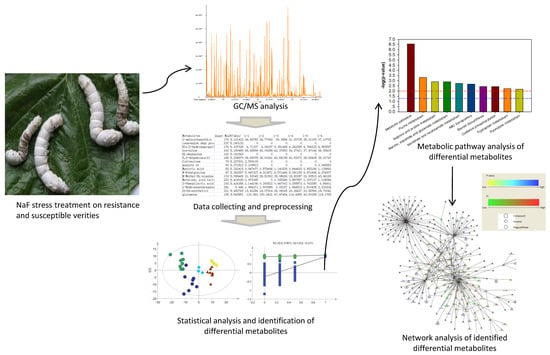
Gas Chromatography-Mass Spectrometry Based Midgut Metabolomics Reveals the Metabolic Perturbations under NaF Stress in Bombyx mori
Abstract
1. Introduction
2. Materials and Methods
2.1. Silkworm Rearing and Material Treatment
2.2. Sample Preparation
2.3. GC-MS Analysis Conditions
2.4. Metabonomics Data Preprocessing
2.5. Multivariate Statistical Analysis
2.6. Screening of Differential Metabolites
2.7. Metabolic Pathway Analysis of Differential Metabolites
3. Results and Analysis
3.1. Analysis of GC-MS Results
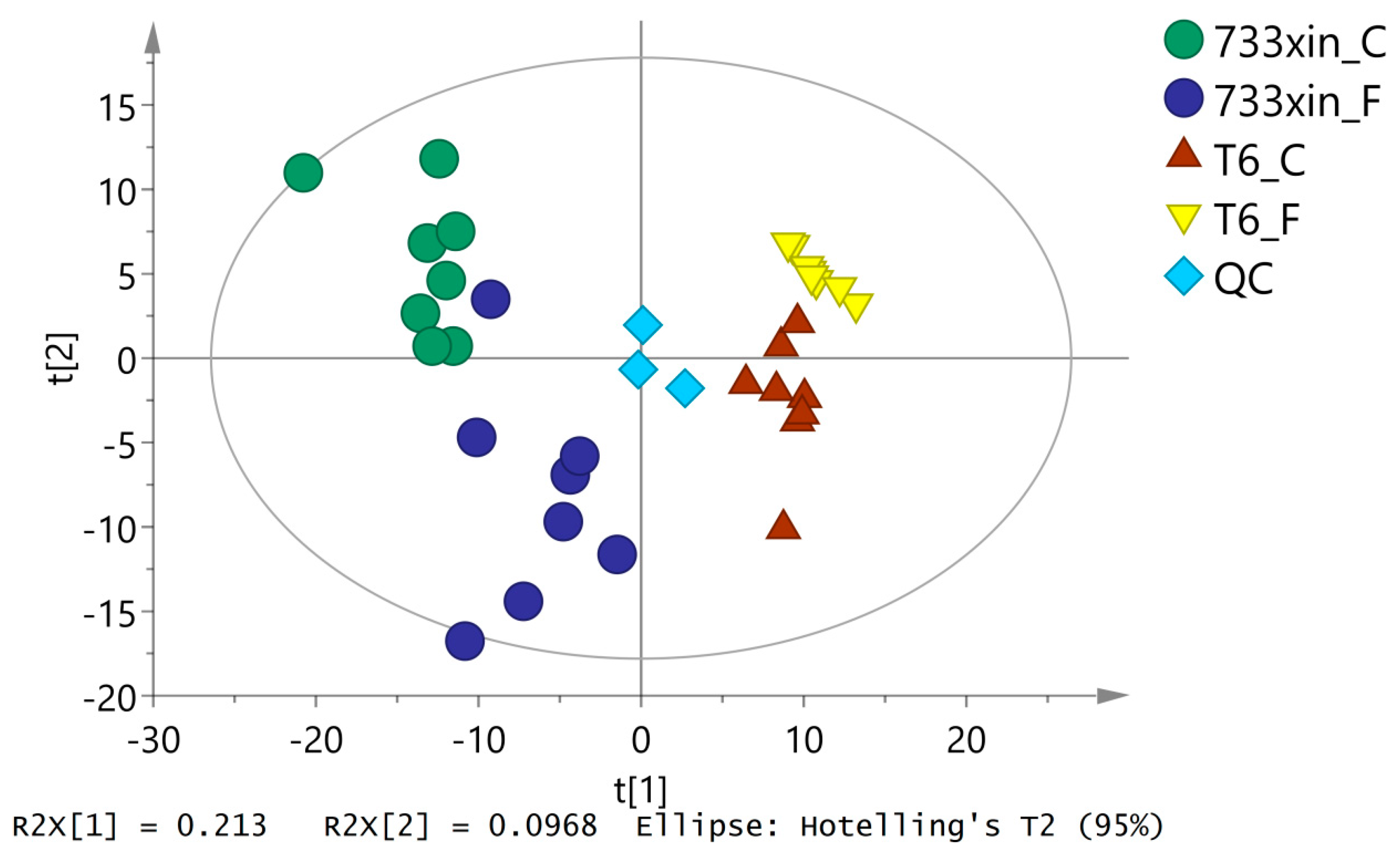
3.2. Overall PCA of Metabolic Phenotypes
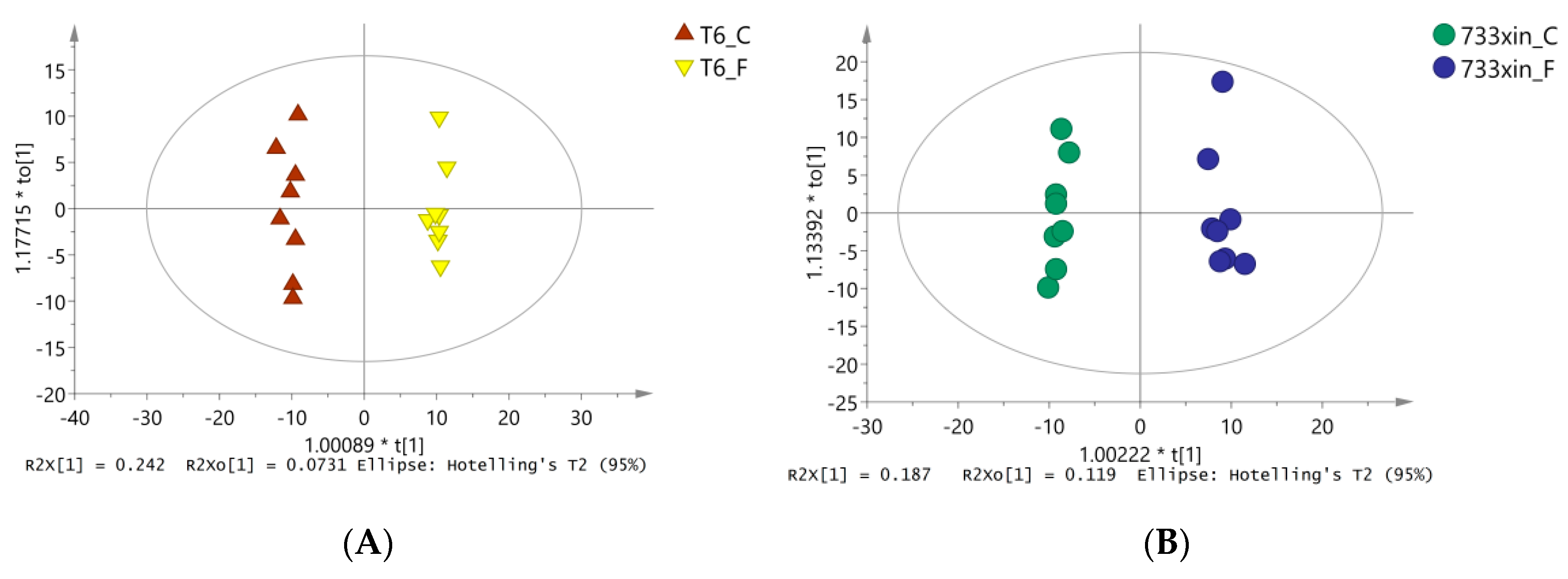
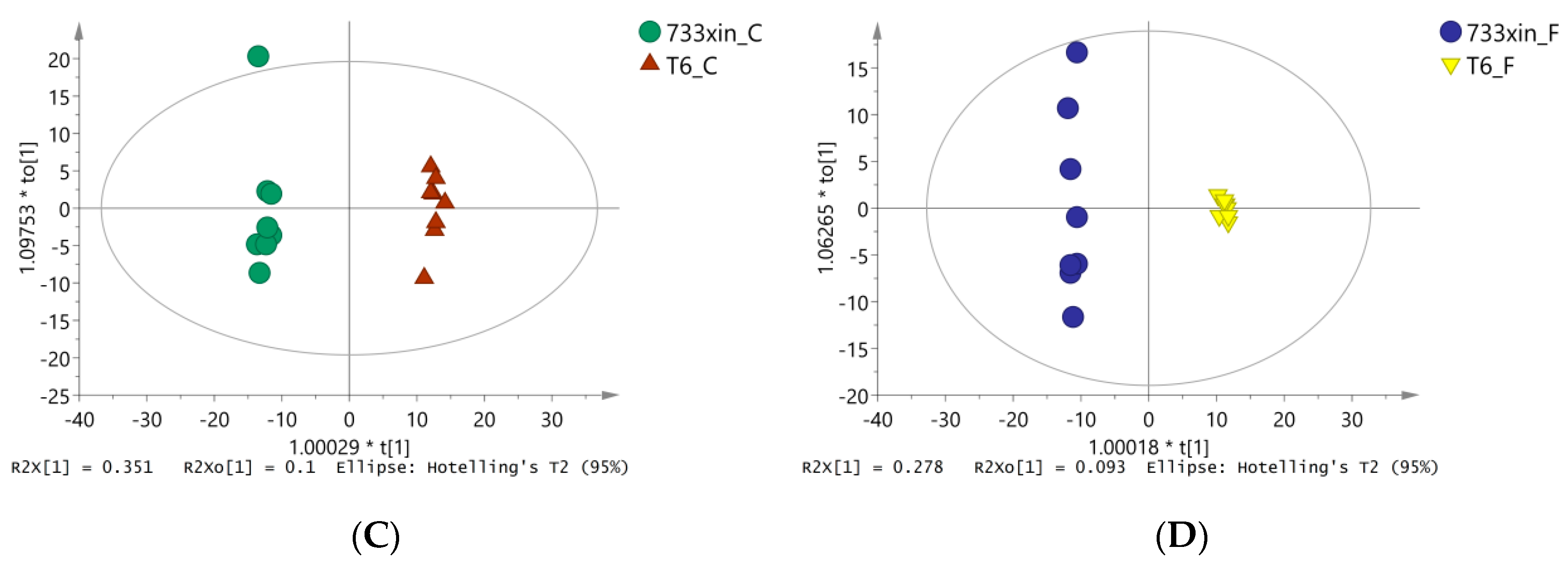
3.3. Orthogonal Partial Least Squares Discriminate (OPLS-DA) Analysis
3.4. Screening of Differential Metabolites
3.5. Metabolic Pathway Analysis of Differential Metabolites
4. Discussion
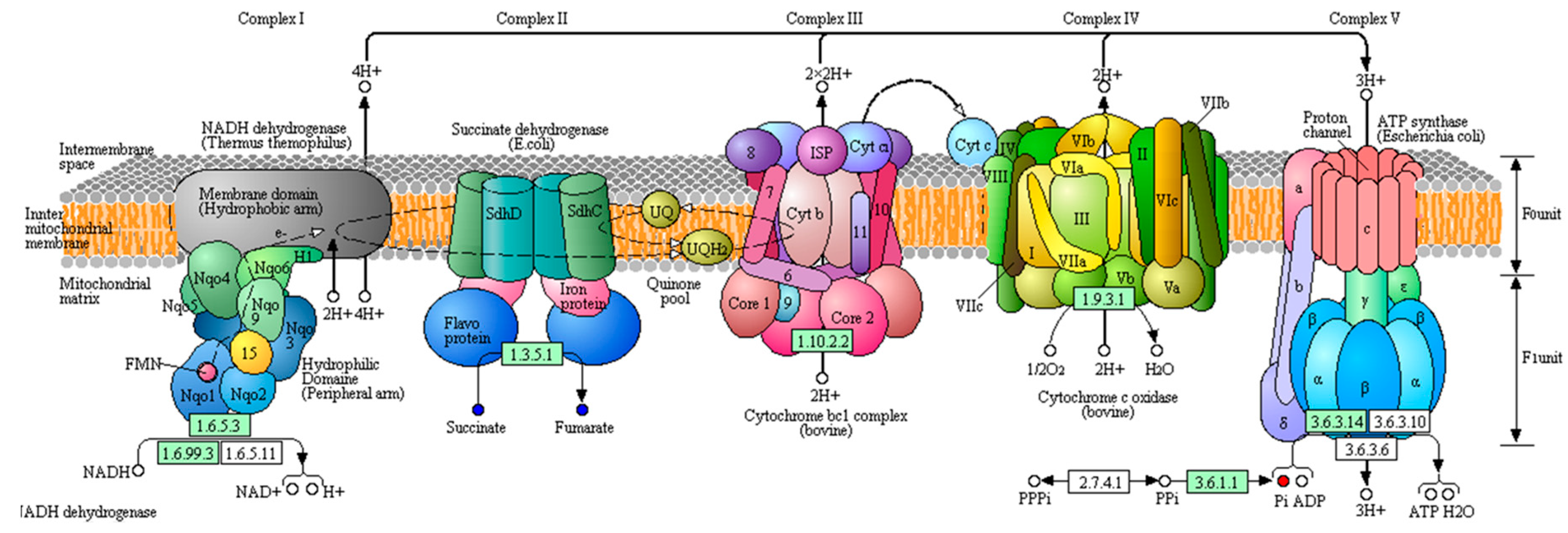
4.1. Energy Metabolism
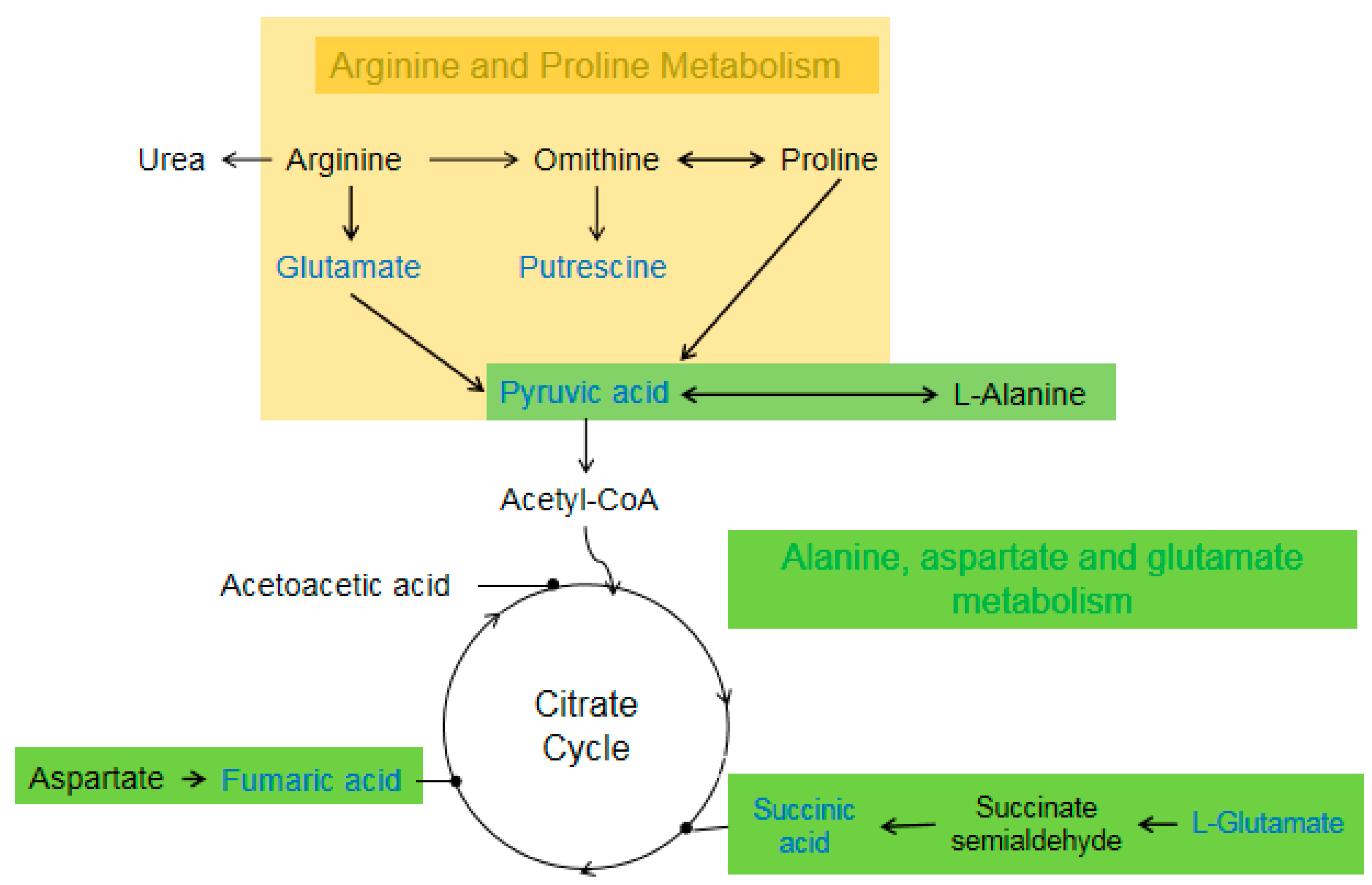
4.2. Amino Acid Metabolism
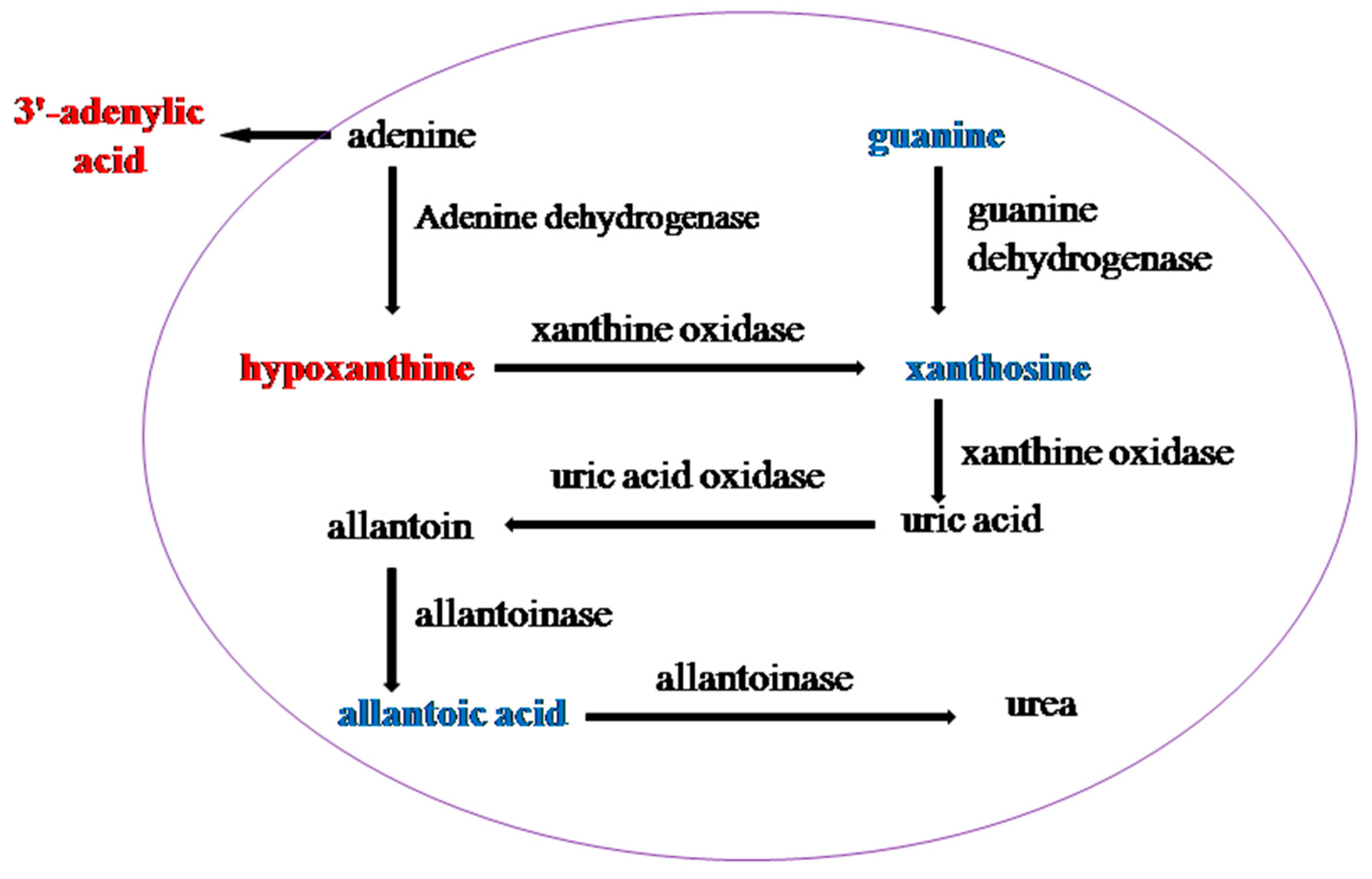
4.3. Nucleotide Metabolism
4.4. Other Metabolism
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chen, Y. Differences in fluoride effects on fecundity among varieties of the silkworm Bombyx mori. Fluoride 2003, 36, 163–169. [Google Scholar]
- Qian, H.; Li, G.; He, Q.; Zhang, H.; Xu, A. Analysis of differentially expressed genes between fluoride-sensitive and fluoride-endurable individuals in midgut of silkworm, Bombyx mori. Gene 2016, 588, 47–53. [Google Scholar] [CrossRef]
