Bioclimatic Modelling Identifies Suitable Habitat for the Establishment of the Invasive European Paper Wasp (Hymenoptera: Vespidae) across the Southern Hemisphere
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
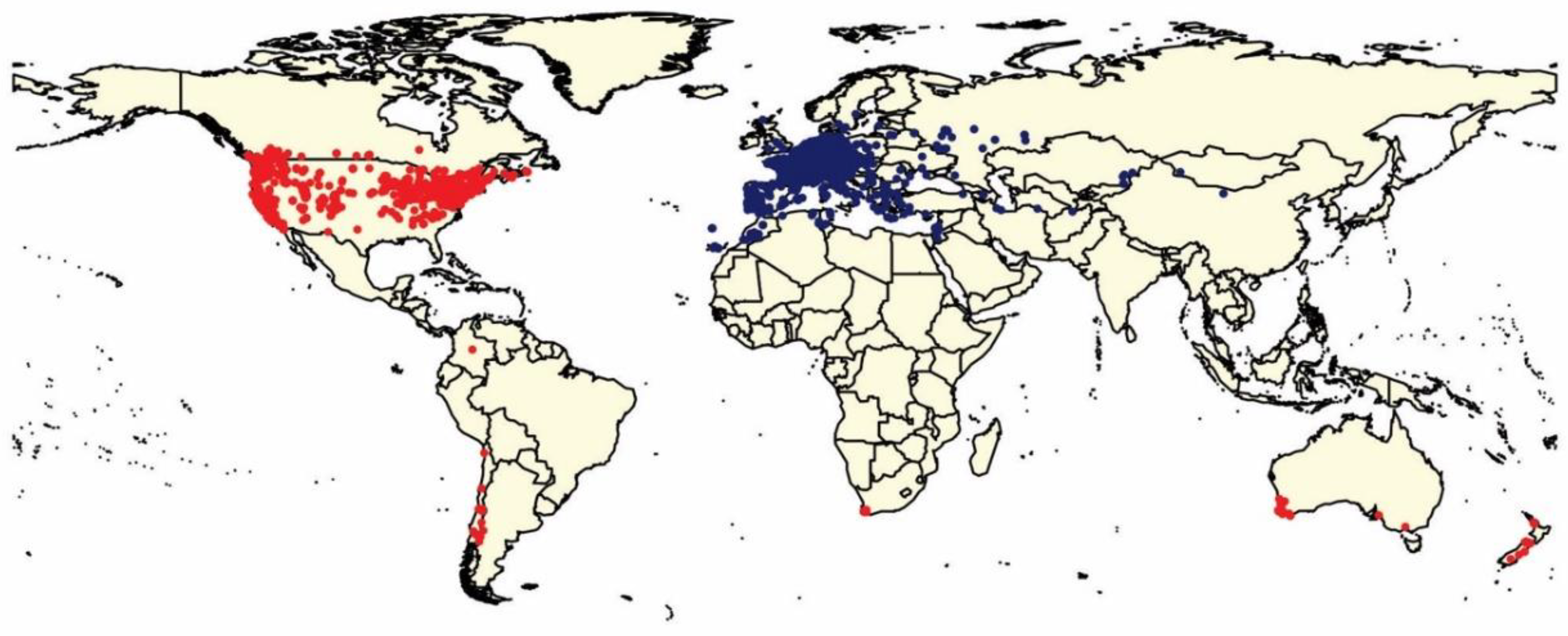
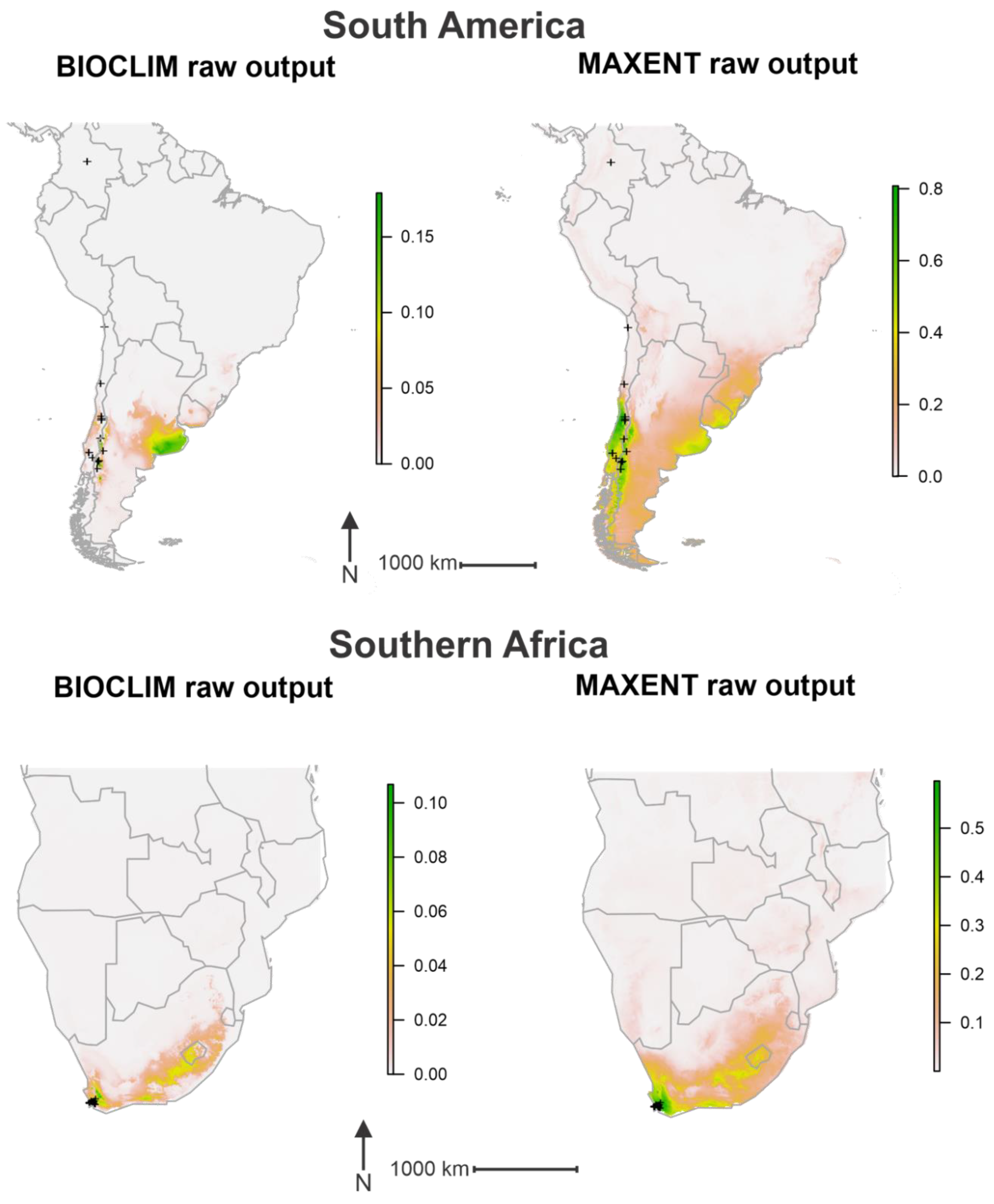
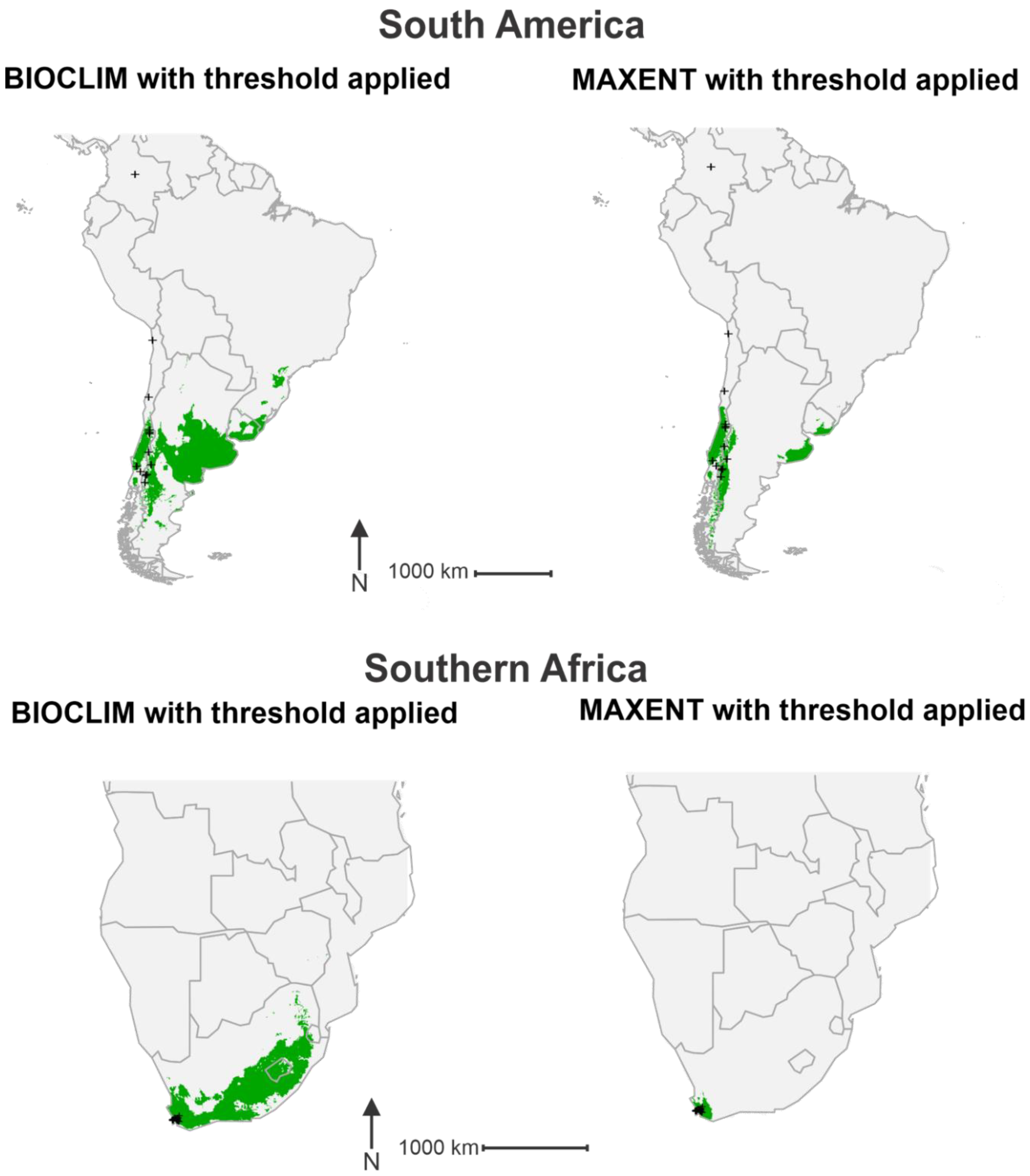
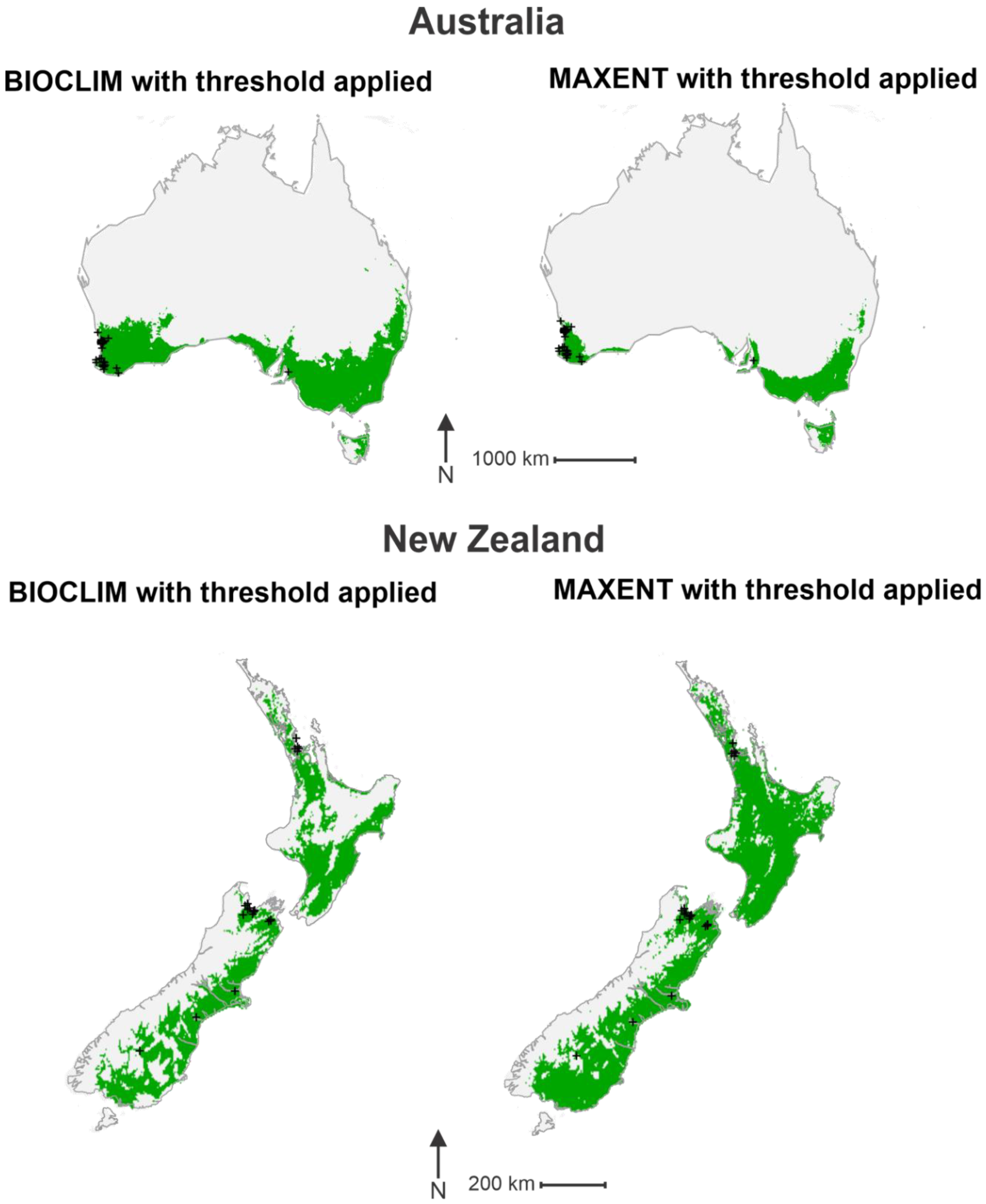
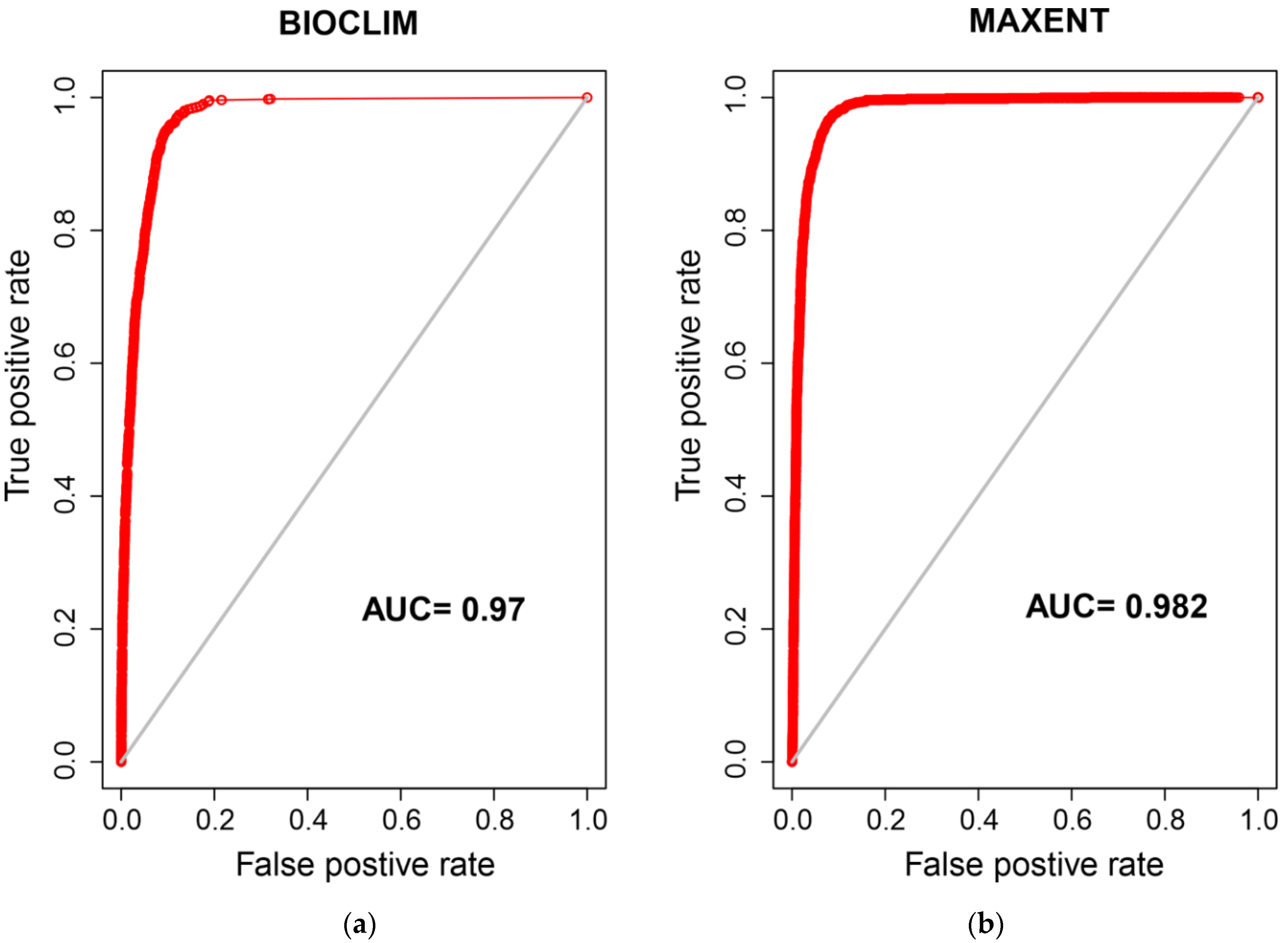
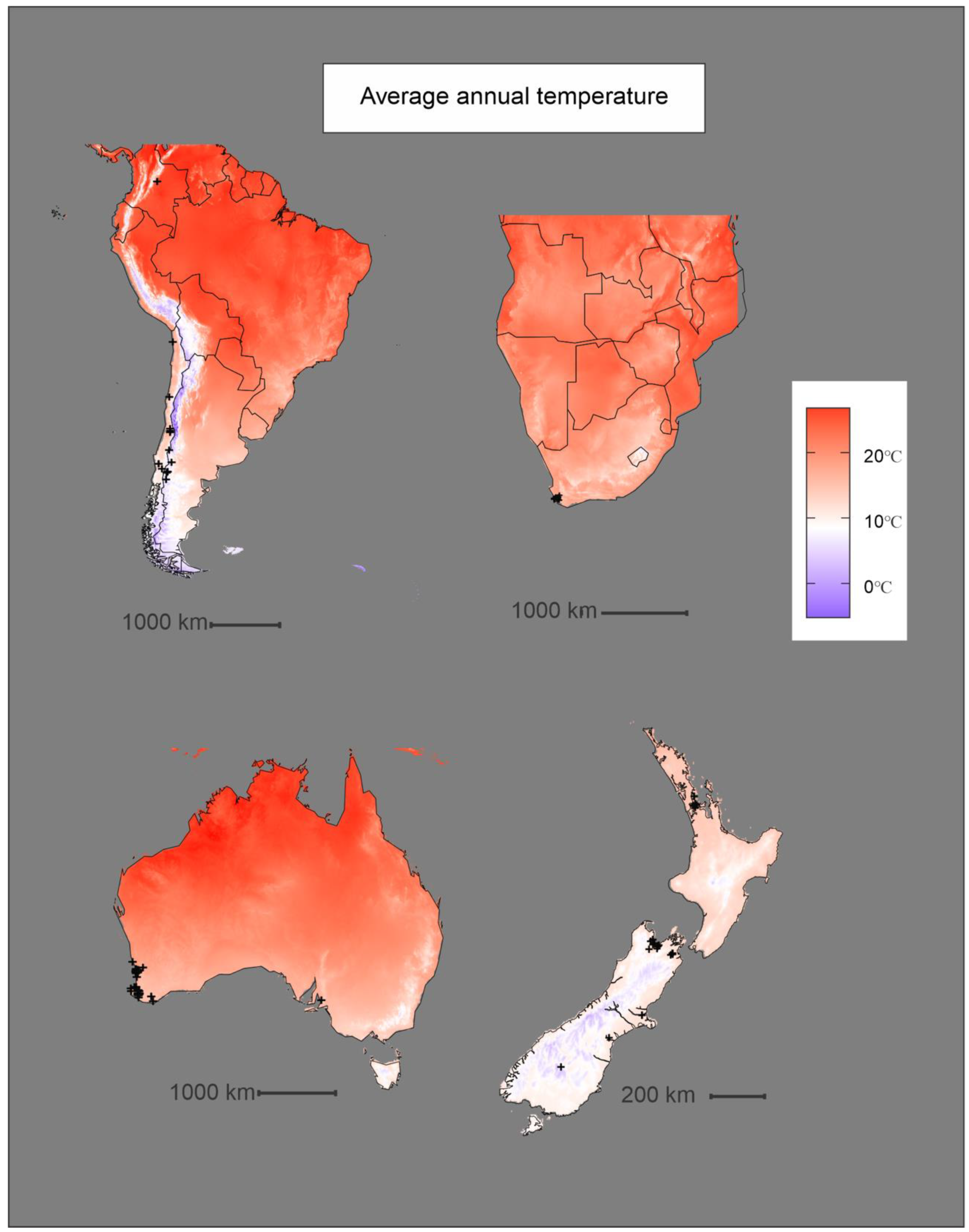
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pearson, R.G. Species’ distribution modeling for conservation educators and practitioners. Lessons Conserv. 2007, 3, 54–89. [Google Scholar]
- Leathwick, J.; Rowe, D.; Richardson, J.; Elith, J.; Hastie, T. Using multivariate adaptive regression splines to predict the distributions of New Zealand’s freshwater diadromous fish. Freshw. Biol. 2005, 50, 2034–2052. [Google Scholar] [CrossRef]
