Gene Sequences of Potential Targets of Insecticidal PF2 Lectin Identified from the Larval De Novo Transcriptome of the Mexican Bean Weevil (Zabrotes Subfasciatus; Boheman 1833)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Culture
2.2. Larvae Sampling and RNA Isolation
2.3. cDNA Library Preparation and Illumina Sequencing
2.4. RNA-Seq Data Processing and De Novo Assembly
2.5. Unigene Annotation and Classification
2.6. Identification and Phylogenetic Analysis of Potential Targets of PF2 Lectin
3. Results
3.1. Sequence Analysis and De Novo Assembly
3.2. Unigene Annotation
3.3. Protein Domains Annotation
3.4. Functional Categorization and Metabolic Pathway Annotation
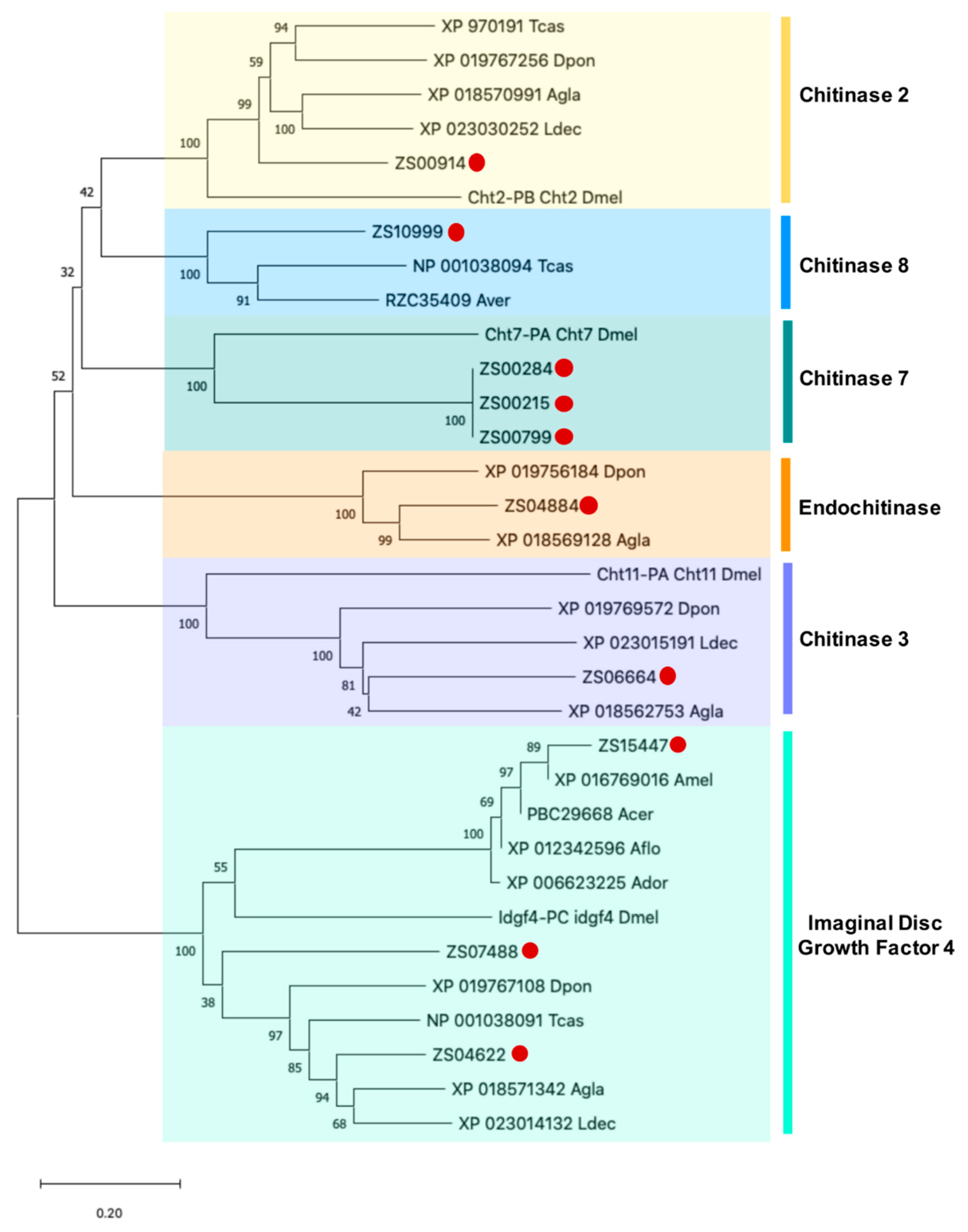
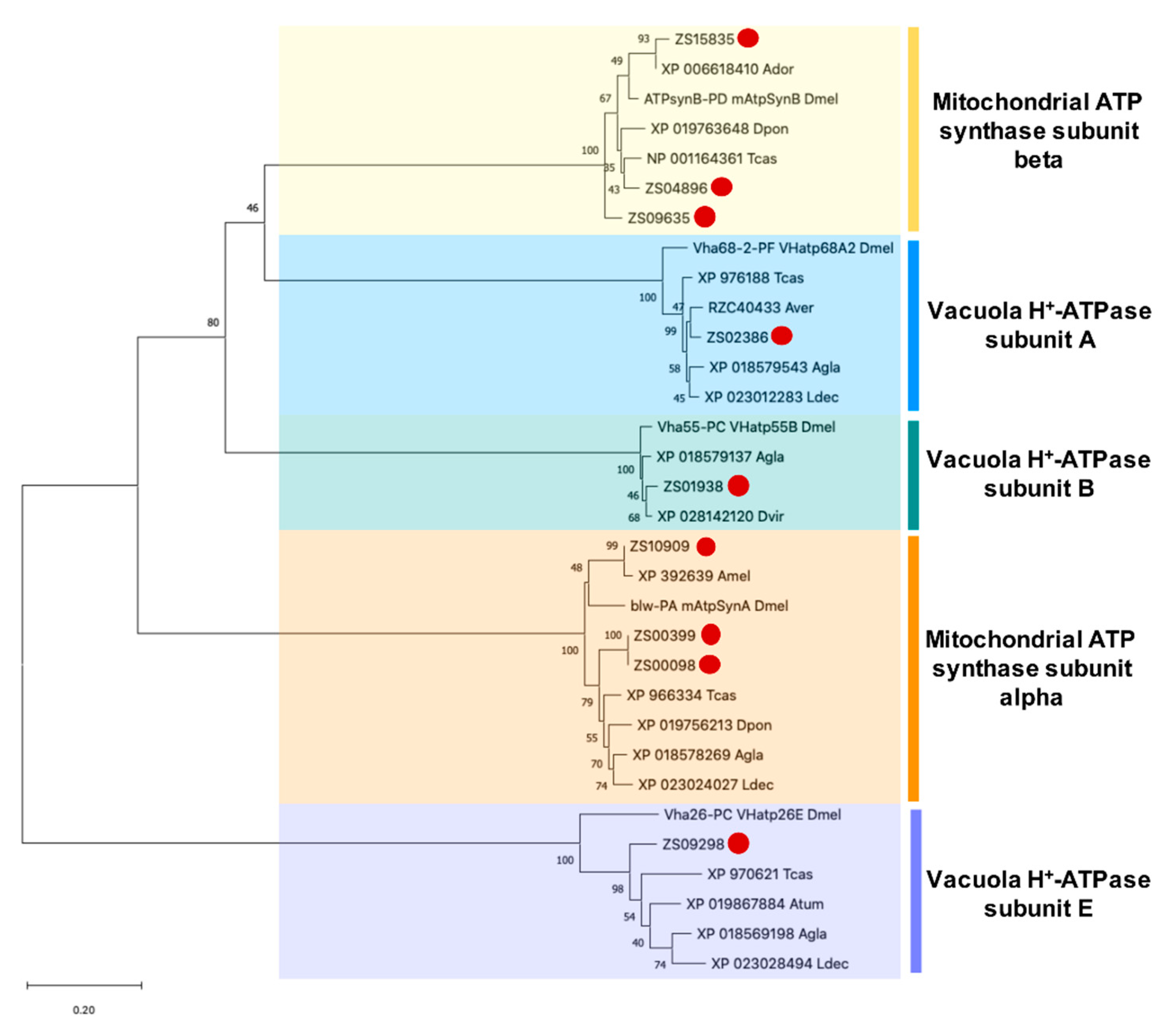
3.5. Identification and Phylogenetic Analysis of Potential PF2 Lectin Target Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guzzo, E.C.; Vendramim, J.D.; Lourenção, A.L.; Chiorato, A.F.; Carbonell, S.A.M.; Corrêa, O.M.B. Adult attractiveness and oviposition preference of Zabrotes subfasciatus toward genotypes of common bean Phaseolus vulgaris. Phytoparasitica 2018, 46, 645–651. [Google Scholar] [CrossRef] [Green Version]
- Bitocchi, E.; Nanni, L.; Bellucci, E.; Rossi, M.; Giardini, A.; Zeuli, P.S.; Logozzo, G.; Stougaard, J.; McClean, P.; Attene, G.; et al. Mesoamerican origin of the common bean (Phaseolus vulgaris; L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 2012, 109, 788. [Google Scholar] [CrossRef] [Green Version]
- Cariño, R.N.R.; Nápoles, J.R.; Graziano, J.V.; Martínez, A.E.; Martínez, N.B.; Morales, J.H.; García, A.A. Demografía de brúquidos (Coleoptera: Bruchidae) asociados con Vigna unguiculata (L.) Walp (Fabaceae). Acta Zool. Mex. 2017, 33, 9–17. [Google Scholar]
- Ortega-Nieblas, M.M.; Robles-Burgueño, M.R.; Vázquez-Moreno, L.; Cortez-Mondaca, E.; González-León, A.; Morales-Trejo, A.; González-Rios, H. Toxic and persistent effect of Oregano’s essential oil against Zabrotes subfasciatus (Coleoptera: Bruchidae) in stored dry beans. Southwest. Entomol. 2014, 39, 147–162. [Google Scholar] [CrossRef]
- Mancillas-Paredes, J.M.; Hernández-Sánchez, H.; Jaramillo-Flores, M.E.; García-Gutiérrez, C. Proteases and chitinases induced in Beauveria bassiana during infection by Zabrotes subfasciatus. Southwest. Entomol. 2019, 44, 125–137. [Google Scholar] [CrossRef]