- Anuradha, C.D.; Kanno, S.; Hirano, S. Oxidative damage to mitochondria is a preliminary step to caspase-3 activation in fluoride-induced apoptosis in HL-60 cells. Free Radic. Boil. Med. 2001, 31, 367–373. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, A.; He, W.; He, P.; Xu, B.; Xia, T.; Chen, X.; Yang, K. Effects of fluoride on the expression of NCAM, oxidative stress, and apoptosis in primary cultured hippocampal neurons. Toxicology 2007, 236, 208–216. [Google Scholar] [CrossRef]
- Li, G.-N.; Xia, X.-J.; Tang, W.-C.; Zhu, Y. Intestinal microecology associated with fluoride resistance capability of the silkworm (Bombyx mori L.). Appl. Microbiol. Biotechnol. 2016, 100, 6715–6724. [Google Scholar] [CrossRef]
- Lin, C.Q.; Mi, Y.D.; Yao, Q.; Wu, D.X.; Wei, Z.J. The discovery of fluoride resistant dominant gene in silkworm. Sci. Seric. 1997, 23, 237–239. [Google Scholar]
- Chen, Y.; Du, X.; Hangzhou, Y.J. Cytochemical evidence for an anomalous dose-response of acid phosphatase activity in the blood but not the midgut of fluoride-treated silkworm larvae, Bombyx mori. Fluoride 2005, 38, 133–138. [Google Scholar]
- Zhou, H.; Chen, K.; Yao, Q.; Gao, L.; Wang, Y. Molecular cloning of Bombyx mori cytochrome P450 gene and its involvement in fluoride resistance. J. Hazard. Mater. 2008, 160, 330–336. [Google Scholar] [CrossRef]
- Chen, L.; Chen, H.; Yao, C.; Chang, C.; Xia, H.; Zhang, C.; Zhou, Y.; Yao, Q.; Chen, K. The toxicity of NaF on BmN cells and a comparative proteomics approach to identify protein expression changes in cells under NaF-stress: Impact of NaF on BmN cells. J. Hazard. Mater. 2015, 286, 624–631. [Google Scholar] [CrossRef]
