Physiological Analysis and Transcriptome Analysis of Asian Honey Bee (Apis cerana cerana) in Response to Sublethal Neonicotinoid Imidacloprid
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects Material
2.2. Pesticide Exposure Experiment
2.3. Climbing Assays
2.4. Sucrose Responsiveness
2.5. Enzyme Activity Assay
2.6. RNA Isolation and RNA Sequencing Analysis
2.7. Bioinformatics Analysis of RNA-seq Data
2.8. Confirmation of RNA-Seq Results by qRT- PCR
2.9. Statistical Analyses
3. Results
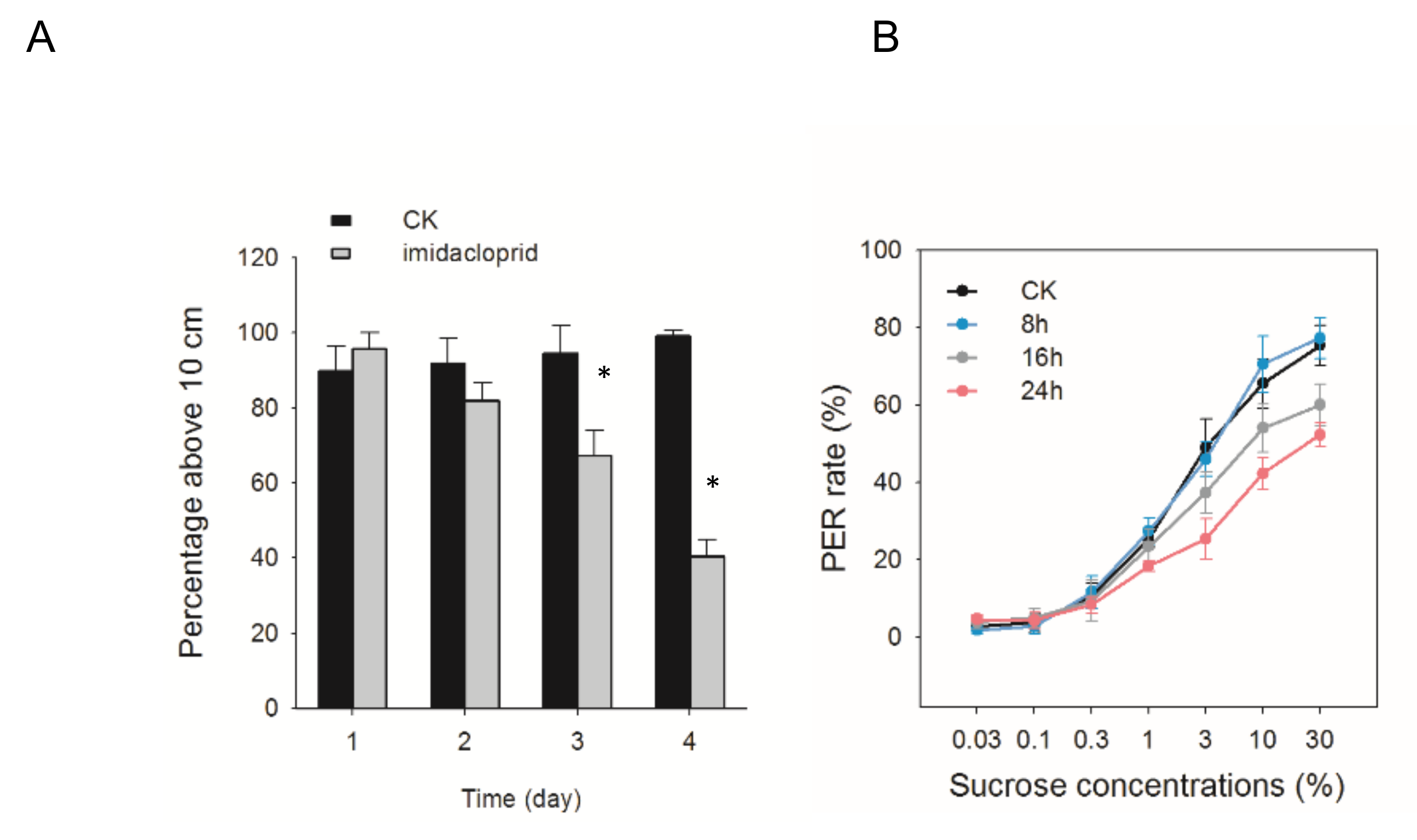
3.1. Climbing Ability, and Sucrose Responsiveness Changes of A. cerana after Exposed to Imidacloprid
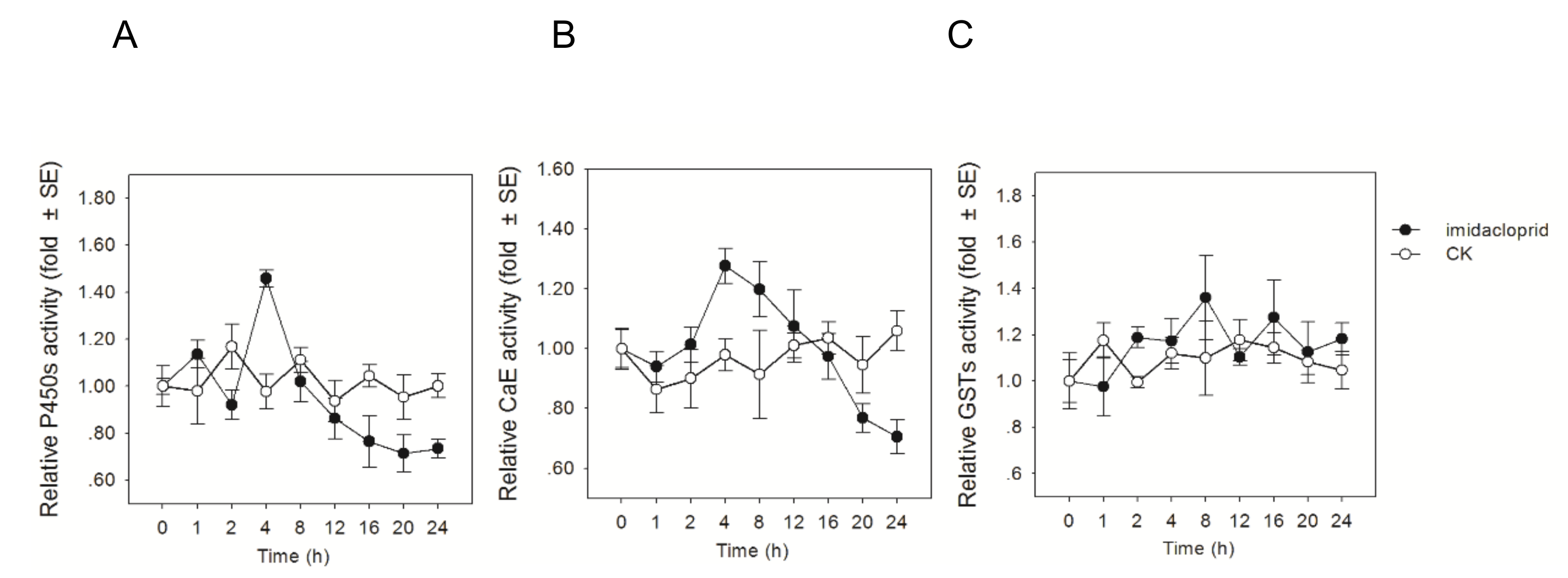
3.2. Influences of Imidacloprid on Detoxification Enzyme Activities
3.3. Illumina Sequencing and Transcriptome Assembly
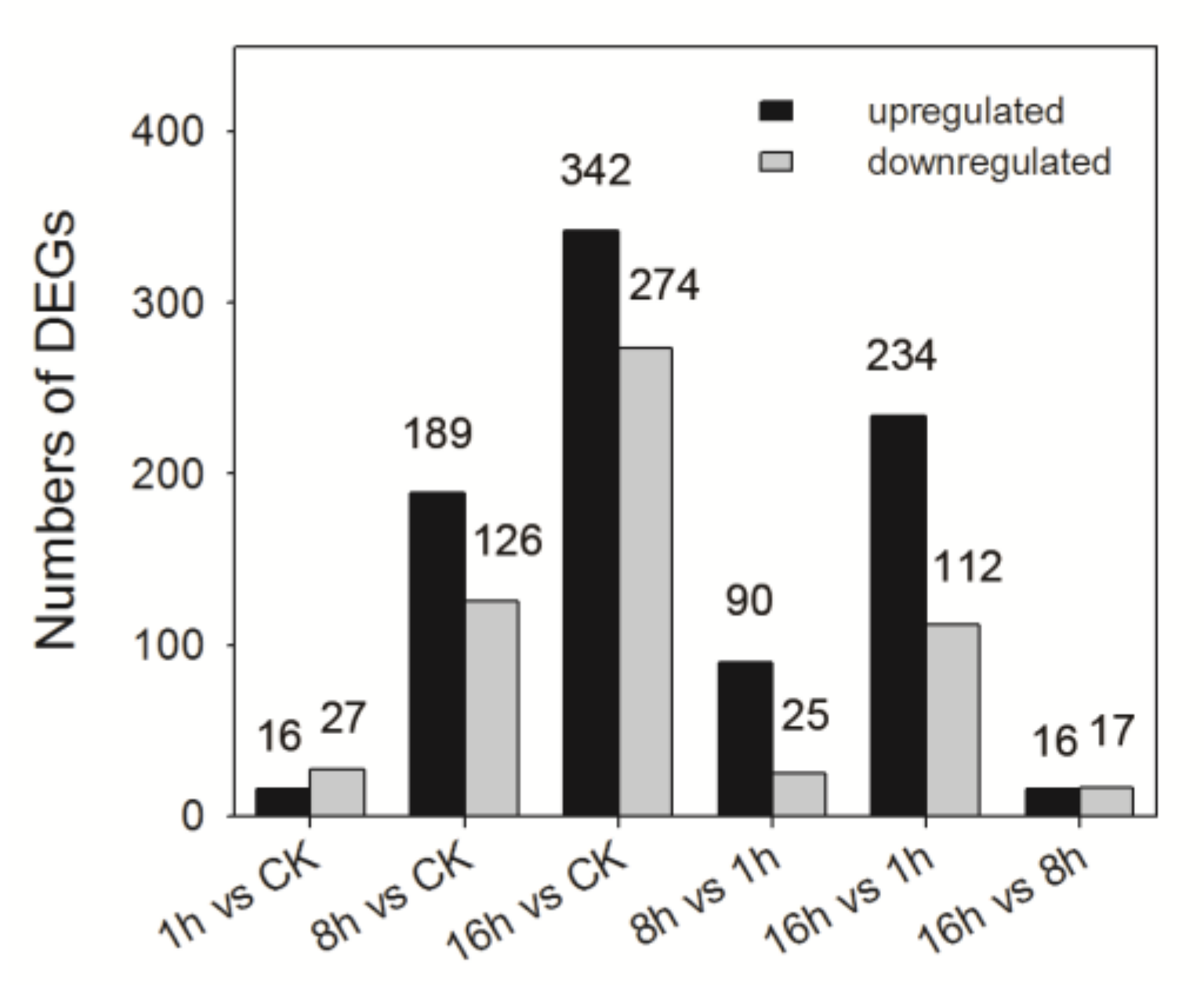
3.4. DEGs in Response to Sublethal Concentration of Imidacloprid
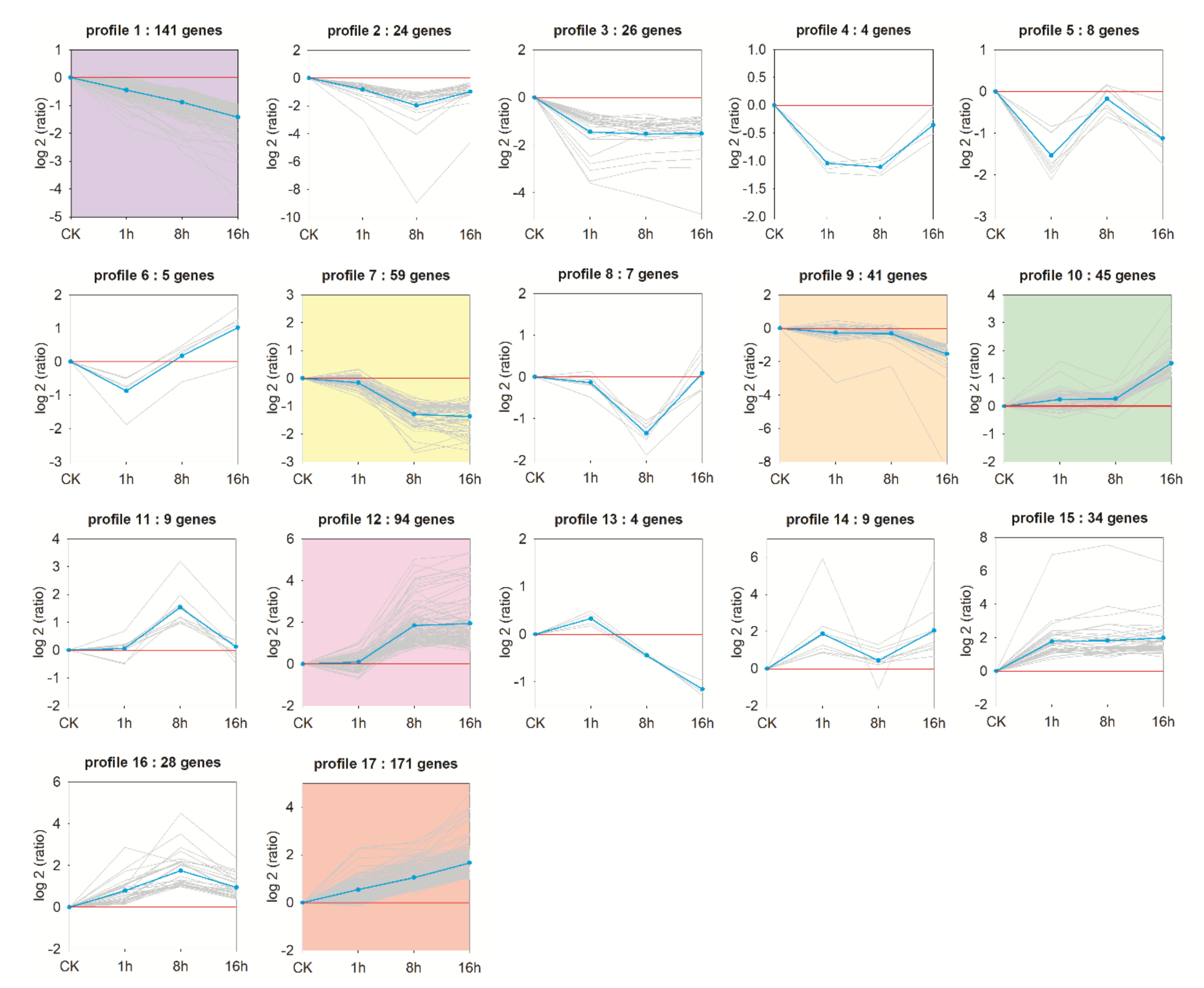
3.5. Trend Analysis of DEGs
3.6. Functional Annotation and Classification of Differentially Expressed Genes
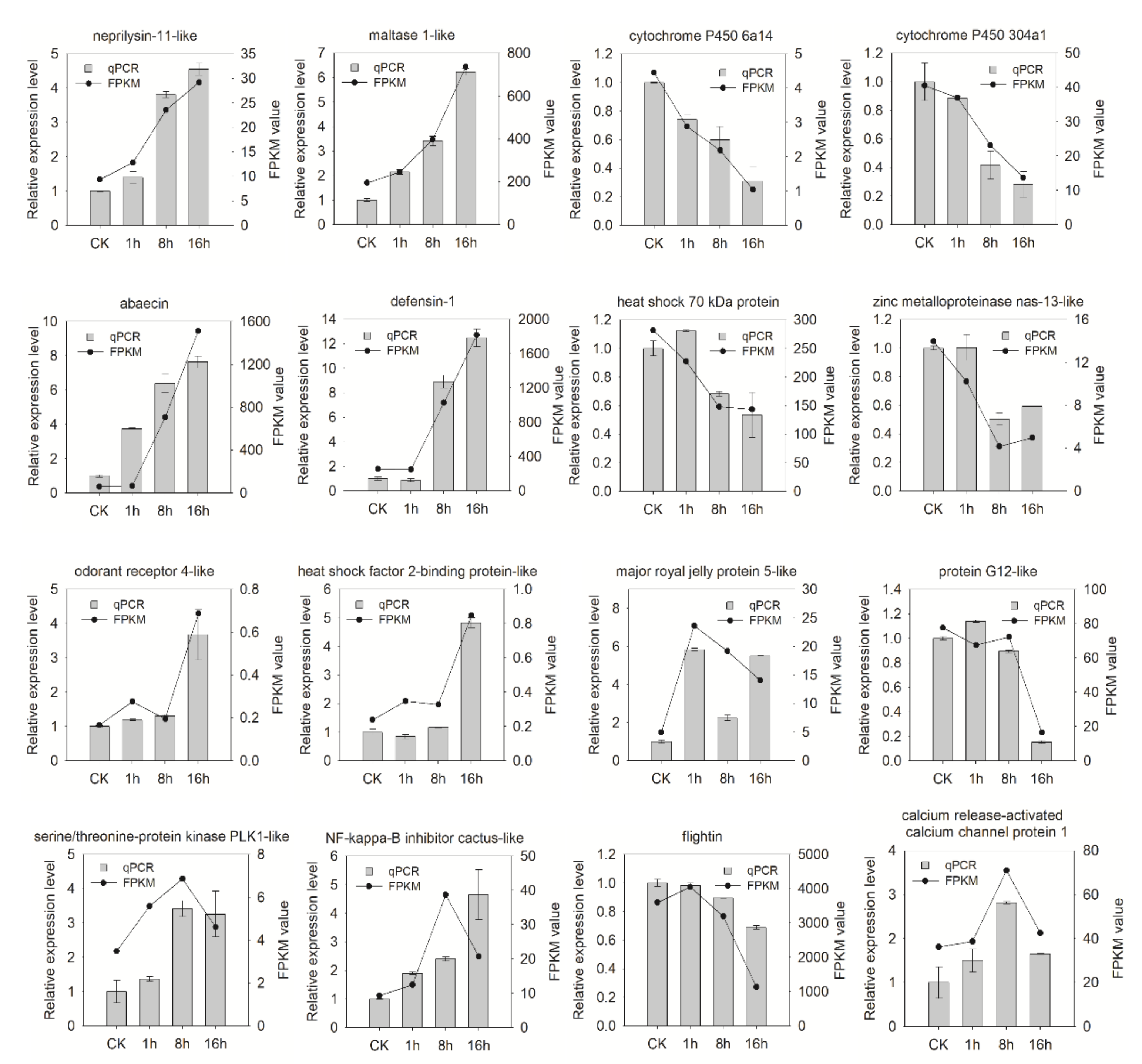
3.7. Validation of RNA-seq Data by Quantitative Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gallai, N.; Salles, J.; Settele, J.; Vaissière, B. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Underwood, R.M.; van Engelsdorp, D. Colony collapse disorder: Have we seen this before? Bee culture. Science 2007, 318, 283–287. [Google Scholar]