- Carlini, C.R.; Grossi-de-Sá, M.F. Plant toxic proteins with insecticidal properties. A review on their potentialities as bioinsecticides. Toxicon 2002, 40, 1515–1539. [Google Scholar] [CrossRef]
- Hamid, R.; Masood, A.; Wani, I.H.; Rafiq, S. Lectins: Proteins with diverse applications. J. Appl. Pharm. Sci. 2013, 3, 93–103. [Google Scholar]
- Lagarda-Diaz, I.; Guzman-Partida, A.; Vazquez-Moreno, L. Legume lectins: Proteins with diverse applications. Int. J. Mol. Sci. 2017, 18, 1242. [Google Scholar] [CrossRef] [Green Version]
- Hernández, G.U. Identificacion de Estructuras Glicosıdicas Reconocidas por la Lectina PF2 en Fetuına y Tejidos Linfoides; Centro de Investigación en Alimentación y Desarrollo, A.C.: Hermosillo, Mexico, 2007. [Google Scholar]
- Lagarda-Diaz, I.; Guzman-Partida, A.M.; Urbano-Hernandez, G.; Ortega-Nieblas, M.M.; Robles-Burgueño, M.R.; Winzerling, J.; Vazquez-Moreno, L. Insecticidal action of PF2 lectin from Olneya tesota (Palo Fierro) against Zabrotes subfasciatus larvae and midgut glycoconjugate binding. J. Agric. Food Chem. 2009, 57, 689–694. [Google Scholar] [CrossRef]
- Lagarda-Diaz, I.; Geiser, D.; Guzman-Partida, A.M.; Winzerling, J.; Vazquez-Moreno, L. Recognition and binding of the PF2 lectin to α-Amylase from Zabrotes subfasciatus (Coleoptera:Bruchidae) larval midgut. J. Insect Sci. 2014, 14. [Google Scholar] [CrossRef] [Green Version]
- Lagarda-Diaz, I.; Guzman-Partida, A.M.; Huerta-Ocampo, J.A.; Winzerling, J.; Vazquez-Moreno, L. Identification of membrane proteins of the midgut of Zabrotes subfasciatus larvae associated with the insecticidal mechanism of PF2 lectin. J. Asia Pacif. Entomol. 2016, 19, 677–682. [Google Scholar] [CrossRef]
- Lagarda-Diaz, I.; Robles-Burgeño, M.R.; Guzman-Partida, A.M.; Geiser, D.; Winzerling, J.; Vazquez-Moreno, L. Binding of PF2 lectin from Olneya tesota to gut proteins of Zabrotes subfasciatus larvae associated with the insecticidal mechanism. J. Agric. Food Chem. 2012, 60, 2398–2402. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Quiroz, M.; Valdez-Carrasco, J.; Vera-Graziano, J.; Castillo-Morales, A. Identificación de ínstares larvales de Zabrotes subfasciatus (BOH.) (Coloptera: Bruchidae) mediante las dimensiones de sus cápsulas cefálicas. Agrociencia 2000, 34, 83–90. [Google Scholar]