- Tao, H.; Li, X.; Qiu, J.-F.; Cui, W.-Z.; Sima, Y.-H.; Xu, S.-Q. Inhibition of expression of the circadian clock gene Period causes metabolic abnormalities including repression of glycometabolism in Bombyx mori cells. Sci. Rep. 2017, 7, 46258. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Chen, Q.; Hou, Y.; Xia, Q.; Zhao, P. Metabolomics Analysis of the Larval Head of the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2016, 17, 1460. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, X.; Zhao, P.; Sun, Y.; Zhao, X.; Xiong, Y.; Xu, G.; Xia, Q. GC/MS-based metabolomic studies reveal key roles of glycine in regulating silk synthesis in silkworm, Bombyx mori. Insect Biochem. Mol. Boil. 2015, 57, 41–50. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Lei, H.; Hao, F.; Liu, X.; Wang, Y.; Tang, H. Simultaneous Quantification of Amino Metabolites in Multiple Metabolic Pathways Using Ultra-High Performance Liquid Chromatography with Tandem-mass Spectrometry. Sci. Rep. 2017, 7, 1423. [Google Scholar] [CrossRef]
- Tang, W.; Zheng, X.; Li, D.; Xiao, Y.; Yang, C.; Shang, S.; Shi, M.; Zhu, Y. Effects of sodium fluoride on the reproductive development of Bombyx mori. Environ. Toxicol. Pharmacol. 2018, 64, 41–47. [Google Scholar] [CrossRef]
- Tang, W.; Xiao, Y.; Li, G.; Zheng, X.; Yin, Y.; Wang, L.; Zhu, Y. Analysis of digital gene expression profiling in the gonad of male silkworms (Bombyx mori) under fluoride stress. Ecotoxicol. Environ. Saf. 2018, 153, 127–134. [Google Scholar] [CrossRef]
