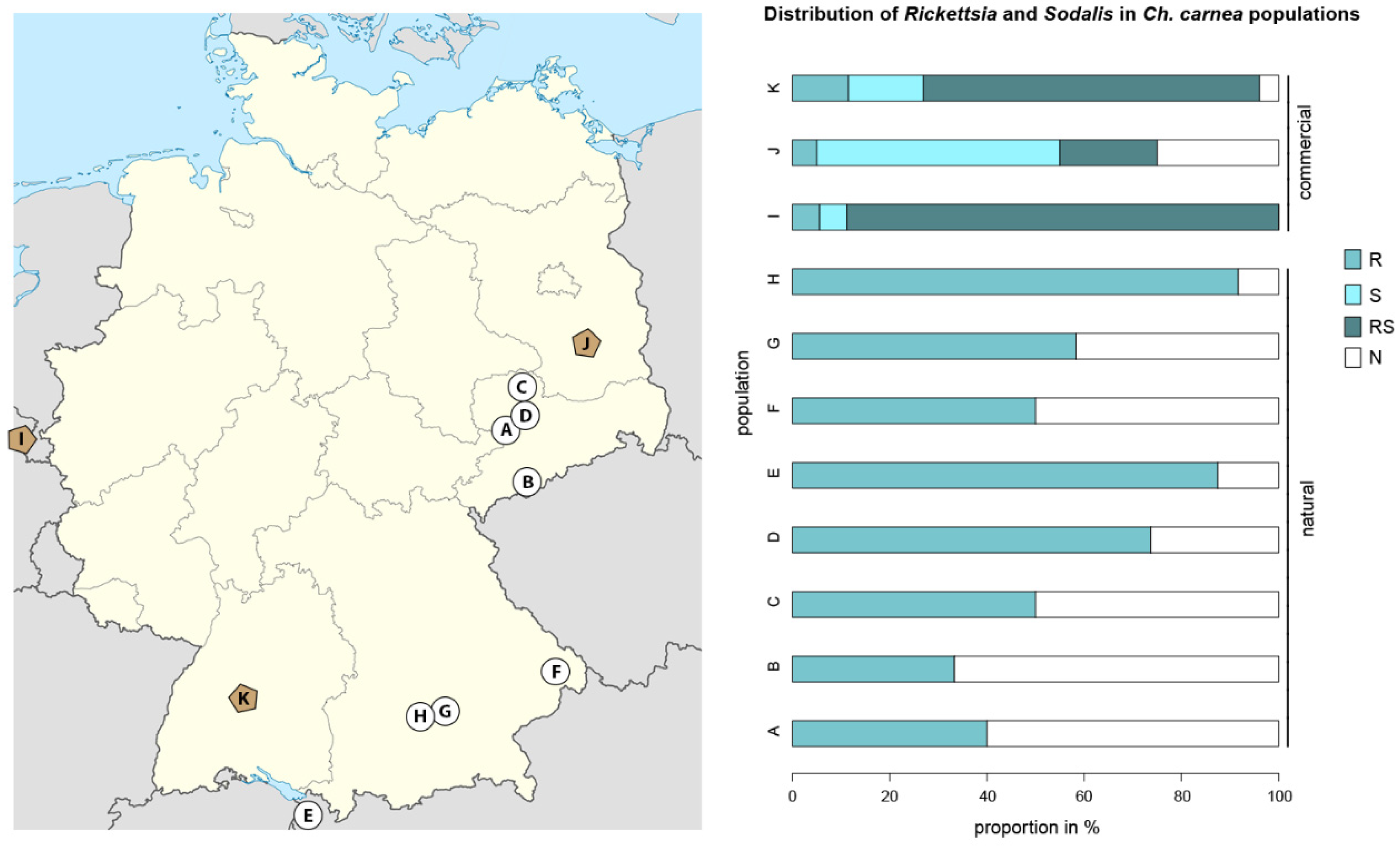
3.1. The Screening of Rickettsia and Sodalis Symbionts on Population Levels in Natural and Laboratory Ch. carnea
3.2. Molecular Phylogenetic Characterization of Endosymbionts
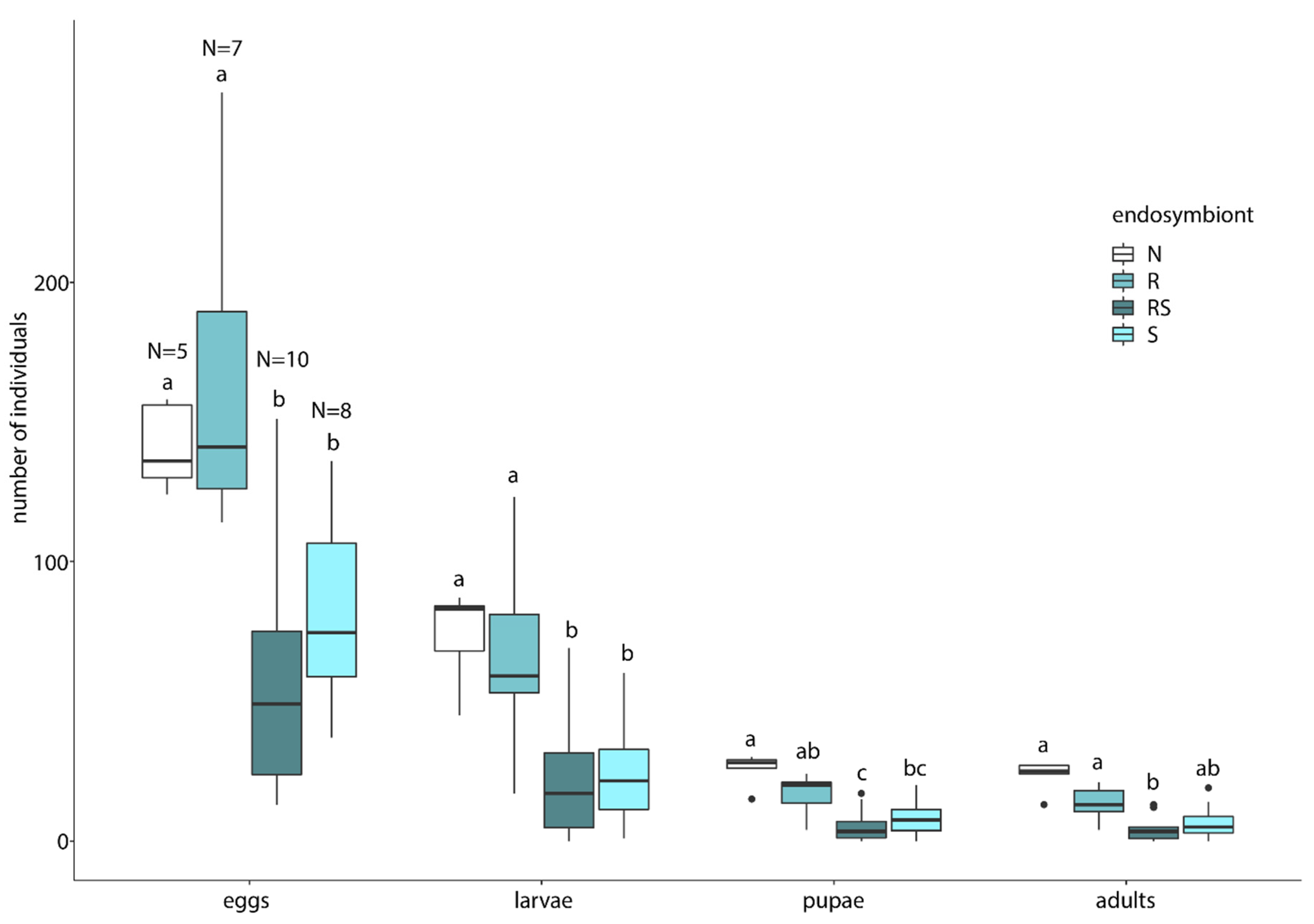
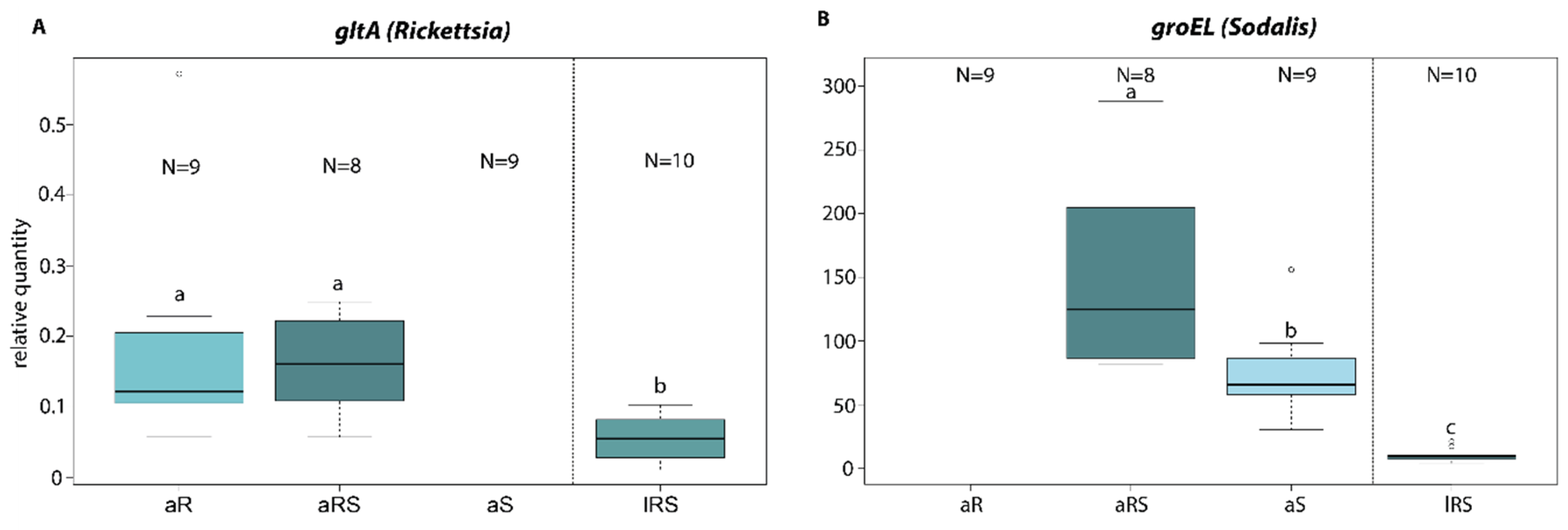
3.3. Endosymbiont Host Interaction
4. Discussion
4.1. Endosymbiont Screening on Population Levels in Natural and Laboratory Ch. carnea
4.2. Molecular Phylogenetic Characterization of Endosymbionts
4.3. Endosymbiont Host Interaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aspöck, U. Phylogeny of the neuropterida (insecta: Holometabola). Zool. Scr. 2002, 31, 51–55. [Google Scholar] [CrossRef]
- Henry, C.S. Sibling species, call differences, and speciation in green lacewings (neuroptera: Chrysopidae: Chrysoperla). Evolution 1985, 39, 965–984. [Google Scholar]
- Henry, C.S.; Brooks, S.J.; Duelli, P.; Johnson, J.B.; Wells, M.M.; Mochizuki, A. Obligatory duetting behaviour in the chrysoperla carnea—Group of cryptic species (neuroptera: Chrysopidae): Its role in shaping evolutionary history. Biol. Rev. 2013, 88, 787–808. [Google Scholar] [CrossRef] [PubMed]
- Canard, M.; Semeria, Y.; New, T. Biology of Chrysopidae; Springer: Dordrecht, The Netherlands, 1984. [Google Scholar]
- McEwen, P.K.; New, T.R.; Whittington, A.E. Lacewings in the Crop Environment; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Hagley, E.; Miles, N. Release of chrysoperla carnea stephens (neuroptera: Chrysopidae) for control of tetranychus urticae koch (acarina: Tetranychidae) on peach grown in a protected environment structure. Can. Entomol. 1987, 119, 205–206. [Google Scholar] [CrossRef]
- Hommen, U.; Baveco, J.; Galic, N.; van den Brink, P.J. Potential application of ecological models in the european environmental risk assessment of chemicals i: Review of protection goals in eu directives and regulations. Integr. Environ. Assess. Manag. 2010, 6, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Bigler, F. Biological control by chrysopids: Intergration with pesticides. In Biology of Chrysopidae; Canard, M., Semeria, Y., New, T., Eds.; Springer: Dordrecht, The Netherlands, 1984; Volume 27, pp. 233–245. [Google Scholar]
- Tsuchida, T.; Koga, R.; Horikawa, M.; Tsunoda, T.; Maoka, T.; Matsumoto, S.; Simon, J.-C.; Fukatsu, T. Symbiotic bacterium modifies aphid body color. Science 2010, 330, 1102–1104. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Aksoy, S. Tsetse–a haven for microorganisms. Parasitol. Today 2000, 16, 114–118. [Google Scholar] [CrossRef]