- Matsuda, K.; Buckingham, S.D.; Kleier, D.; Rauh, J.J.; Grauso, M.; Sattelle, D.B. Neonicotinoids: Insecticides acting on insect nicotinic acetylcholine receptors. Trends Pharmacol. Sci. 2001, 22, 573–580. [Google Scholar] [CrossRef]
- Lundin, O.; Rundlof, M.; Smith, H.G.; Fries, I.; Bommarco, R. Neonicotinoid insecticides and their impacts on bees: A systematic review of research approaches and identification of knowledge gaps. PLoS ONE 2015, 10, e0136928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatjina, F.; Papaefthimiou, C.; Charistos, L.; Dogaroglu, T.; Bouga, M.; Emmanouil, C.; Arnold, G. Sublethal doses of imidacloprid decreased size of hypopharyngeal glands and respiratory rhythm of honeybees in vivo. Apidologie 2013, 44, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Goulson, D. Review: An overview of the environmental risks posed by neonicotinoid insecticides. J. Appl. Ecol. 2013, 50, 977–987. [Google Scholar] [CrossRef]
- Bonmatin, J.-M.; Giorio, C.; Girolami, V.; Goulson, D.; Kreutzweiser, D.P.; Krupke, C.; Liess, M.; Long, E.; Marzaro, M.; Mitchell, E.A.D.; et al. Environmental fate and exposure; neonicotinoids and fipronil. Environ. Sci. Pollut. Res. 2015, 22, 35–67. [Google Scholar] [CrossRef]
- Kessler, S.C.; Tiedeken, E.J.; Simcock, K.L.; Derveau, S.; Mitchell, J.; Softley, S.; Stout, J.C.; Wright, G.A. Bees prefer foods containing neonicotinoid pesticides. Nature 2015, 521, 74–76. [Google Scholar] [CrossRef]