- Li, G.; Shi, M.; Zhao, S.; Li, D.; Long, Y.; Yang, C.; Zhu, Y. RNA-Seq comparative analysis reveals the response of Enterococcus faecalis TV4 under fluoride exposure. Gene 2019, 144197. [Google Scholar] [CrossRef]
- O’Hagan, S.; Dunn, W.B.; Broadhurst, D.; Ellis, D.I.; Brown, M.; Halsall, A.; Spasic, I.; Tseng, A.; Kell, D.B. A GC-TOF-MS study of the stability of serum and urine metabolomes during the UK Biobank sample collection and preparation protocols. Int. J. Epidemiol. 2008, 37, 23–30. [Google Scholar]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Boil. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Miao, Y.G.; Jiang, L.J.; Bharathi, D. Biochemical effects of fluoride on haemolymph of the silkworm, Bombyx mori L. Fluoride 2004, 37, 117–124. [Google Scholar]
- Chen, Z.W.; Jia, X.Y. Effects of fluoride on activities of phosphatase in hemolymph and midgut tissues of silkworm. Agro-Environ. Prot. 2002, 21, 130–133. [Google Scholar]
- Li, Z.; Wang, Y.; Wang, L.; Zhou, Z. Molecular and biochemical responses in the midgut of the silkworm, Bombyx mori, infected with Nosema bombycis. Parasites Vectors 2018, 11, 147. [Google Scholar] [CrossRef] [PubMed]
- Glavinas, H.; Krajcsi, P.; Cserepes, J.; Sarkadi, B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr. Drug Deliv. 2004, 1, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Cheng, T.; Wang, G.; Duan, J.; Niu, W.; Xia, Q. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Mol. Boil. Rep. 2012, 39, 7281–7291. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, C.; Sugimura, M.; Shinbo, H. Recycling of urea associated with the host plant urease in the silkworm larvae, Bombyx mori. J. Insect Physiol. 1999, 45, 15–20. [Google Scholar] [CrossRef]