- Sandström, J.P.; Russell, J.A.; White, J.P.; Moran, N.A. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 2001, 10, 217–228. [Google Scholar] [CrossRef]
- Baldo, L.; Ayoub, N.A.; Hayashi, C.Y.; Russell, J.A.; Stahlhut, J.K.; Werren, J.H. Insight into the routes of wolbachia invasion: High levels of horizontal transfer in the spider genus agelenopsis revealed by wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 2008, 17, 557–569. [Google Scholar] [CrossRef]
- Weinert, L.A.; Araujo-Jnr, E.V.; Ahmed, M.Z.; Welch, J.J. The Incidence of Bacterial Endosymbionts in Terrestrial Arthropods. Proc. R. Soc. 2015, 282, 20150249. [Google Scholar] [CrossRef] [PubMed]
- Weinert, L.A.; Werren, J.H.; Aebi, A.; Stone, G.N.; Jiggins, F.M. Evolution and diversity of rickettsia bacteria. BMC Biol. 2009, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Cowdry, E.V. The distribution of rickettsia in the tissues of insects and arachnids. J. Exp. Med. 1923, 37, 431–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, L. How charles nicolle of the pasteur institute discovered that epidemic typhus is transmitted by lice: Reminiscences from my years at the pasteur institute in paris. Proc. Natl. Acad. Sci. USA 1996, 93, 10539–10540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.J.; Ying, Z.; Brunner, B.R.; Pantoja, A.; Ferwerda, F.H. Rickettsial relative associated with papaya bunchy top disease. Curr. Microbiol. 1998, 36, 80–84. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Sameshima, S.; Kitade, O.; Kojima, J.; Fukatsu, T. Novel clade of rickettsia spp. From leeches. Appl. Environ. Microbiol. 2002, 68, 999–1004. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.W.; Lohr, K.E.; Cameron, C.M.; Williams, D.E.; Peters, E.C. Disease dynamics and potential mitigation among restored and wild staghorn coral, acropora cervicornis. PeerJ 2014, 2, e541. [Google Scholar] [CrossRef] [Green Version]
- Modeo, L.; Salvetti, A.; Rossi, L.; Castelli, M.; Szokoli, F.; Krenek, S.; Serra, V.; Sabaneyeva, E.; Di Giuseppe, G.; Fokin, S.I. “Candidatus trichorickettsia mobilis”, a rickettsiales bacterium, can be transiently transferred from the unicellular eukaryote paramecium to the planarian dugesia japonica. PeerJ 2020, 8, e8977. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Hurst, G.; Zhang, W.; Breeuwer, J.; Stouthamer, R.; Majerus, M. Rickettsial relative associated with male killing in the ladybird beetle (adalia bipunctata). J. Bacteriol. 1994, 176, 388–394. [Google Scholar] [CrossRef] [Green Version]