- Moor, H.; Hylander, K.; Norberg, J. Predicting climate change effects on wetland ecosystem services using species distribution modeling and plant functional traits. Ambio 2015, 44, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Curry, P.A.; Yeung, N.W.; Hayes, K.A.; Cowie, R.H. The potential tropical island distribution of a temperate invasive snail, Oxychilus alliarius, modeled on its distribution in Hawaii. Biol. Invasions 2020, 22, 307–327. [Google Scholar] [CrossRef]
- Buck, M.; Marshall, S.A.; Cheung, D.K. Identification atlas of the Vespidae (Hymenoptera, Aculeata) of the northeastern Nearctic region. Can. J. Arthropod Identif. 2008, 5, 1–492. [Google Scholar] [CrossRef]
- Carpenter, J.M. Distributional checklist of species of the genus Polistes (Hymenoptera: Vespidae; Polistinae, Polistini). Am. Mus. Novit. 1996, 3188, 1–39. [Google Scholar]
- Ministry for Primary Industries. Pest Watch. In Surveillance; Ministry for Primary Industries: Wellington, New Zealand, 2016; Volume 43, p. 26. ISSN 1176-5305. [Google Scholar]
- Brown, R.; Payne, A.; Graham, K.K.; Starks, P.T. Prey capture and caste-specific payload capacities in the European paper wasp Polistes dominulus. Insectes Sociaux 2012, 59, 519–525. [Google Scholar] [CrossRef]
- Baker, A.M.; Potter, D.A. Invasive paper wasp turns urban pollinator gardens into ecological traps for monarch butterfly larvae. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jeon, M.G.; Kim, T.G.; Jung, J.C.; Choi, M.B. Prey diversity of Polistes rothneyi koreanus in different landscapes using DNA barcoding. J. Appl. Entomol. 2019, 143, 1052–1063. [Google Scholar] [CrossRef]