- Cresswell, J.E.; Desneux, N.; van Engelsdorp, D. Dietary traces of neonicotinoid pesticides as a cause of population declines in honey bees: An evaluation by hill’s epidemiological criteria. Pest Manag. Sci. 2012, 68, 819–827. [Google Scholar] [CrossRef]
- Yang, E.C.; Chuang, Y.C.; Chen, Y.L.; Chang, L.H. Abnormal foraging behavior induced by sublethal dosage of imidacloprid in the honey bee (hymenoptera: Apidae). J. Econ. Entomol. 2008, 101, 1743–1748. [Google Scholar] [CrossRef]
- Williamson, S.M.; Baker, D.D.; Wright, G.A. Acute exposure to a sublethal dose of imidacloprid and coumaphos enhances olfactory learning and memory in the honeybee apis mellifera. Invertebr. Neurosci. 2013, 13, 63–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laycock, I.; Lenthall, K.M.; Barratt, A.T.; Cresswell, J.E. Effects of imidacloprid, a neonicotinoid pesticide, on reproduction in worker bumble bees (Bombus terrestris). Ecotoxicology 2012, 21, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef] [Green Version]
- Alburaki, M.; Boutin, S.; Mercier, P.L.; Loublier, Y.; Chagnon, M.; Derome, N. Neonicotinoid-coated zea mays seeds indirectly affect honeybee performance and pathogen susceptibility in field trials. PLoS ONE 2015, 10, e0125790. [Google Scholar] [CrossRef] [Green Version]
- Alaux, C.; Brunet, J.-L.; Dussaubat, C.; Mondet, F.; Tchamitchan, S.; Cousin, M.; Brillard, J.; Baldy, A.; Belzunces, L.P.; Le Conte, Y. Interactions between nosema microspores and a neonicotinoid weaken honeybees (Apis mellifera). Environ. Microbiol. 2010, 12, 774–782. [Google Scholar] [CrossRef] [Green Version]