- Bolger, A.M.; Usadel, B.; Lohse, M. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.T.; Howe, A.; Zhang, Q.; Pyrkosz, A.B.; Brom, T.H. A reference-free algorithm for computational normalization of shotgun sequencing data. Genomics 2012, arXiv:1203.4802. [Google Scholar]
- Brown, C.T.; Scott, C.; Crusoe, M.S.; Leigh, S.; Rosenthal, J.; Howe, A. Khmer-Protocols Documentation. 2013. Available online: https://khmer-protocols.readthedocs.io/en/v0.8.4/ (accessed on 22 October 2020).
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.-J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef] [Green Version]
- Keeling, C.I.; Yuen, M.M.S.; Liao, N.Y.; Docking, T.R.; Chan, S.K.; Taylor, G.A.; Palmquist, D.L.; Jackman, S.D.; Nguyen, A.; Li, M.; et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013, 14, R27. [Google Scholar] [CrossRef] [Green Version]
- Schoville, S.D.; Chen, Y.H.; Andersson, M.N.; Benoit, J.B.; Bhandari, A.; Bowsher, J.H.; Brevik, K.; Cappelle, K.; Chen, M.-J.M.; Childers, A.K.; et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribolium-Genome-Sequencing-Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkin, L.C.; Oppert, B. Gene expression in Tribolium castaneum life stages: Identifying a species-specific target for pest control applications. PeerJ 2019, 7, e6946. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Congiu, L.; Lindström, L.; Piiroinen, S.; Vidotto, M.; Grapputo, A. Sequencing, de novo assembly and annotation of the Colorado potato beetle, Leptinotarsa decemlineata, transcriptome. PLoS ONE 2014, 9, e86012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walski, T.; De Schutter, K.; Van Damme, E.J.M.; Smagghe, G. Diversity and functions of protein glycosylation in insects. Insect Biochem. Mol. Biol. 2017, 83, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yamaguchi, S.; Fujii-Taira, I.; Iijima, R.; Natori, S.; Homma, K.J. Involvement of insect-derived growth factor (IDGF) in the cell growth of an embryonic cell line of flesh fly. Biochem. Biophys. Res. Commun. 2006, 350, 334–338. [Google Scholar] [CrossRef]
- Zurovcova, M.; Benes, V.; Zurovec, M.; Kucerova, L. Expansion of imaginal disc growth factor gene family in diptera reflects the evolution of novel functions. Insects 2019, 10, 365. [Google Scholar] [CrossRef] [Green Version]