- Ward, D.F.; Ramón-Laca, A. Molecular identification of the prey range of the invasive Asian paper wasp. Ecol. Evol. 2013, 3, 4408–4414. [Google Scholar] [CrossRef] [PubMed]
- Kasper, M.L.; Reeson, A.F.; Cooper, S.J.; Perry, K.D.; Austin, A.D. Assessment of prey overlap between a native (Polistes humilis) and an introduced (Vespula germanica) social wasp using morphology and phylogenetic analyses of 16S rDNA. Mol. Ecol. 2004, 13, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Cervo, R.; Zacchi, F.; Turillazzi, S. Polistes dominulus (Hymenoptera, Vespidae) invading North America: Some hypotheses for its rapid spread. Insectes Sociaux 2000, 47, 155–157. [Google Scholar] [CrossRef]
- Schenk, D.; Bacher, S. Functional response of a generalist insect predator to one of its prey species in the field. J. Anim. Ecol. 2002, 71, 524–531. [Google Scholar] [CrossRef]
- Roets, F.; Benadé, P.; Samways, M.; Veldtman, R. Better colony performance, not natural enemy release, explains numerical dominance of the exotic Polistes dominula wasp over a native congener in South Africa. Biol. Invasions 2019, 21, 925–933. [Google Scholar] [CrossRef]
- Pickett, K.; Wenzel, J. High productivity in haplometrotic colonies of the introduced paper wasp Polistes dominulus (Hymenoptera: Vespidae; Polistinae). J. N. Y. Entomol. Soc. 2000, 108, 314–325. [Google Scholar] [CrossRef]
- Gamboa, G.; Greig, E.; Thom, M. The comparative biology of two sympatric paper wasps, the native Polistes fuscatus and the invasive Polistes dominulus (Hymenoptera, Vespidae). Insectes Sociaux 2002, 49, 45–49. [Google Scholar] [CrossRef]
- GBIF.org. GBIF Occurence Download. 2020. Available online: https://doi.org/10.15468/dl.jzezgu (accessed on 28 February 2020).