- Boas Balieira, K.V.; Mazzo, M.; Veiga Bizerra, P.F.; de Jesus Santos Guimares, A.R.; Nicodemo, D.; Mingatto, F.E. Imidacloprid-induced oxidative stress in honey bees and the antioxidant action of caffeine. Apidologie 2018, 49, 562–572. [Google Scholar] [CrossRef] [Green Version]
- Nicodemo, D.; Maioli, M.A.; Medeiros, H.C.D.; Guelfi, M.; Balieira, K.V.B.; De Jong, D.; Mingatto, F.E. Fipronil and imidacloprid reduce honeybee mitochondrial activity. Environ. Toxicol. Chem. 2014, 33, 2070–2075. [Google Scholar] [CrossRef]
- Christen, V.; Kunz, P.Y.; Fent, K. Endocrine disruption and chronic effects of plant protection products in bees: Can we better protect our pollinators? Environ. Pollut. 2018, 243, 1588–1601. [Google Scholar] [CrossRef]
- Nicodemo, D.; De Jong, D.; Reis, L.G.; de Almeida, J.M.V.; dos Santos, A.A.; Lisboa, L.A.M. Transgenic corn decreased total and key storage and lipid transport protein levels in honey bee hemolymph while seed treatment with imidacloprid reduced lipophorin levels. J. Apic. Res. 2018, 57, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Arena, M.; Sgolastra, F. A meta-analysis comparing the sensitivity of bees to pesticides. Ecotoxicology 2014, 23, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Luo, S.; Liu, J.; Wu, J. Apis cerana is less sensitive to most neonicotinoids, despite of their smaller body mass. J. Econ. Entomol. 