- Pesch, Y.-Y.; Riedel, D.; Patil, K.R.; Loch, G.; Behr, M. Chitinases and Imaginal disc growth factors organize the extracellular matrix formation at barrier tissues in insects. Sci. Rep. 2016, 6, 18340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruttero, L.L.; Leyria, J.; Moyetta, N.R.; Ramos, F.O.; Settembrini, B.P.; Canavoso, L.E. The fat body of the hematophagous insect, Panstrongylus megistus (Hemiptera: Reduviidae): Histological features and participation of the β-chain of ATP synthase in the lipophorin-mediated lipid transfer. J. Insect Sci. 2019, 19. [Google Scholar] [CrossRef]
- Eyraud, V.; Balmand, S.; Karaki, L.; Rahioui, I.; Sivignon, C.; Delmas, A.F.; Royer, C.; Rahbé, Y.; Da Silva, P.; Gressent, F. The interaction of the bioinsecticide PA1b (Pea Albumin 1 subunit b) with the insect V-ATPase triggers apoptosis. Sci. Rep. 2017, 7, 4902. [Google Scholar] [CrossRef]
- Vandenborre, G.; Smagghe, G.; Ghesquière, B.; Menschaert, G.; Rao, R.N.; Gevaert, K.; Van Damme, E.J.M. Diversity in protein glycosylation among insect species. PLoS ONE 2011, 6, e16682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merzendorfer, H.; Huss, M.; Schmid, R.; Harvey, W.R.; Wieczorek, H. A novel insect V-ATPase subunit M9.7 is glycosylated extensively. J. Biol. Chem. 1999, 274, 17372–17378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Lage, J.-L. The amylases of insects. Int. J. Insect Sci. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Mach, J.; Poliak, P.; Matušková, A.; Žárský, V.; Janata, J.; Lukeš, J.; Tachezy, J. An advanced system of the mitochondrial processing peptidase and core protein family in Trypanosoma brucei and multiple origins of the core I subunit in eukaryotes. Genome Biol. Evol. 2013, 5, 860–875. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.B.; Smith, B.S.; Kitada, S.; Kojima, K.; Miyaura, H.; Otwinowski, Z.; Ito, A.; Deisenhofer, J. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 2001, 9, 615–625. [Google Scholar] [CrossRef] [Green Version]
- VenkatRao, V.; Chaitanya, R.K.; Kumar, D.N.; Bramhaiah, M.; Dutta-Gupta, A. Developmental and hormone-induced changes of mitochondrial electron transport chain enzyme activities during the last instar larval development of maize stem borer, Chilo partellus (Lepidoptera: Crambidae). Gen. Comp. Endocrinol. 2016, 239, 32–39. [Google Scholar] [CrossRef]