- Liebert, A.E.; Gamboa, G.J.; Stamp, N.E.; Curtis, T.R.; Monnet, K.M.; Turillazzi, S.; Starks, P.T. Genetics, behavior and ecology of a paper wasp invasion: Polistes dominulus in North America. Ann. Zool. Fenn. 2006, 43, 595–624. [Google Scholar]
- Taylor, R.W.; Brown, D.R.; Cardale, J.C. Hymenoptera: Formicoidea, Vespoidea and Sphecoidea; Australian Government Publishing Service: Canberra, Australia, 1985; Volume 2, ISSN 0644039221.
- González, R.H. Insectos y Ácaros de Importancia Agrícola y Cuarentenaria en Chile; Universidad de Chile Santiago: Santiago, Chile, 1989; ISBN 9567049016. [Google Scholar]
- Elgueta, M. Antecedentes sobre especies de reciente introducción a Chile (Diptera: Stratiomyidae; Hymenoptera: Vespidae). Rev. Chil. Entomol. 1989, 17, 97–98. [Google Scholar]
- Sackmann, P.; Villacide, J.M.; Corley, J. Presencia de una nueva avispa social exótica, Polistes dominulus (Hymenoptera: Vespidae) en la Patagonia Argentina. Rev. Soc. Entomológica Argent. 2003, 62, 68–70. [Google Scholar]
- Eardley, C.; Koch, F.; Wood, A. Polistes dominulus (Christ, 1791)(Hymenoptera: Polistinae: Vespidae) newly recorded from South Africa. Afr. Entomol. 2009, 17, 226–227. [Google Scholar] [CrossRef]
- Stringer, I.; Hitchmough, R. Assessing the conservation status of New Zealand’s native terrestrial invertebrates. N. Z. Entomol. 2012, 35, 77–84. [Google Scholar] [CrossRef]
- Lester, P.; Brown, S.; Edwards, E.; Holwell, G.; Pawson, S.; Ward, D.; Watts, C. Critical issues facing New Zealand entomology. N. Z. Entomol. 2014, 37, 1–13. [Google Scholar] [CrossRef]
- Sands, D.P.A. Important issues facing insect conservation in Australia: Now and into the future. Austral Entomol. 2018, 57, 150–172. [Google Scholar] [CrossRef] [Green Version]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. bioclim: The first species distribution modelling package, its early applications and relevance to most current MaxEnt studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Nix, H.A. A biogeographic analysis of Australian elapid snakes. In Atlas of Elapid Snakes of Australia; Longmore, R., Ed.; Australian Government Publishing Service: Canberra, Australian, 1986; Volume 7, pp. 16–77. [Google Scholar]
- Hijmans, R.J.; Elith, J. Species Distribution Modeling with R. Available online: http://cran.r-project.org/web/packages/dismo/vignettes/dm.pdf (accessed on 8 February 2020).
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Duan, R.-Y.; Kong, X.-Q.; Huang, M.-Y.; Fan, W.-Y.; Wang, Z.-G. The predictive performance and stability of six species distribution models. PLoS ONE 2014, 9, e112764. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. Dismo: Species distribution modeling. In R Package Version 1.1-4; 2017; Available online: https://CRAN.R-project.org/package=dismo (accessed on 8 February 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Sumner, S.; Bevan, P.; Hart, A.G.; Isaac, N.J.B. Mapping species distributions in 2 weeks using citizen science. Insect Conserv. Divers. 2019, 12, 382–388. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Beckham, J.L.; Atkinson, S. An updated understanding of Texas bumble bee (Hymenoptera: Apidae) species presence and potential distributions in Texas, USA. PeerJ 2017, 5, e3612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, D.F. Modelling the potential geographic distribution of invasive ant species in New Zealand. Biol. Invasions 2007, 9, 723–735. [Google Scholar] [CrossRef]
- Rodder, D.; Schmidtlein, S.; Veith, M.; Lotters, S. Alien invasive slider turtle in unpredicted habitat: A matter of niche shift or of predictors studied? PLoS ONE 2009, 4, e7843. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, R.K.; Peterson, A.T.; Cobos, M.E.; Ganta, R.; Foley, D. Current and future distribution of the lone star tick, Amblyomma americanum (L.)(Acari: Ixodidae) in North America. PLoS ONE 2019, 14, e0209082. [Google Scholar] [CrossRef]
- Dudaniec, R.Y.; Yong, C.J.; Lancaster, L.T.; Svensson, E.I.; Hansson, B. Signatures of local adaptation along environmental gradients in a range-expanding damselfly (Ischnura elegans). Mol. Ecol. 2018, 27, 2576–2593. [Google Scholar] [CrossRef] [Green Version]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. In R Package Version 3.3-13; 2020; Available online: https://CRAN.R-project.org/package=raster (accessed on 8 February 2020).