- Lawson, E.T.; Mousseau, T.A.; Klaper, R.; Hunter, M.D.; Werren, J.H. Rickettsia associated with male-killing in a buprestid beetle. Heredity 2001, 86, 497–505. [Google Scholar] [CrossRef] [Green Version]
- Hagimori, T.; Abe, Y.; Date, S.; Miura, K. The first finding of a rickettsia bacterium associated with parthenogenesis induction among insects. Curr. Microbiol. 2006, 52, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Giorgini, M.; Bernardo, U.; Monti, M.; Nappo, A.; Gebiola, M. Rickettsia symbionts cause parthenogenetic reproduction in the parasitoid wasp pnigalio soemius (hymenoptera: Eulophidae). Appl. Environ. Microbiol. 2010, 76, 2589–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiel, E.; Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Katzir, N.; Inbar, M.; Ghanim, M. Biotype-dependent secondary symbiont communities in sympatric populations of bemisia tabaci. Bull. Entomol. Res. 2007, 97, 407–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Nomura, M.; Kageyama, D. Rapid comeback of males: Evolution of male-killer suppression in a green lacewing population. Proc. R. Soc. B 2018, 285, 20180369. [Google Scholar] [CrossRef]
- Hayashi, M.; Watanabe, M.; Yukuhiro, F.; Nomura, M.; Kageyama, D. A nightmare for males? A maternally transmitted male-killing bacterium and strong female bias in a green lacewing population. PLoS ONE 2016, 11, e0155794. [Google Scholar] [CrossRef]
- Gerth, M.; Wolf, R.; Bleidorn, C.; Richter, J.; Sontowski, R.; Unrein, J.; Schlegel, M.; Gruppe, A. Green lacewings (neuroptera: Chrysopidae) are commonly associated with a diversity of rickettsial endosymbionts. Zool. Lett. 2017, 3, 12. [Google Scholar] [CrossRef]
- Williams, K.P.; Gillespie, J.J.; Sobral, B.W.; Nordberg, E.K.; Snyder, E.E.; Shallom, J.M.; Dickerman, A.W. Phylogeny of gammaproteobacteria. J. Bacteriol. 2010, 192, 2305–2314. [Google Scholar] [CrossRef] [Green Version]
- Aksoy, S.; Chen, X.-A.; Hypsa, V. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (diptera: Glossinidae). Insect Mol. Biol. 1997, 6, 183–190. [Google Scholar] [CrossRef]
- Kaiwa, N.; Hosokawa, T.; Kikuchi, Y.; Nikoh, N.; Meng, X.Y.; Kimura, N.; Ito, M.; Fukatsu, T. Primary gut symbiont and secondary, sodalis-allied symbiont of the scutellerid stinkbug cantao ocellatus. Appl. Environ. Microbiol. 2010, 76, 3486–3494. [Google Scholar] [CrossRef] [Green Version]