- Tabunoki, H. Can the silkworm (Bombyx mori) be used as a human disease model? Int. Congr. Entomol. 2016, 10, 3–8. [Google Scholar]
- Liang, D.G.; Tu, Z.L.; Lu, S.L. Effects of glutathione on oxygen free radicals and malondialdehyde levels in haemolymph of fluorosis silkworm. China Seric. 2006, 27, 22–24. [Google Scholar]
- Nebert, D.W.; Vasiliou, V. Analysis of the glutathione S-transferase (GST) gene family. Hum. Genom. 2004, 1, 460–464. [Google Scholar] [CrossRef]
- Frova, C. Glutathione transferases in the genomics era: New insights and perspectives. Biomol. Eng. 2006, 23, 149–169. [Google Scholar] [CrossRef]
- Cheng, X.; Hu, J.; Li, J.; Chen, J.; Wang, H.; Mao, T.; Xue, B.; Li, B. The silk gland damage and the transcriptional response to detoxifying enzymes-related genes of Bombyx mori under phoxim exposure. Chemosphere 2018, 209, 964–971. [Google Scholar] [CrossRef]
- Tan, X.; Hu, X.-M.; Zhong, X.-W.; Chen, Q.-M.; Xia, Q.-Y.; Zhao, P. Antenna-Specific Glutathione S-Transferase in Male Silkmoth Bombyx mori. Int. J. Mol. Sci. 2014, 15, 7429–7443. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, X.; Weng, X.; Qian, H.; Xu, A. Glutathione S-transferase may participate in fluoride resistance in silkworm. Sci. Seric. 2018, 44, 85–93. [Google Scholar]