2018, 111, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, S.; Yan, Z.; Liu, F.; Diao, Q.; Dai, P. Effects of three common pesticides on survival, food consumption and midgut bacterial communities of adult workers Apis cerana and Apis mellifera. Environ. Pollut. 2019, 249, 860–867. [Google Scholar] [CrossRef]
- Cresswell, J.E.; Page, C.J.; Uygun, M.B.; Holmbergh, M.; Li, Y.; Wheeler, J.G.; Laycock, I.; Pook, C.J.; De Ibarra, N.H.; Smirnoff, N.; et al. Differential sensitivity of honey bees and bumble bees to a dietary insecticide (imidacloprid). Zoology 2012, 115, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; He, J.; Zhao, X.; Chaimanee, V.; Huang, W.-F.; Nie, H.; Zhao, Y.; Su, S. Differential physiological effects of neonicotinoid insecticides on honey bees: A comparison between Apis mellifera and Apis cerana. Pestic. Biochem. Physiol. 2017, 140, 1–8. [Google Scholar] [CrossRef]
- Li, Z.G.; Li, M.; Huang, J.N.; Ma, C.S.; Xiao, L.C.; Huang, Q.; Zhao, Y.Z.; Nie, H.Y.; Su, S.K. Effects of sublethal concentrations of chlorpyrifos on olfactory learning and memory performances in two bee species, apis mellifera and apis cerana. Sociobiology 2017, 64, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Laurino, D.; Manino, A.; Patetta, A.; Porporato, M. Toxicity of neonicotinoid insecticides on different honey bee genotypes. Bull. Insectol. 2013, 66, 119–126. [Google Scholar]
- European Commission. Commission implementing regulation (eu). Off. J. Eur. Union 2018, L132, 35–39.