- Nováková, E.; Hypša, V. A new sodalis lineage from bloodsucking fly craterina melbae (diptera, hippoboscoidea) originated independently of the tsetse flies symbiont sodalis glossinidius. FEMS Microbiol. Lett. 2007, 269, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Masson, F.; Vallier, A.; Balmand, S.; Rey, M.; Vincent-Monégat, C.; Aksoy, E.; Aubailly-Giraud, E.; Zaidman-Rémy, A.; Heddi, A. Insects recycle endosymbionts when the benefit is over. Curr. Biol. 2014, 24, 2267–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukatsu, T.; Koga, R.; Smith, W.A.; Tanaka, K.; Nikoh, N.; Sasaki-Fukatsu, K.; Yoshizawa, K.; Dale, C.; Clayton, D.H. Bacterial endosymbiont of the slender pigeon louse, columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 2007, 73, 6660–6668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosokawa, T.; Kaiwa, N.; Matsuura, Y.; Kikuchi, Y.; Fukatsu, T. Infection prevalence of sodalis symbionts among stinkbugs. Zool. Lett. 2015, 1, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šochová, E.; Husník, F.; Nováková, E.; Halajian, A.; Hypša, V. Arsenophonus and sodalis replacements shape evolution of symbiosis in louse flies. PeerJ 2017, 5, e4099. [Google Scholar] [CrossRef] [Green Version]
- Dale, C.; Welburn, S. The endosymbionts of tsetse flies: Manipulating host–parasite interactions. Int. J. Parasitol. 2001, 31, 628–631. [Google Scholar] [CrossRef]
- Enomoto, S.; Chari, A.; Clayton, A.L.; Dale, C. Quorum sensing attenuates virulence in sodalis praecaptivus. Cell Host Microbe. 2017, 21, 629–636.e625. [Google Scholar] [CrossRef] [Green Version]
- Gottlieb, Y.; Ghanim, M.; Chiel, E.; Gerling, D.; Portnoy, V.; Steinberg, S.; Tzuri, G.; Horowitz, A.R.; Belausov, E.; Mozes-Daube, N. Identification and localization of a rickettsia sp. In bemisia tabaci (homoptera: Aleyrodidae). Appl. Environ. Microbiol. 2006, 72, 3646–3652. [Google Scholar] [CrossRef] [Green Version]
- Ayoubi, A.; Talebi, A.A.; Fathipour, Y.; Mehrabadi, M. Coinfection of the secondary symbionts, hamiltonella defensa and arsenophonus sp. Contribute to the performance of the major aphid pest, aphis gossypii (hemiptera: Aphididae). Insect Sci. 2018, 27, 86–98. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, 2010, pdb. prot5448. [Google Scholar] [CrossRef]
- Kircher, M. Analysis of high-throughput ancient DNA sequencing data. Anc. DNA Methods Protoc. 2012, 197–228. [Google Scholar]