- Merow, C.; Smith, M.J.; Silander, J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.; Uriarte, M.; Anderson, R.P. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for ecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Liu, C.; Newell, G.; White, M. On the selection of thresholds for predicting species occurrence with presence-only data. Ecol. Evol. 2016, 6, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; White, M.; Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Assessing the fit of the model. In Applied Logistic Regression, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 153–226. [Google Scholar]
- Benadé, P.C.; Veldtman, R.; Samways, M.J.; Roets, F. Rapid range expansion of the invasive wasp Polistes dominula (Hymenoptera: Vespidae: Polistinae) and first record of parasitoids on this species and the native Polistes marginalis in the Western Cape province of South Africa. Afr. Entomol. 2014, 22, 220–225. [Google Scholar] [CrossRef]
- Beggs, J.R.; Rees, J.S. Restructuring of Lepidoptera communities by introduced Vespula wasps in a New Zealand beech forest. Oecologia 1999, 119, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Toft, R.J.; Rees, J.S. Reducing predation of orb-web spiders by controlling common wasps (Vespula vulgaris) in a New Zealand beech forest. Ecol. Entomol. 1998, 23, 90–95. [Google Scholar] [CrossRef]
- Potter-Craven, J.; Kirkpatrick, J.B.; McQuillan, P.B.; Bell, P. The effects of introduced vespid wasps (Vespula germanica and V. vulgaris) on threatened native butterfly (Oreixenica ptunarra) populations in Tasmania. J. Insect Conserv. 2018, 22, 521–532. [Google Scholar] [CrossRef]
- Macarra, G.R. The climate and weather of West Coast. In NIWA Science and Technology Series; NIWA: Christchurch, New Zealand, 2016; ISSN 1173-0382. [Google Scholar]
- Beaumont, L.J.; Hughes, L.; Poulsen, M. Predicting species distributions: Use of climatic parameters in BIOCLIM and its impact on predictions of species’ current and future distributions. Ecol. Model. 2005, 186, 251–270. [Google Scholar] [CrossRef]
- Stamp, N.; Bowers, M. Direct and indirect effects of predatory wasps (Polistes sp.: Vespidae) on gregarious caterpillars (Hemileuca lucina: Saturniidae). Oecologia 1988, 75, 619–624. [Google Scholar] [CrossRef]
- Pilowsky, J.; Starks, P. Displacement and replacement in real time: Polistes dominula’s impact on P. fuscatus in the northeastern U.S. Biol. Invasions 2018, 20, 1161–1169. [Google Scholar] [CrossRef]
- Parent, C.E.; Peck, S.B.; Causton, C.E.; Roque-Albelo, L.; Lester, P.J.; Bulgarella, M. Polistes versicolor (Hymenoptera: Vespidae), an introduced wasp in the Galapagos Islands: Its life cycle and ecological impact. Environ. Entomol. 2020. [Google Scholar] [CrossRef]
- Beggs, J.R.; Brockerhoff, E.G.; Corley, J.C.; Kenis, M.; Masciocchi, M.; Muller, F.; Rome, Q.; Villemant, C. Ecological effects and management of invasive alien Vespidae. BioControl 2011, 56, 505–526. [Google Scholar] [CrossRef]
- Lefort, M.-C.; Beggs, J.R.; Glare, T.R.; Saunders, T.E.; Doyle, E.J.; Stephane, B. A Molecular Approach to Studying Hymenoptera Diets Using Polistine Wasps. Available online: https://www.biorxiv.org/content/10.1101/2020.04.06.024422v1 (accessed on 17 August 2020).
- Stringer, I.; Hitchmough, R.; Dugdale, J.; Edwards, E.; Hoare, R.; Patrick, B. The conservation status of New Zealand Lepidoptera. N. Z. Entomol. 2012, 35, 120–127. [Google Scholar] [CrossRef]
- Höcherl, N.; Tautz, J. Nesting behavior of the paper wasp Polistes dominula in Central Europe-a flexible system for expanding into new areas. Ecosphere 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Schapheer, C.; Sandoval, G.; Villagra, C.A. Pest Cockroaches May Overcome Environmental Restriction Due to Anthropization. J. Med Entomol. 2018, 55, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Cranshaw, W.S.; Larsen, H.J.; Zimmerman, R.J. Notes on fruit damage by the European paper wasp, Polistes dominula (Christ) (Hymenoptera: Vespidae). Southwest. Entomol. 2011, 36, 103–105. [Google Scholar] [CrossRef]
- Wisz, M.S.; Pottier, J.; Kissling, W.D.; Pellissier, L.; Lenoir, J.; Damgaard, C.F.; Dormann, C.F.; Forchhammer, M.C.; Grytnes, J.A.; Guisan, A. The role of biotic interactions in shaping distributions and realised assemblages of species: Implications for species distribution modelling. Biol. Rev. 2013, 88, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Giannini, T.C.; Chapman, D.S.; Saraiva, A.M.; Alves-dos-Santos, I.; Biesmeijer, J.C. Improving species distribution models using biotic interactions: A case study of parasites, pollinators and plants. Ecography 2013, 36, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Godsoe, W.; Harmon, L.J. How do species interactions affect species distribution models? Ecography 2012, 35, 811–820. [Google Scholar] [CrossRef]