- Lu, C.; Chang, C.-H.; Palmer, C.; Zhao, M.; Zhang, Q. Neonicotinoid residues in fruits and vegetables: An integrated dietary exposure assessment approach. Environ. Sci. Technol. 2018, 52, 3175–3184. [Google Scholar] [CrossRef]
- Wang, X.R.; Goulson, D.; Chen, L.Z.; Zhang, J.Z.; Zhao, W.; Jin, Y.; Yang, S.P.; Li, Y.; Zhou, J.H. Occurrence of neonicotinoids in chinese apiculture and a corresponding risk exposure assessment. Environ. Sci. Technol. 2020, 54, 5021–5030. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Luo, Q.H.; Hou, C.S.; Wang, Q.; Dai, P.L.; Gao, J.; Liu, Y.J.; Diao, Q.Y. Sublethal effects of imidacloprid on targeting muscle and ribosomal protein related genes in the honey bee apis mellifera l. Sci. Rep. 2017, 7, 15943. [Google Scholar] [CrossRef]
- Johnson, R.M.; Ellis, M.D.; Mullin, C.A.; Frazier, M. Pesticides and honey bee toxicity—USA. Apidologie 2010, 41, 312–331. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Kuang, H.; Wang, S.; Wang, J.; Liu, W.; Wu, Z.; Tian, Y.; Huang, Z.Y.; Miao, X. Comparative sucrose responsiveness in apis mellifera and a. Cerana foragers. PLoS ONE 2013, 8, e79026. [Google Scholar] [CrossRef] [Green Version]
- Pankiw, T.; Waddington, K.D.; Page, R.E. Modulation of sucrose response thresholds in honey bees (Apis mellifera l.): Influence of genotype, feeding, and foraging experience. J. Comp. Physiol. A 2001, 187, 293–301. [Google Scholar] [CrossRef]
- Tang, S.S.; Chang, G.G. Kinetic characterization of the endogenous glutathione transferase activity of octopus lens s-crystallin. J. Biochem. 1996, 119, 1182–1188. [Google Scholar] [CrossRef]
- Chanda, S.M.; Mortensen, S.R.; Moser, V.C.; Padilla, S. Tissue-specific effects of chlorpyrifos on carboxylesterase and cholinesterase activity in adult rats: An in vitro and in vivo comparison. Fundam. Appl. Toxicol. 1997, 38, 148–157. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Wakamiya, N.; Ueng, Y.F.; Guengerich, F.P.; Inui, Y. Characterization of microsomal cytochrome p450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal livers and adult lungs. Drug Metab. Dispos. 1996, 24, 515–522. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. Wego: A web tool for plotting go annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. Stem: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [Green Version]
- Teeters, B.S.; Johnson, R.M.; Ellis, M.D.; Siegfried, B.D. Using video-tracking to assess sublethal effects of pesticides on honey bees (Apis mellifera L.). Environ. Toxicol. Chem. 2012, 31, 1349–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cresswell, J.E. A meta-analysis of experiments testing the effects of a neonicotinoid insecticide (imidacloprid) on honey bees. Ecotoxicology 2011, 20, 149–157. [Google Scholar] [CrossRef]
- Yang, E.C.; Chang, H.C.; Wu, W.Y.; Chen, Y.W. Impaired olfactory associative behavior of honeybee workers due to contamination of imidacloprid in the larval stage. PLoS ONE 2012, 7, e49472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medrzycki, P.; Montanari, R.; Bortolotti, L.; Sabatini, A.G.; Maini, S.; Porrini, C. Effects of imidacloprid administered in sublethal doses on honey bee behaviour. Laboratory tests. Bull. Insectol. 2003, 56, 59–62. [Google Scholar]
- Williamson, S.M.; Willis, S.J.; Wright, G.A. Exposure to neonicotinoids influences the motor function of adult worker honeybees. Ecotoxicology 2014, 23, 1409–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christen, V.; Schirrmann, M.; Frey, J.E.; Fent, K. Global transcriptomic effects of environmentally relevant concentrations of the neonicotinoids clothianidin, lmidacloprid, and thiamethoxam in the brain of honey bees (Apis mellifera). Environ. Sci. Technol. 2018, 52, 7534–7544. [Google Scholar] [CrossRef]
- Derecka, K.; Blythe, M.J.; Malla, S.; Genereux, D.P.; Guffanti, A.; Pavan, P.; Moles, A.; Snart, C.; Ryder, T.; Ortori, C.A.; et al. Transient exposure to low levels of insecticide affects metabolic networks of honeybee larvae. PLoS ONE 2013, 8, e68191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Meng, Z.Y.; Tian, S.N.; Teng, M.M.; Yan, J.; Jia, M.; Li, R.S.; Zhou, Z.Q.; Zhu, W.T. Neonicotinoid insecticides exposure cause amino acid metabolism disorders, lipid accumulation and oxidative stress in icr mice. Chemosphere 2020, 246, 125661. [Google Scholar] [CrossRef]
- Lee, M.J.; Goldsworthy, G.J. Chloromethyl ketones are insulin-like stimulators of lipid synthesis in locust fat body. Arch. Insect Biochem. Physiol. 1998, 39, 9–17. [Google Scholar] [CrossRef]
- Wilson, D.F.; Matschinsky, F.M. Metabolic homeostasis: Oxidative phosphorylation and the metabolic requirements of higher plants and animals. J. Appl. Physiol. 2018, 125, 1183–1192. [Google Scholar] [CrossRef]