- Renaud, G.; Kircher, M.; Stenzel, U.; Kelso, J. Freeibis: An efficient basecaller with calibrated quality scores for illumina sequencers. Bioinformatics 2013, 29, 1208–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. Idba-ud: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlazeck, F.J.; Rescheneder, P.; von Haeseler, A. Nextgenmap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 2013, 29, 2790–2791. [Google Scholar] [CrossRef] [Green Version]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. Guidance2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Mao, J.; Zeng, F. Chrysopa septempunctata (neuroptera: Chrysopidae) vitellogenin functions through effects on egg production and hatching. J. Econ. Entomol. 2015, 108, 2779–2788. [Google Scholar] [CrossRef] [PubMed]
- Caspi-Fluger, A.; Inbar, M.; Steinberg, S.; Friedmann, Y.; Freund, M.; Mozes-Daube, N.; Zchori-Fein, E. Characterization of the symbiont rickettsia in the mirid bug nesidiocoris tenuis (reuter)(heteroptera: Miridae). Bull. Entomol. Res. 2014, 104, 681–688. [Google Scholar] [CrossRef]
- Dale, C.; Maudlin, I. Sodalis gen. Nov. And sodalis glossinidius sp. Nov., a microaerophilic secondary endosymbiont of the tsetse fly glossina morsitans morsitans. Int. J. Syst. Evol. Microbiol. 1999, 49, 267–275. [Google Scholar] [CrossRef]
- Koga, R.; Moran, N.A. Swapping symbionts in spittlebugs: Evolutionary replacement of a reduced genome symbiont. ISME J. 2014, 8, 1237–1246. [Google Scholar] [CrossRef] [Green Version]
- Saeed, A.; White, J.A. Surveys for maternally-inherited endosymbionts reveal novel and variable infections within solitary bee species. J. Invertebr. Pathol. 2015, 132, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef] [PubMed]
- Doudoumis, V.; Tsiamis, G.; Wamwiri, F.; Brelsfoard, C.; Alam, U.; Aksoy, E.; Dalaperas, S.; Abd-Alla, A.; Ouma, J.; Takac, P. Detection and characterization of wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus glossina). BMC Microbiol. 2012, 12, S3. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.A.; Turelli, M.; Harshman, L.G. Factors affecting the distribution of cytoplasmic incompatibility in drosophila simulans. Genetics 1990, 126, 933–948. [Google Scholar] [PubMed]
- Prado, S.S.; Golden, M.; Follett, P.A.; Daugherty, M.P.; Almeida, R.P. Demography of gut symbiotic and aposymbiotic nezara viridula l.(hemiptera: Pentatomidae). Environ. Entomol. 2009, 38, 103–109. [Google Scholar] [CrossRef]