- Jobgen, W.S.; Fried, S.K.; Fu, W.J.; Meininger, C.J.; Wu, G. Regulatory role for the arginine–nitric oxide pathway in metabolism of energy substrates. J. Nutr. Biochem. 2006, 17, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Banno, Y. Identification of a novel function of the silkworm integument in nitrogen metabolism: Uric acid is synthesized within the epidermal cells in B. mori. Insect Biochem. Mol. Boil. 2019, 105, 43–50. [Google Scholar] [CrossRef] [PubMed]
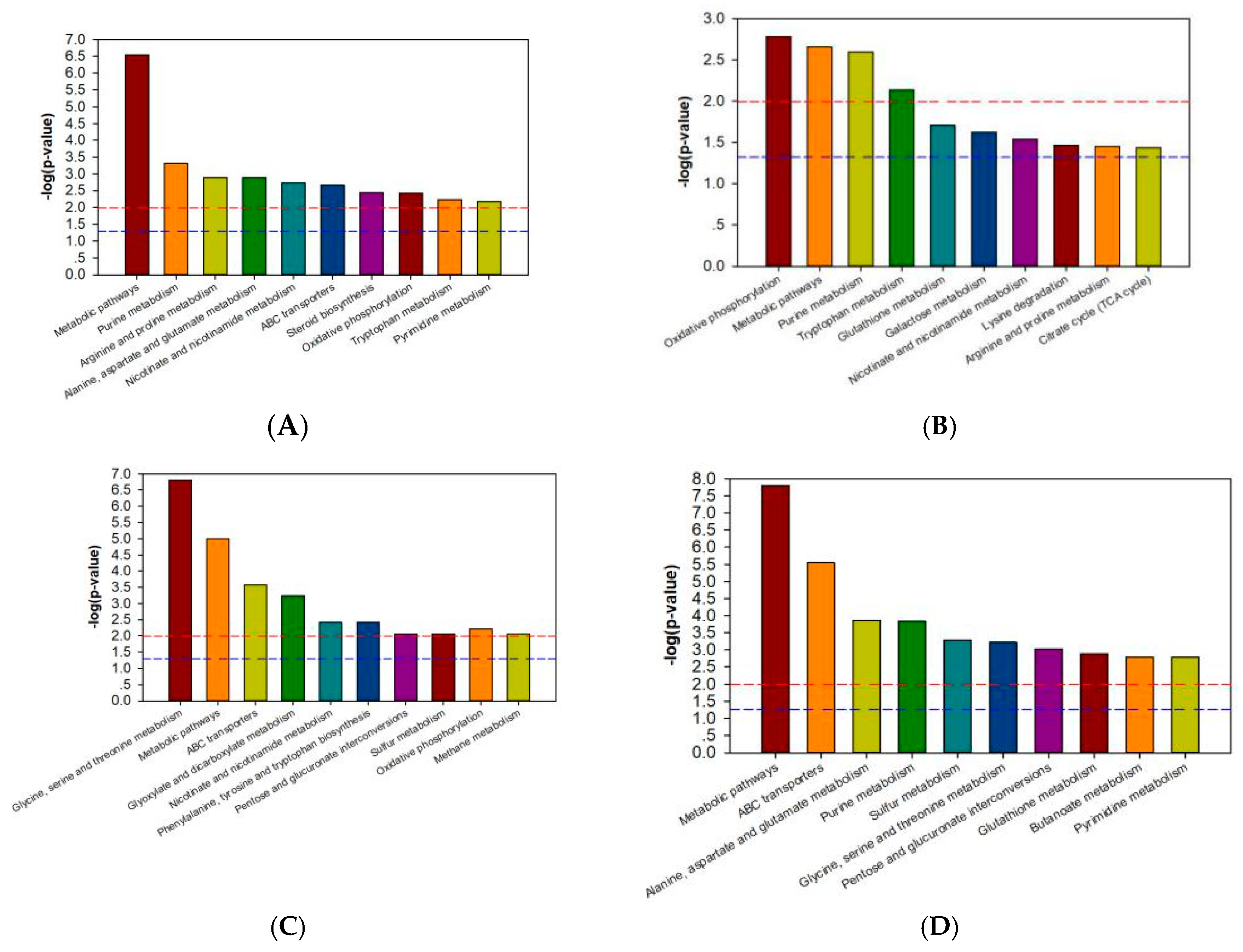
| Metabolite | 733xin-F vs. 733xin-C | T6-F vs. T6-C | T6-C vs. 733xin-C | T6-F vs. 733xin-F | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIP | p | FC | VIP | p | FC | VIP | p | FC | VIP | p | FC | |
| raffinose | 1.9730 | <0.0001 | 2.6702 | 1.1615 | 0.0270 | 0.7653 | 1.2283 | 0.0008 | 1.9180 | 1.6267 | <0.0001 | 0.5497 |
| tricetin | 1.9949 | <0.0001 | 0.4461 | 1.1013 | 0.0379 | 1.3371 | 1.2692 | 0.0008 | 0.6576 | 1.4342 | 0.0009 | 1.9712 |
| arachidic acid | 1.8459 | 0.0001 | 0.6691 | 1.3336 | 0.0054 | 1.1608 | 1.6063 | <0.0001 | 0.4817 | 1.1006 | 0.0224 | 0.8358 |
| androstanediol | 1.7021 | 0.0001 | 0.6277 | 1.3489 | 0.0016 | 1.3559 | 1.2781 | <0.0001 | 0.6977 | 1.2835 | 0.0010 | 1.5070 |
| prostaglandin E2 | 1.7541 | 0.0005 | 1.2606 | 1.1868 | 0.0145 | 0.7736 | 1.0471 | 0.0082 | 1.3228 | 1.3324 | 0.0018 | 0.8118 |
| N-methyl-DL-alanine | 1.6075 | 0.0008 | 1.5624 | 1.3262 | 0.0092 | 1.1077 | 1.2269 | 0.0002 | 1.6073 | 1.1278 | 0.0161 | 1.1396 |
| nicotinamide | 1.2384 | 0.0028 | 0.5651 | 1.0728 | 0.0489 | 1.0777 | 1.1909 | 0.0043 | 1.2455 | 1.3247 | <0.0001 | 2.3752 |