- Hansford, R.G.; Lehninger, A.L. The effect of the coupled oxidation of substrate on the permeability of blowfly flight-muscle mitochondria to potassium and other cations. Biochem. J. 1972, 126, 689–700. [Google Scholar] [CrossRef]
- Novák, F.; Nováková, O.; Kubišta, V. Heterogeneity of insect flight muscle mitochondria as demonstrated by different phospholipid turnover in vivo. Insect Biochem. 1979, 9, 389–396. [Google Scholar] [CrossRef]
- Ande, S.R.; Moulik, S.; Mishra, S. Interaction between O-GlcNAc modification and tyrosine phosphorylation of prohibitin: Implication for a novel binary switch. PLoS ONE 2009, 4, e4586. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Murphy, L.C.; Nyomba, B.L.G.; Murphy, L.J. Prohibitin: A potential target for new therapeutics. Trends Mol. Med. 2005, 11, 192–197. [Google Scholar] [CrossRef]
- Thuaud, F.; Ribeiro, N.; Nebigil, C.G.; Désaubry, L. Prohibitin ligands in cell death and survival: Mode of action and therapeutic potential. Chem. Biol. 2013, 20, 316–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
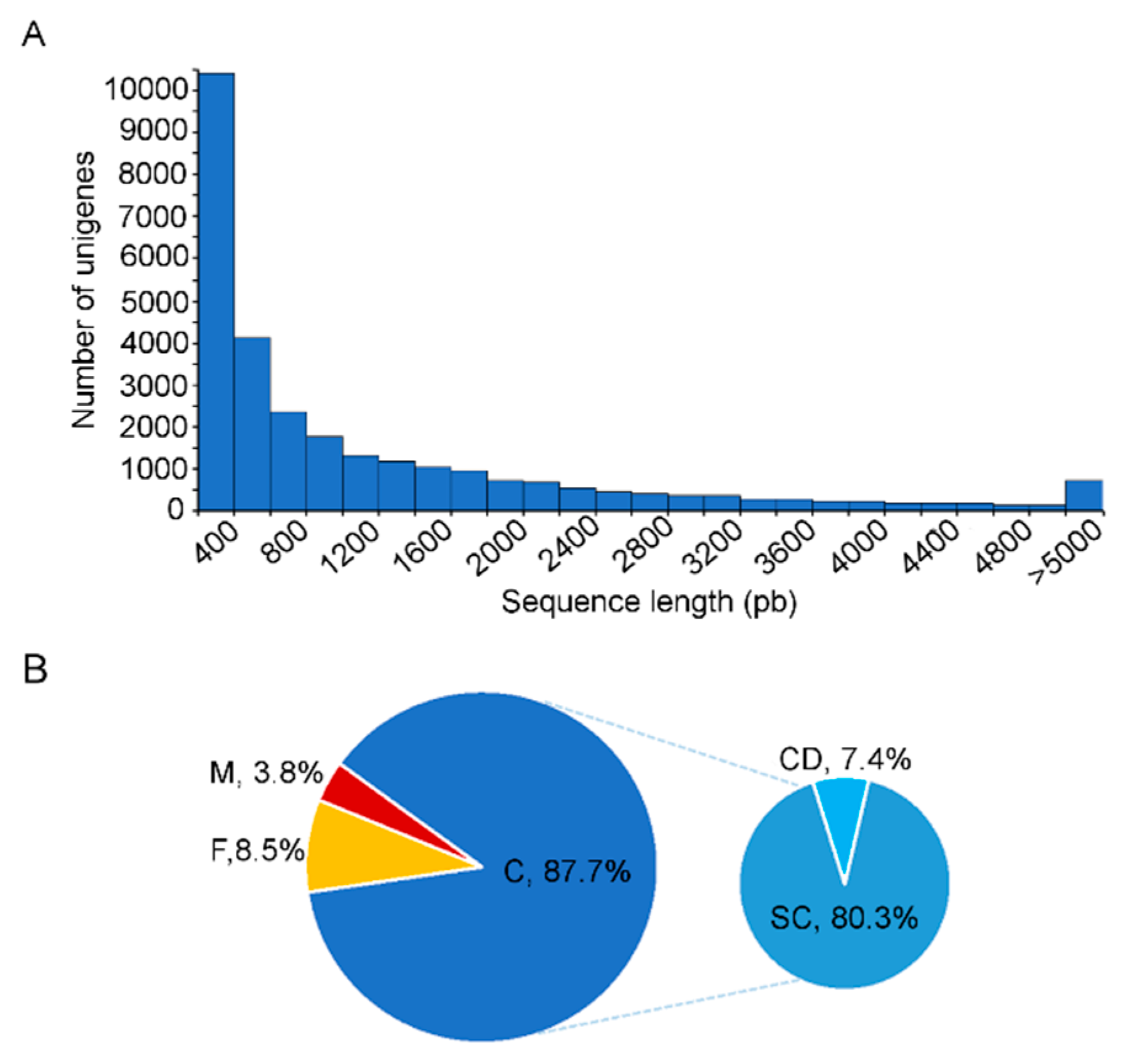
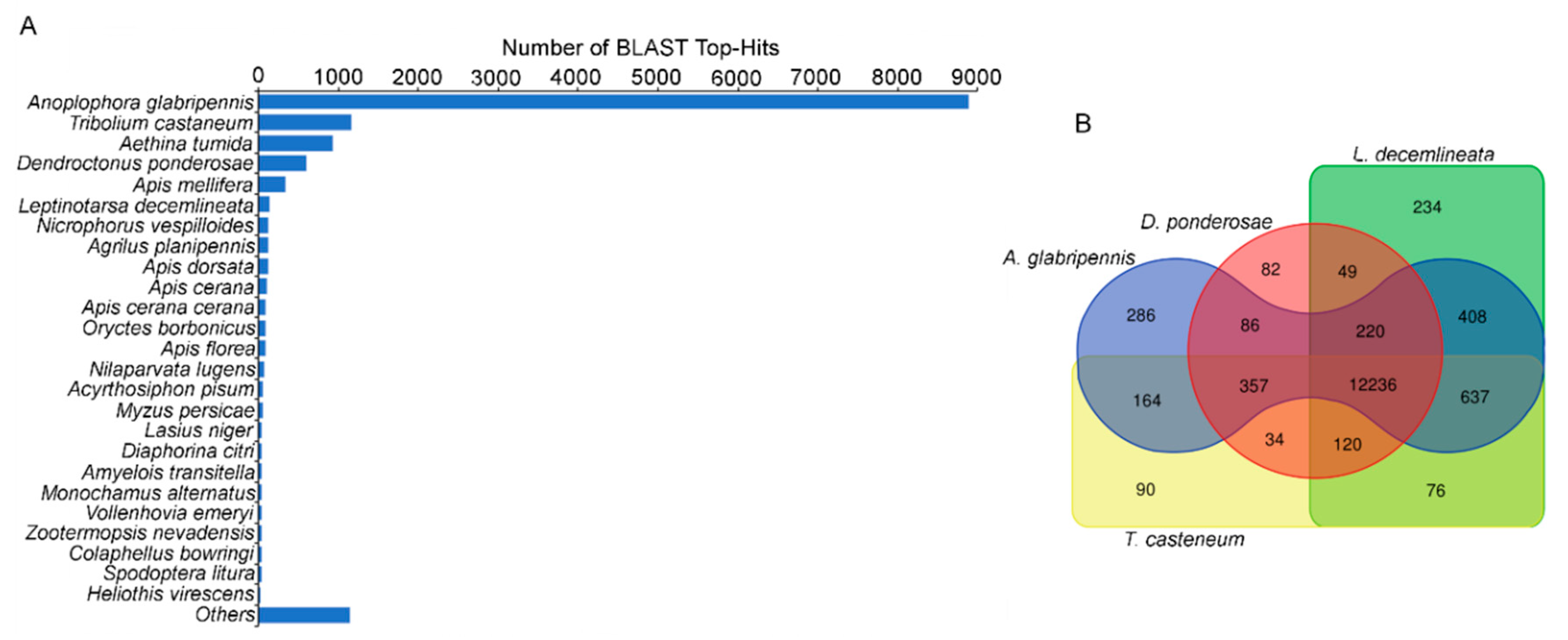
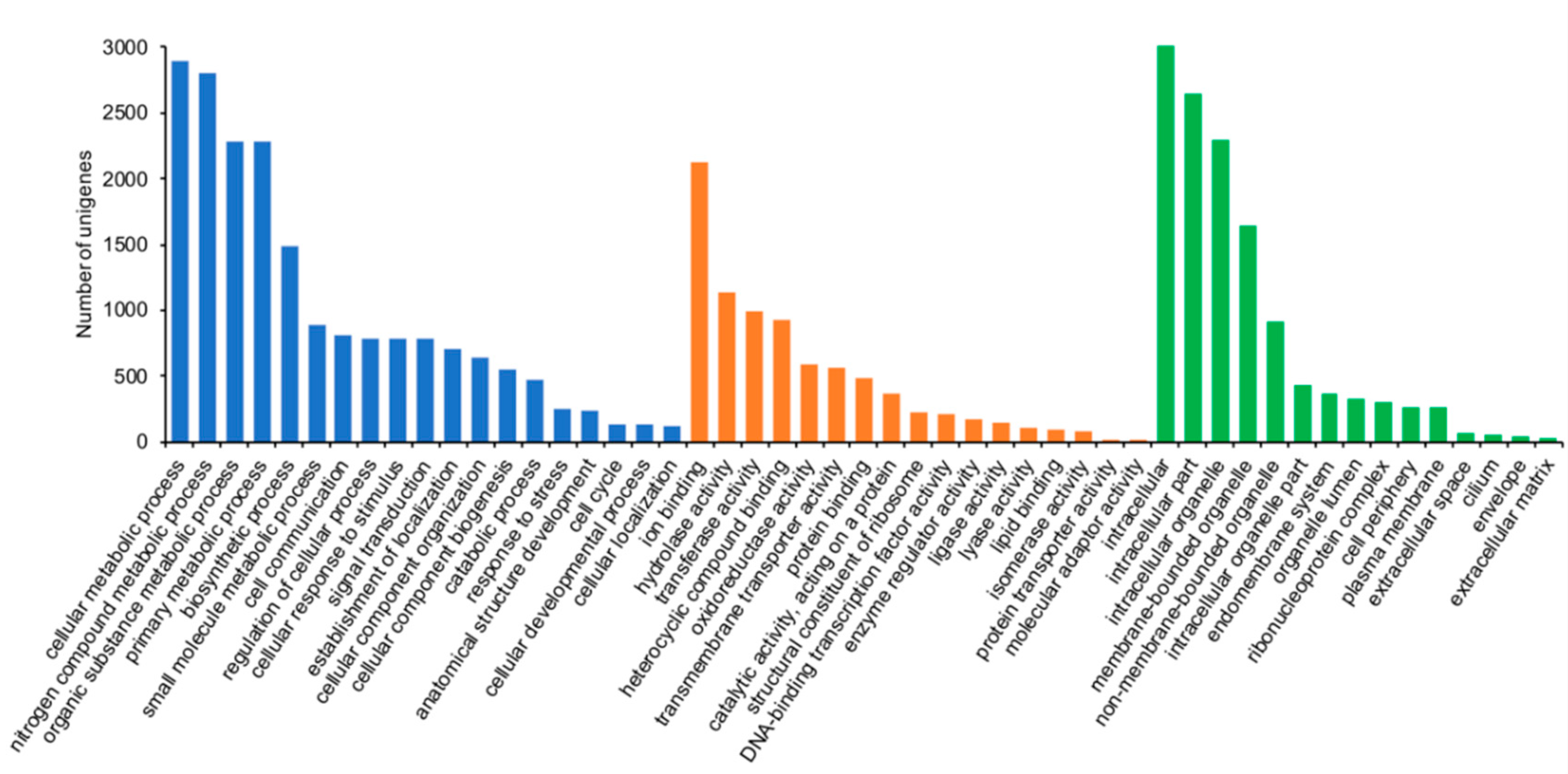


| Total NUMBER of Assembled Unigenes | 29,029 |
|---|---|
| Total bases pairs (Mbp) | 33.9 |
| Smallest unigene (bp) | 200 |
| Largest unigene (bp) | 15,454 |
| Mean length unigene (bp) | 1168.9 |
| Unigenes over 1 kbp | 10,379 |
| Unigenes over 10 kbp | 23 |
| Unigenes with open reading frames | 21,380 |
| N90 | 443 |
| N70 | 1285 |
| N50 | 2196 |
| GC percentage (%) | 39 |
| Proportion of N bases (%) | 0.05 |
| InterPro Domains | Number of Unigenes |
|---|---|
| (IPR013087) Zinc finger Cys2His2-type | 440 |
| (IPR000719) Protein kinase domain | 195 |
| (IPR020846) Major facilitator superfamily domain | 137 |
| (IPR017986) beta-transducin-repeat-containing domain | 135 |
| (IPR000504) RNA recognition motif domain | 132 |
| (IPR001841) Zinc finger, RING (Really Interesting New Gene)-type | 94 |
| (IPR007110) Immunoglobulin-like domain | 94 |
| (IPR020683) Ankyrin repeat-containing domain | 91 |
| (IPR002048) EF-hand domain | 80 |
| (IPR013026) Tetratricopeptide repeat-containing domain | 71 |
| (IPR001254) Serine proteases, trypsin domain | 71 |
| (IPR014001) Helicase superfamily 1/2, ATP-binding domain | 71 |
| (IPR001650) Helicase, C-terminal | 70 |