- Toju, H.; Fukatsu, T. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: Relevance of local climate and host plants. Mol. Ecol. 2011, 20, 853–868. [Google Scholar] [CrossRef]
- Snyder, A.K.; McMillen, C.M.; Wallenhorst, P.; Rio, R.V. The phylogeny of sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol. Lett. 2011, 317, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Santos-Garcia, D.; Silva, F.J.; Morin, S.; Dettner, K.; Kuechler, S.M. The all-rounder sodalis: A new bacteriome-associated endosymbiont of the lygaeoid bug henestaris halophilus (heteroptera: Henestarinae) and a critical examination of its evolution. Genome Biol. Evol. 2017, 9, 2893–2910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chari, A.; Oakeson, K.F.; Enomoto, S.; Jackson, D.G.; Fisher, M.A.; Dale, C. Phenotypic characterization of sodalis praecaptivus sp. Nov., a close non-insect-associated member of the sodalis-allied lineage of insect endosymbionts. Int. J. Syst. Evol. Microbiol. 2015, 65, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.L.; Oakeson, K.F.; Gutin, M.; Pontes, A.; Dunn, D.M.; von Niederhausern, A.C.; Weiss, R.B.; Fisher, M.; Dale, C. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 2012, 8, e1002990. [Google Scholar] [CrossRef] [PubMed]
- Oakeson, K.F.; Gil, R.; Clayton, A.L.; Dunn, D.M.; von Niederhausern, A.C.; Hamil, C.; Aoyagi, A.; Duval, B.; Baca, A.; Silva, F.J.; et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 2014, 6, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Toh, H.; Weiss, B.L.; Perkin, S.A.; Yamashita, A.; Oshima, K.; Hattori, M.; Aksoy, S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of sodalis glossinidius in the tsetse host. Genome Res. 2006, 16, 149–156. [Google Scholar] [CrossRef] [Green Version]
- Rosas-Pérez, T.; de León, A.V.-P.; Rosenblueth, M.; Ramírez-Puebla, S.T.; Rincón-Rosales, R.; Martínez-Romero, J.; Dunn, M.F.; Kondorosi, É.; Martínez-Romero, E. The symbiome of llaveia cochineals (hemiptera: Coccoidea: Monophlebidae) includes a gammaproteobacterial cosymbiont sodalis tme1 and the known candidatus walczuchella monophlebidarum. In Insect Physiology and Ecology; IntechOpen: London, UK, 2017. [Google Scholar]
- Silva, F.J.; Latorre, A.; Gómez-Valero, L.; Moya, A. Genomic changes in bacteria: From free-living to endosymbiotic life. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 149–165. [Google Scholar]