- Bulgarella, M.; Trewick, S.A.; Minards, N.A.; Jacobson, M.J.; Morgan-Richards, M. Shifting ranges of two tree weta species (Hemideina spp.): Competitive exclusion and changing climate. J. Biogeogr. 2014, 41, 524–535. [Google Scholar] [CrossRef]
- Mpakairi, K.S.; Ndaimani, H.; Tagwireyi, P.; Gara, T.W.; Zvidzai, M.; Madhlamoto, D. Missing in action: Species competition is a neglected predictor variable in species distribution modelling. PLoS ONE 2017, 12, e0181088. [Google Scholar] [CrossRef]
- Pellissier, L.; Anne Bråthen, K.; Pottier, J.; Randin, C.F.; Vittoz, P.; Dubuis, A.; Yoccoz, N.G.; Alm, T.; Zimmermann, N.E.; Guisan, A. Species distribution models reveal apparent competitive and facilitative effects of a dominant species on the distribution of tundra plants. Ecography 2010, 33, 1004–1014. [Google Scholar] [CrossRef]
- Gamboa, G.; Noble, M.; Thom, M.; Togal, J.; Srinivasan, R.; Murphy, B. The comparative biology of two sympatric paper wasps in Michigan, the native Polistes fuscatus and the invasive Polistes dominulus (Hymenoptera, Vespidae). Insectes Sociaux 2004, 51, 153–157. [Google Scholar] [CrossRef]
- Miller, G.L.; Donnelly, C.R.; Gamboa, G.J. A ten-year comparative study of the population dynamics and parasitoidism in the native paper wasp Polistes fuscatus and the invasive P. dominulus. Insectes Sociaux 2013, 60, 49–56. [Google Scholar] [CrossRef]
- Kearney, M.R.; Wintle, B.A.; Porter, W.P. Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conserv. Lett. 2010, 3, 203–213. [Google Scholar] [CrossRef]



| Variable Code | Variable Title | Unit | Inclusion in Final Model |
|---|---|---|---|
| Bio1 | Annual Mean Temperature | °C | ✓ |
| Bio2 | Mean Diurnal Range (mean of monthly (max temp-min temp)) | °C | ✓ |
| Bio3 | Isothermality ((Bio2/Bio7) × 100) | % | ✓ |
| Bio4 | Temperature Seasonality (standard deviation × 100) | °C | ✓ |
| Bio5 | Max Temperature of Warmest Month | °C | ✓ |
| Bio6 | Min Temperature of Coldest Month | °C | |
| Bio7 | Temperature Annual Range (Bio5-Bio6) | °C | ✓ |
| Bio8 | Mean Temperature of Wettest Quarter | °C | ✓ |
| Bio9 | Mean Temperature of Driest Quarter | °C | ✓ |
| Bio10 | Mean Temperature of Warmest Quarter | °C | ✓ |
| Bio11 | Mean Temperature of Coldest Quarter | °C | ✓ |
| Bio12 | Annual Precipitation | mm | |
| Bio13 | Precipitation of Wettest Month | mm | ✓ |
| Bio14 | Precipitation of Driest Month | mm | ✓ |
| Bio15 | Precipitation Seasonality (coefficient of variation) | % | |
| Bio16 | Precipitation of Wettest Quarter | mm | ✓ |
| Bio17 | Precipitation of Driest Quarter | mm | |
| Bio18 | Precipitation of Warmest Quarter | mm | ✓ |
| Bio19 | Precipitation of Coldest Quarter | mm | ✓ |
| Variable Code | Variable Title | Permutation Importance |
|---|---|---|
| Bio1 | Annual Mean Temperature | 37.9% |
| Bio19 | Precipitation of Coldest Quarter | 15.9% |
| Bio4 | Temperature Seasonality (standard deviation × 100) | 11.4% |
| Bio10 | Mean Temperature of Warmest Quarter | 9.0% |
| Bio16 | Precipitation of Wettest Quarter | 4.7% |
| Bio18 | Precipitation of Warmest Quarter | 4.2% |
| Bio11 | Mean Temperature of Coldest Quarter | 3.8% |
| Bio3 | Isothermality ((Bio2/Bio7) × 100) | 2.8% |
| Bio9 | Mean Temperature of Driest Quarter | 2.7% |
| Bio14 | Precipitation of Driest Month | 2.3% |
| Bio2 | Mean Diurnal Range (mean of monthly (max temp–min temp)) | 1.8% |
| Bio5 | Max Temperature of Warmest Month | 1.4% |
| Bio13 | Precipitation of Wettest Month | 1.1% |
| Bio7 | Temperature Annual Range (Bio5-Bio6) | 0.5% |
| Bio8 | Mean Temperature of Wettest Quarter | 0.4% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howse, M.W.F.; Haywood, J.; Lester, P.J. Bioclimatic Modelling Identifies Suitable Habitat for the Establishment of the Invasive European Paper Wasp (Hymenoptera: Vespidae) across the Southern Hemisphere. Insects 2020, 11, 784. https://doi.org/10.3390/insects11110784
Howse MWF, Haywood J, Lester PJ. Bioclimatic Modelling Identifies Suitable Habitat for the Establishment of the Invasive European Paper Wasp (Hymenoptera: Vespidae) across the Southern Hemisphere. Insects. 2020; 11(11):784. https://doi.org/10.3390/insects11110784
Chicago/Turabian StyleHowse, Matthew W. F., John Haywood, and Philip J. Lester. 2020. "Bioclimatic Modelling Identifies Suitable Habitat for the Establishment of the Invasive European Paper Wasp (Hymenoptera: Vespidae) across the Southern Hemisphere" Insects 11, no. 11: 784. https://doi.org/10.3390/insects11110784
APA StyleHowse, M. W. F., Haywood, J., & Lester, P. J. (2020). Bioclimatic Modelling Identifies Suitable Habitat for the Establishment of the Invasive European Paper Wasp (Hymenoptera: Vespidae) across the Southern Hemisphere. Insects, 11(11), 784. https://doi.org/10.3390/insects11110784