| (IPR027806) Harbinger transposase-derived nuclease domain | 67 |
| (IPR005225) Small GTP-binding protein domain | 62 |
| (IPR001849) Pleckstrin homology domain | 61 |
| (IPR001478) PDZ domain | 61 |
| (IPR001452) SH3 domain | 60 |
| (IPR003961) Fibronectin type III | 57 |
| (IPR000742) EGF-like domain | 55 |
| (IPR000210) BTB/POZ domain | 55 |
| (IPR001356) Homeobox domain | 54 |
| (IPR003439) ABC transporter-like | 52 |
| (IPR000477) Reverse transcriptase domain | 50 |
| (IPR013098) Immunoglobulin I-set | 50 |
| (IPR002557) Chitin binding domain | 50 |
| (IPR011545) DEAD/DEAH box helicase domain | 48 |
| (IPR001245) Serine-threonine/tyrosine-protein kinase, catalytic domain | 42 |
| (IPR017452) GPCR, rhodopsin-like, 7TM | 41 |
| (IPR003959) ATPase, AAA-type, core | 38 |
| Unigen ID | Length (pb) | Open Reading Frame (aa) | BLASTX Best Hit (Accession Number/Description) | Identity (%) | E-Value | Gly Sites a | Full Length b |
|---|---|---|---|---|---|---|---|
| Alpha amylase | |||||||
| ZS18820 | 393 | 128 | AAM20738.1 | AF259649_1alpha-amylase [Apis mellifera mellifera] | 100.0 | 3.78−62 | 2 | No |
| ZS13222 | 698 | 104 | AAM20738.1 | AF259649_1alpha-amylase [Apis mellifera mellifera] | 100.0 | 2.57−105 | 1 | No |
| IDGF4 (Imaginal Disc Growth Factor 4) | |||||||
| ZS15447 | 541 | 179 | XP_016769016.1 | PREDICTED: chitinase-like protein Idgf4 isoform X2 [Apis mellifera] | 100.0 | 1.08−126 | 1 | No |
| ZS04622 | 2177 | 351 | XP_018571342.1 | PREDICTED: chitinase-like protein Idgf4 isoform X2 [Anoplophora glabripennis] | 92.7 | 0 | 1 | No |
| ZS07488 | 1479 | 245 | XP_018571343.1 | PREDICTED: chitinase-like protein Idgf4 [Anoplophora glabripennis] | 89.9 | 4.63−109 | 1 | No |
| Chitinase | |||||||
| ZS00914 | 4653 | 426 | XP_018570991.1 | PREDICTED: probable chitinase 2 [Anoplophora glabripennis] | 81.9 | 0 | - | No |
| ZS10999 | 928 | 196 | XP_018570656.1 | PREDICTED: acidic mammalian chitinase-like [Anoplophora glabripennis] | 78.1 | 2.53−58 | - | No |
| ZS00284 | 6248 | 935 | XP_018575205.1 | probable chitinase 10 [Anoplophora glabripennis] | 94.5 | 0 | - | No |
| ZS00215 | 6614 | 935 | XP_018575205.1 | probable chitinase 10 [Anoplophora glabripennis] | 94.5 | 0 | - | No |
| ZS00799 | 4836 | 935 | XP_018575205.1 | probable chitinase 10 [Anoplophora glabripennis] | 94.5 | 0 | - | No |