- Kondo, N.; Shimada, M.; Fukatsu, T. Infection density of wolbachia endosymbiont affected by co-infection and host genotype. Biol. Lett. 2005, 1, 488–491. [Google Scholar] [CrossRef] [Green Version]
- Goto, S.; Anbutsu, H.; Fukatsu, T. Asymmetrical interactions between wolbachia and spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 2006, 72, 4805–4810. [Google Scholar] [CrossRef] [Green Version]
- Balmand, S.; Lohs, C.; Aksoy, S.; Heddi, A. Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 2013, 112, S116–S122. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Aksoy, S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 1999, 8, 125–132. [Google Scholar] [CrossRef]
- Chen, D.Q.; Montllor, C.B.; Purcell, A.H. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, acyrthosiphon pisum, and the blue alfalfa aphid, a. Kondoi. Entomol. Exp. Et Appl. 2000, 95, 315–323. [Google Scholar] [CrossRef]
- Sakurai, M.; Koga, R.; Tsuchida, T.; Meng, X.-Y.; Fukatsu, T. Rickettsia symbiont in the pea aphid acyrthosiphon pisum: Novel cellular tropism, effect on host fitness, and interaction with the essential symbiont buchnera. Appl. Environ. Microbiol. 2005, 71, 4069–4075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farikou, O.; Njiokou, F.; Mbida, J.A.M.; Njitchouang, G.R.; Djeunga, H.N.; Asonganyi, T.; Simarro, P.P.; Cuny, G.; Geiger, A. Tripartite interactions between tsetse flies, sodalis glossinidius and trypanosomes—An epidemiological approach in two historical human african trypanosomiasis foci in cameroon. Infect. Genet. Evol. 2010, 10, 115–121. [Google Scholar] [CrossRef]
- Welburn, S.; Maudlin, I. Tsetse–trypanosome interactions: Rites of passage. Parasitol. Today 1999, 15, 399–403. [Google Scholar] [CrossRef]
- Liu, X.-D.; Lei, H.-X.; Chen, F.-F. Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment-dependent. Sci. Rep. 2019, 9, 4013. [Google Scholar] [CrossRef]
- Zytynska, S.E.; Thighiouart, K.; Frago, E. A meta-analysis on the benefits and costs of hosting secondary endosymbionts in sap-sucking insects. bioRxiv 2019, 563031. [Google Scholar] [CrossRef]
- Boyd, B.M.; Allen, J.M.; Koga, R.; Fukatsu, T.; Sweet, A.D.; Johnson, K.P.; Reed, D.L. Two bacterial genera, sodalis and rickettsia, associated with the seal louse proechinophthirus fluctus (phthiraptera: Anoplura). Appl. Environ. Microbiol. 2016, 82, 3185–3197. [Google Scholar] [CrossRef] [Green Version]