| indole-3-acetamide | 1.7442 | 0.0075 | 2.5556 | 1.7658 | 0.0001 | 0.5501 | 1.4274 | 0.0001 | 1.9831 | 1.3437 | 0.0099 | 0.4269 |
| xylose | 1.4202 | 0.0144 | 0.3171 | 1.5321 | 0.0013 | 0.4759 | 1.5303 | <0.0001 | 8.0269 | 1.7642 | <0.0001 | 12.0456 |
| phosphate | 1.3474 | 0.0149 | 2.0608 | 1.4143 | 0.0063 | 1.4409 | 1.3344 | 0.0002 | 2.1808 | 1.0803 | 0.0229 | 1.5248 |
| D-alanyl-D-alanine | 1.3563 | 0.0160 | 1.1455 | 1.3713 | 0.0093 | 1.0976 | 1.2162 | 0.0014 | 1.1518 | 1.0003 | 0.0402 | 1.1036 |
| N-methyl-L-glutamic acid | 1.4813 | 0.0164 | 2.0773 | 1.8072 | <0.0001 | 0.4503 | 1.3420 | 0.0007 | 1.7325 | 1.3931 | 0.0048 | 0.3756 |
| glucosaminic acid | 1.3853 | 0.0191 | 1.3027 | 1.6018 | 0.0004 | 0.7394 | 1.2018 | 0.0025 | 1.3776 | 1.1969 | 0.0084 | 0.7820 |
| guanine | 1.4256 | 0.0207 | 1.5516 | 1.6091 | 0.0010 | 0.6318 | 1.1917 | 0.0091 | 1.6934 | 1.4074 | 0.0014 | 0.6895 |
| 5-methoxyindole-3-acetic acid | 1.3218 | 0.0268 | 1.4926 | 1.0875 | 0.0289 | 1.4566 | 1.2071 | 0.0010 | 0.5024 | 1.4787 | 0.0011 | 0.4903 |
| stigmasterol | 1.2893 | 0.0412 | 0.6565 | 1.2993 | 0.0055 | 1.5196 | 1.2808 | 0.0015 | 1.9553 | 1.7558 | <0.0001 | 4.5257 |
| melatonin | 1.1751 | 0.0416 | 1.1508 | 1.3098 | 0.0166 | 1.3538 | 1.2652 | 0.0010 | 1.2440 | 1.3180 | 0.0061 | 1.4634 |
| methyl-beta-D-galactopyranoside | 1.1474 | 0.0426 | 1.7359 | 1.7925 | <0.0001 | 0.5967 | 1.0187 | <0.0001 | 8.9987 | 1.8350 | <0.0001 | 3.0935 |
| alpha-D-glucosamine 1-phosphate | 1.2684 | 0.0449 | 1.5270 | 1.8635 | <0.0001 | 0.3464 | 1.2883 | 0.0012 | 1.7887 | 1.5467 | 0.0013 | 0.4058 |