| ZS04884 | 2088 | 551 | XP_018569128.1 | PREDICTED: endochitinase [Anoplophora glabripennis] | 84.5 | 0 | - | No |
| ZS06664 | 1647 | 479 | XP_018562753.1 | PREDICTED: chitinase-3-like protein 2 [Anoplophora glabripennis] | 74.7 | 0 | - | No |
| Mitochondrial-processing peptidase subunit | |||||||
| ZS06108 | 1764 | 545 | XP_018566230.1 | PREDICTED: mitochondrial-processing peptidase subunit alpha [Anoplophora glabripennis] | 87.5 | 0 | 2 | Yes |
| ZS03356 | 2689 | 208 | XP_018560943.1 | PREDICTED: mitochondrial-processing peptidase subunit beta [Anoplophora glabripennis] | 95.2 | 1.40−158 | - | No |
| ZS04132 | 2348 | 376 | XP_018560943.1 | PREDICTED: mitochondrial-processing peptidase subunit beta [Anoplophora glabripennis] | 95.2 | 1.19−158 | - | No |
| NADH-ubiquinone oxidoreductase subunit mictochondrial | |||||||
| ZS03487 | 2626 | 423 | XP_018567499.1 | PREDICTED: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial [Anoplophora glabripennis] | 92.0 | 0 | - | No |
| Prohibitin 1 | |||||||
| ZS28283 | 208 | 51 | XP_006559390.1 | PREDICTED: protein l(2)37Cc [Apis mellifera] | 100.0 | 1.00−16 | - | No |
| ZS00843 | 4761 | 276 | XP_018567952.1 | PREDICTED: protein l(2)37Cc [Anoplophora glabripennis] | 98.9 | 1.70−163 | 1 | Yes |
| Prohibitin 2 | |||||||
| ZS07780 | 1423 | 209 | XP_974101.1 | PREDICTED: prohibitin-2 isoform X1 [Tribolium castaneum] | 94.9 | 1.41−133 | 2 | No |
| ZS09582 | 1115 | 309 | AEE63210.1 | unknown [Dendroctonus ponderosae] | 90.6 | 3.31−180 | 4 | Yes |
| Band7 / SPFH-like | |||||||
| ZS05334 | 1961 | 291 | XP_019868837.1 | PREDICTED: band 7 protein AGAP004871 isoform X1 [Aethina tumida] | 96.6 | 0 | - | No |
| Mitochondrial ATP synthase subunit alpha | |||||||
| ZS10909 | 937 | 296 | XP_016919804.1 | PREDICTED: ATP synthase subunit alpha, mitochondrial [Apis cerana] | 100.0 | 0 | - | No |
| ZS00399 | 5790 | 556 | XP_966334.1 | PREDICTED: ATP synthase subunit alpha, mitochondrial [Tribolium castaneum] | 95.6 | 0 | - | Yes |
| ZS00098 | 7710 | 556 | XP_966334.1 | PREDICTED: ATP synthase subunit alpha, mitochondrial [Tribolium castaneum] | 95.6 | 0 | - | Yes |
| Mitochondrial ATP synthase subunit beta | |||||||
| ZS15835 | 519 | 173 | KOB78881.1 | ATP synthase subunit beta [Operophtera brumata] | 97.7 | 1.25−117 | - | No |