- Skaljac, M.; Zanic, K.; Ban, S.G.; Kontsedalov, S.; Ghanim, M. Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol. 2010, 10, 142. [Google Scholar] [CrossRef] [Green Version]

| Rickettsia | Sodalis | |
|---|---|---|
| Number of offspring infected with only one endosymbiont | 3 (2.9%) | 11 (10.6%) |
| Number of offspring infected with both endosymbionts | 90 (86.5%) | 90 (86.5%) |
| Total number of infected offspring | 93 (89.4%) | 101 (97.1%) |
| R-N | RS-N | S-N | R-RS | R-S | RS-S | ||
|---|---|---|---|---|---|---|---|
| Eggs | t p-value | 0.678 0.504 | −3.066 0.005 ** | −2.063 0.049 * | −3.739 0.001 ** | −2.730 0.012 * | 0.90 0.382 |
| Larvae | t p-value | −0.327 0.747 | −3.212 0.004 ** | −2.941 0.007 ** | −2.877 0.009 ** | −2.614 0.016 * | 0.069 0.945 |
| Pupae | t p-value | −1.502 0.145 | −4.276 <0.001 *** | −3.340 0.003 ** | −2.856 0.009 ** | −1.914 0.070 | 0.868 0.398 |
| Adults | t p-value | −1.864 0.074 | −4.518 <0.001 *** | −3.606 0.001 ** | −2.809 0.010 * | −1.863 0.080 | 0.883 0.390 |
| Test | Lines | gltA(Rickettsia) | groEL (Sodalis) |
|---|---|---|---|
| ANOVA | F(2/24) = 9.744 | F(2/24) = 146.18 | |
| p < 0.001 *** | p < 0.001 *** | ||
| Tukey post-hoc | aS-aRS | - | p = 0.021 * |
| aR-aRS | p = 0.997 | - | |
| lRS-aRS | p = 0.002 ** | p < 0.001 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sontowski, R.; Gerth, M.; Richter, S.; Gruppe, A.; Schlegel, M.; van Dam, N.M.; Bleidorn, C. Infection Patterns and Fitness Effects of Rickettsia and Sodalis Symbionts in the Green Lacewing Chrysoperla carnea. Insects 2020, 11, 867. https://doi.org/10.3390/insects11120867
Sontowski R, Gerth M, Richter S, Gruppe A, Schlegel M, van Dam NM, Bleidorn C. Infection Patterns and Fitness Effects of Rickettsia and Sodalis Symbionts in the Green Lacewing Chrysoperla carnea. Insects. 2020; 11(12):867. https://doi.org/10.3390/insects11120867
Chicago/Turabian StyleSontowski, Rebekka, Michael Gerth, Sandy Richter, Axel Gruppe, Martin Schlegel, Nicole M. van Dam, and Christoph Bleidorn. 2020. "Infection Patterns and Fitness Effects of Rickettsia and Sodalis Symbionts in the Green Lacewing Chrysoperla carnea" Insects 11, no. 12: 867. https://doi.org/10.3390/insects11120867
APA StyleSontowski, R., Gerth, M., Richter, S., Gruppe, A., Schlegel, M., van Dam, N. M., & Bleidorn, C. (2020). Infection Patterns and Fitness Effects of Rickettsia and Sodalis Symbionts in the Green Lacewing Chrysoperla carnea. Insects, 11(12), 867. https://doi.org/10.3390/insects11120867