- Badiou-Beneteau, A.; Carvalho, S.M.; Brunet, J.-L.; Carvalho, G.A.; Bulete, A.; Giroud, B.; Belzunces, L.P. Development of biomarkers of exposure to xenobiotics in the honey bee apis mellifera: Application to the systemic insecticide thiamethoxam. Ecotoxicol. Environ. Saf. 2012, 82, 22–31. [Google Scholar] [CrossRef]
- Berenbaum, M.R.; Johnson, R.M. Xenobiotic detoxification pathways in honey bees. Curr. Opin. Insect Sci. 2015, 10, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Diao, Q. Current knowledge of detoxification mechanisms of xenobiotic in honey bees. Ecotoxicology 2017, 26, 1–12. [Google Scholar] [CrossRef]
- Wu, M.; Chang, Y.; Kuanghui, L.; Encheng, Y. Gene expression changes in honey bees induced by sublethal imidacloprid exposure during the larval stage. Insect Biochem. Mol. Biol. 2017, 88, 12–22. [Google Scholar] [CrossRef]
- Claudianos, C.; Ranson, H.; Johnson, R.M.; Biswas, S.; Schuler, M.A.; Berenbaum, M.R.; Feyereisen, R.; Oakeshott, J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef] [Green Version]
- Manjon, C.; Troczka, B.J.; Zaworra, M.; Beadle, K.; Randall, E.; Hertlein, G.; Singh, K.S.; Zimmer, C.T.; Homem, R.A.; Lueke, B.; et al. Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 2018, 28, 1137–1143. [Google Scholar] [CrossRef] [Green Version]
- De Smet, L.; Hatjina, F.; Ioannidis, P.; Hamamtzoglou, A.; Schoonvaere, K.; Francis, F.; Meeus, I.; Smagghe, G.; de Graaf, D.C. Stress indicator gene expression profiles, colony dynamics and tissue development of honey bees exposed to sub-lethal doses of imidacloprid in laboratory and field experiments. PLoS ONE 2017, 12, e0171529. [Google Scholar] [CrossRef] [Green Version]
- Alptekin, S.; Bass, C.; Nicholls, C.; Paine, M.J.I.; Clark, S.J.; Field, L.; Moores, G.D. Induced thiacloprid insensitivity in honeybees (Apis mellifera L.) is associated with up-regulation of detoxification genes. Insect Mol. Biol. 2016, 25, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Tarek, H.; Hamiduzzaman, M.M.; Morfin, N.; Guzman-Novoa, E. Sub-lethal doses of neonicotinoid and carbamate insecticides reduce the lifespan and alter the expression of immune health and detoxification related genes of honey bees (Apis mellifera). Genet. Mol. Res. 2018, 17, 16039908. [Google Scholar] [CrossRef]
- Simmons, W.R.; Angelini, D.R. Chronic exposure to a neonicotinoid increases expression of antimicrobial peptide genes in the bumblebee bombus impatiens. Sci. Rep. 2017, 7, 44773. [Google Scholar] [CrossRef] [Green Version]
- Christen, V.; Mittner, F.; Fent, K. Molecular effects of neonicotinoids in honey bees (Apis mellifera). Environ. Sci. Technol. 2016, 50, 4071–4081. [Google Scholar] [CrossRef] [PubMed]
- Diao, Q.; Sun, L.; Zheng, H.; Zeng, Z.; Wang, S.; Xu, S.; Zheng, H.; Chen, Y.; Shi, Y.; Wang, Y.; et al. Genomic and transcriptomic analysis of the asian honeybee apis cerana provides novel insights into honeybee biology. Sci. Rep. 2018, 8, 822. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- James, R.R.; Xu, J. Mechanisms by which pesticides affect insect immunity. J. Invertebr. Pathol. 2012, 109, 175–182. [Google Scholar] [CrossRef]
- Vieira, C.S.; Moreira, O.C.; Batista, K.K.S.; Ratcliffe, N.A.; Castro, D.P.; Azambuja, P. The nf-kappa b inhibitor, imd-0354, affects immune gene expression, bacterial microbiota and trypanosoma cruzi infection in rhodnius prolixus midgut. Front. Physiol. 2018, 9, 1189. [Google Scholar] [CrossRef]
- Li, Z.; Yu, T.; Chen, Y.; Heerman, M.; He, J.; Huang, J.; Nie, H.; Su, S. Brain transcriptome of honey bees (Apis mellifera) exhibiting impaired olfactory learning induced by a sublethal dose of imidacloprid. Pestic. Biochem. Physiol. 2019, 156, 36–43. [Google Scholar] [CrossRef]
- Shi, J.; Yang, H.; Yu, L.; Liao, C.; Liu, Y.; Jin, M.; Yan, W.; Wu, X. Sublethal acetamiprid doses negatively affect the lifespans and foraging behaviors of honey bee (Apis mellifera L.) workers. Sci. Total Environ. 2020, 738, 139924. [Google Scholar] [CrossRef]
- Du, Y.; Xu, K.; Zhao, H.; Liu, Y.; Niu, Q.; Jiang, Y. Research progress in odorant binding proteins of insects. Acta Entomol. Sin. 2020, 63, 365–380. [Google Scholar]
- Li, H.L.; Wu, F.; Zhao, L.; Tan, J.; Jiang, H.T.; Hu, F.L. Neonicotinoid insecticide interact with honeybee odorant-binding protein: Implication for olfactory dysfunction. nt. J. Biol. Macromol. 2015, 81, 624–630. [Google Scholar] [CrossRef]
- Shi, T.-F.; Wang, Y.-F.; Liu, F.; Qi, L.; Yu, L.-S. Sublethal effects of the neonicotinoid insecticide thiamethoxam on the transcriptome of the honey bees (hymenoptera: Apidae). J. Econ. Entomol. 2017, 110, 2283–2289. [Google Scholar] [CrossRef]