| ZS09635 | 1106 | 357 | XP_018562273.1 | PREDICTED: ATP synthase subunit beta, mitochondrial [Anoplophora glabripennis] | 97.4 | 0 | - | No |
| ZS04896 | 2083 | 517 | NP_001164361.1 | ATP synthase subunit beta, mitochondrial [Tribolium castaneum] | 94.5 | 0 | 2 | Yes |
| Vacuola H+-ATPase subunit A | |||||||
| ZS02386 | 3225 | 614 | XP_018579543.1 | PREDICTED: V-type proton ATPase catalytic subunit A [Anoplophora glabripennis] | 98.7 | 0 | 3 | Yes |
| Vacuola H+-ATPase subunit B | |||||||
| ZS01938 | 3568 | 496 | XP_018579137.1 | PREDICTED: V-type proton ATPase subunit B [Anoplophora glabripennis] | 99.2 | 0 | 3 | Yes |
| Vacuola H+-ATPase subunit E | |||||||
| ZS09298 | 1162 | 189 | XP_018569198.1 | PREDICTED: V-type proton ATPase subunit E [Anoplophora glabripennis] | 84.1 | 4.25−99 | - | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagarda-Diaz, I.; Hernández-Oñate, M.Á.; Huerta-Ocampo, J.Á.; Guzmán-Partida, A.M.; Winzerling, J.; Geiser, D.; Vázquez-Moreno, L. Gene Sequences of Potential Targets of Insecticidal PF2 Lectin Identified from the Larval De Novo Transcriptome of the Mexican Bean Weevil (Zabrotes Subfasciatus; Boheman 1833). Insects 2020, 11, 736. https://doi.org/10.3390/insects11110736
Lagarda-Diaz I, Hernández-Oñate MÁ, Huerta-Ocampo JÁ, Guzmán-Partida AM, Winzerling J, Geiser D, Vázquez-Moreno L. Gene Sequences of Potential Targets of Insecticidal PF2 Lectin Identified from the Larval De Novo Transcriptome of the Mexican Bean Weevil (Zabrotes Subfasciatus; Boheman 1833). Insects. 2020; 11(11):736. https://doi.org/10.3390/insects11110736
Chicago/Turabian StyleLagarda-Diaz, Irlanda, Miguel Ángel Hernández-Oñate, José Ángel Huerta-Ocampo, Ana M. Guzmán-Partida, Joy Winzerling, Dawn Geiser, and Luz Vázquez-Moreno. 2020. "Gene Sequences of Potential Targets of Insecticidal PF2 Lectin Identified from the Larval De Novo Transcriptome of the Mexican Bean Weevil (Zabrotes Subfasciatus; Boheman 1833)" Insects 11, no. 11: 736. https://doi.org/10.3390/insects11110736
APA StyleLagarda-Diaz, I., Hernández-Oñate, M. Á., Huerta-Ocampo, J. Á., Guzmán-Partida, A. M., Winzerling, J., Geiser, D., & Vázquez-Moreno, L. (2020). Gene Sequences of Potential Targets of Insecticidal PF2 Lectin Identified from the Larval De Novo Transcriptome of the Mexican Bean Weevil (Zabrotes Subfasciatus; Boheman 1833). Insects, 11(11), 736. https://doi.org/10.3390/insects11110736