- Kang, Z.-W.; Liu, F.-H.; Pang, R.-P.; Tian, H.-G.; Liu, T.-X. Effect of sublethal doses of lmidacloprid on the biological performance of aphid endoparasitoid aphidius gifuensis (hymenoptera: Aphidiidae) and influence on its related gene expression. Front. Physiol. 2018, 9, 1729. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhao, H.T.; Jiang, Y.S. Expressional and functional interactions of two apis cerana cerana olfactory receptors. Peerj 2018, 6, e5005. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Nichols, A.S.; Walden, K.K.O.; Brockmann, A.; Luetje, C.W.; Robertson, H.M. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc. Natl. Acad. Sci. USA 2007, 104, 14383–14388. [Google Scholar] [CrossRef] [Green Version]


| Gene Name | Pathway | Gene Description | Profile |
|---|---|---|---|
| LOC108002218 | AMP | abaecin | 12 |
| LOC108000468 | AMP | apidaecins type 22 | 12 |
| LOC107993803 | AMP | defensin-1 | 12 |
| LOC108000415 | AMP | defensin-2 | 12 |
| LOC107993492 | AMP | hymenoptaecin | 12 |
| LOC108001747 | AMP | hymenoptaecin pseudogene | 12 |
| LOC108004481 | Jak/STAT | E3 ubiquitin-protein ligase TRIM71-like | 17 |
| LOC108000724 | Jak/STAT | E3 ubiquitin-protein ligase MARCH2-like | 1 |
| LOC108003398 | Ribosomal Biosynthesis | 60S ribosomal protein L35 | 1 |
| LOC107992528 | Ribosomal Biosynthesis | 60S ribosomal protein L37a | 1 |
| LOC108000385 | Ribosomal Biosynthesis | 60S ribosomal protein L39 | 2 |
| LOC108001255 | Ribosomal Biosynthesis | 60S ribosomal protein L28-like | 2 |
| LOC107994696 | Ribosomal Biosynthesis | ribosomal RNA processing protein 1 homolog | 7 |
| LOC108002849 | Ribosomal Biosynthesis | 60S ribosomal protein L37 | 8 |
| LOC108003508 | Ribosomal Biosynthesis | histone H2A-like | 1 |
| LOC107998816 | Ribosomal Biosynthesis | histone H2A.Z-specific chaperone CHZ1-like | 7 |
| LOC107996866 | Toll/TLR | toll-like receptor 4 | 7 |
| LOC107998162 | Toll/TLR | toll-like receptor 13 | 17 |
| LOC108001172 | Toll/TLR | protein toll-like | 11 |
| LOC107995659 | Toll/TLR | protein toll | 12 |
| LOC107996104 | Toll/TLR | NF-kappa-B inhibitor cactus-like | 16 |
| LOC108002391 | Toll/TLR | NF-kappa-B inhibitor cactus-like | 16 |
| LOC107998380 | Toll/TLR | serine protease inhibitor 3-like | 10 |
| LOC107994873 | Toll/TLR | serine protease gd-like | 12 |
| LOC108002098 | Toll/TLR | serine protease snake-like | 12 |
| LOC108004289 | Toll/TLR | serine/threonine-protein kinase 32B | 12 |
| LOC107999607 | Toll/TLR | serine/threonine-protein kinase PLK1-like | 16 |
| LOC108003108 | Toll/TLR | serine/threonine-protein kinase Nek5-like | 17 |
| LOC107996922 | IG Superfamily Genes | lachesin-like | 12 |
| LOC107995322 | leukocyte | leukocyte elastase inhibitor-like | 12 |
| LOC107996108 | Lysozyme | lysosomal alpha-mannosidase | 9 |
| LOC107999764 | Scav. Receptor B | scavenger receptor class B member 1 | 1 |
| Gene Name | Gene Description | Profile |
|---|---|---|
| LOC107996029 | odorant-binding protein 59a-like | 1 |
| LOC108002840 | general odorant-binding protein 69a-like (ASP1) | 4 |
| LOC107999620 | general odorant-binding protein 56d-like (OBP13) | 9 |
| LOC107996745 | odorant receptor 4-like | 10 |
| LOC107996764 | odorant receptor 4-like | 12 |
| LOC107996743 | odorant receptor 4-like | 14 |
| LOC107996763 | odorant receptor 4-like | 15 |
| LOC107996720 | odorant receptor 4-like | 7 |
| LOC108000540 | odorant receptor 13a-like | 5 |
| LOC107994426 | odorant receptor 13a-like | 17 |
| LOC107996760 | odorant receptor 22c-like | 10 |
| LOC107999581 | odorant receptor 63a-like | 15 |
| LOC107996752 | odorant receptor 49b-like | 17 |
| LOC108002046 | glutamate receptor ionotropic, kainate 2 | 17 |
| LOC108000903 | glutamate receptor ionotropic, NMDA 2C-like | 3 |
| LOC107997799 | gustatory receptor for sugar taste 64f-like | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Jin, S.-S.; He, Y.; Luo, J.-H.; Xu, C.-Q.; Wu, Y.-Y.; Hou, C.-S.; Wang, Q.; Diao, Q.-Y. Physiological Analysis and Transcriptome Analysis of Asian Honey Bee (Apis cerana cerana) in Response to Sublethal Neonicotinoid Imidacloprid. Insects 2020, 11, 753. https://doi.org/10.3390/insects11110753
Gao J, Jin S-S, He Y, Luo J-H, Xu C-Q, Wu Y-Y, Hou C-S, Wang Q, Diao Q-Y. Physiological Analysis and Transcriptome Analysis of Asian Honey Bee (Apis cerana cerana) in Response to Sublethal Neonicotinoid Imidacloprid. Insects. 2020; 11(11):753. https://doi.org/10.3390/insects11110753
Chicago/Turabian StyleGao, Jing, San-Sheng Jin, Yan He, Jin-Hong Luo, Chun-Qin Xu, Yan-Yan Wu, Chun-Shen Hou, Qiang Wang, and Qing-Yun Diao. 2020. "Physiological Analysis and Transcriptome Analysis of Asian Honey Bee (Apis cerana cerana) in Response to Sublethal Neonicotinoid Imidacloprid" Insects 11, no. 11: 753. https://doi.org/10.3390/insects11110753
APA StyleGao, J., Jin, S.-S., He, Y., Luo, J.-H., Xu, C.-Q., Wu, Y.-Y., Hou, C.-S., Wang, Q., & Diao, Q.-Y. (2020). Physiological Analysis and Transcriptome Analysis of Asian Honey Bee (Apis cerana cerana) in Response to Sublethal Neonicotinoid Imidacloprid. Insects, 11(11), 753. https://doi.org/10.3390/insects11110753