| 5-hydroxyindole-2-carboxylic acid | 1.8575 | 0.0041 | 0.3829 | 1.4027 | 0.0182 | 0.5151 | 1.3143 | 0.0051 | 0.3682 | 1.8083 | <0.0001 | 0.4954 |
| nicotinic acid | 1.4744 | 0.0064 | 1.3137 | 1.7465 | <0.0001 | 0.6829 | 1.1216 | 0.0043 | 0.7923 | 1.8168 | <0.0001 | 0.4119 |
| cytidine-monophosphate | 1.8111 | 0.0068 | 0.4009 | 1.3625 | 0.0285 | 0.4958 | 1.2371 | 0.0124 | 0.4089 | 1.8005 | <0.0001 | 0.5057 |
| Metabolic Pathway | Metabolite | 733xin-F vs. 733xin-C | T6-F vs. T6-C | ||||
|---|---|---|---|---|---|---|---|
| VIP | p | FC | VIP | p | FC | ||
| Purine metabolism | 3′-adenylic acid | 1.4021 | 0.0146 | 1.5897 | 1.1089 | 0.0260 | 1.2742 |
| guanine | 1.4256 | 0.0207 | 1.5516 | 1.6091 | 0.0010 | 0.6318 | |
| xanthosine | 1.0500 | 0.0224 | 0.6018 | 1.1667 | 0.0279 | 0.8304 | |
| guanosine | 1.5426 | 0.0010 | 0.6137 | ||||
| allantoic acid | 1.4377 | 0.0034 | 0.6627 | ||||
| adenosine | 1.4556 | 0.0038 | 0.7421 | ||||
| hypoxanthine | 1.4200 | 0.0048 | 1.1328 | ||||
| Arginine and proline metabolism | N-acetyl-L-glutamic acid | 1.6081 | 0.0167 | 1.8397 | 1.8881 | <0.0001 | 0.3537 |
| glutamic acid | 1.8040 | <0.0001 | 0.7483 | ||||
| putrescine | 1.5495 | 0.0007 | 0.8533 | ||||
| pyruvic acid | 1.3513 | 0.0167 | 0.5493 | ||||
| fumaric acid | 2.0018 | <0.0001 | 1.2941 | 1.018 | 0.0414 | 0.9117 | |
| Urea | 1.7167 | 0.0004 | 1.7151 | ||||
| Alanine, aspartate and glutamate metabolism | L-glutamic acid | 1.8040 | <0.0001 | 0.7483 | |||
| succinic acid | 1.4135 | 0.0076 | 0.6709 | ||||
| pyruvic acid | 1.3513 | 0.0167 | 0.5493 | ||||
| fumaric acid | 2.0018 | <0.0001 | 1.2941 | 1.018 | 0.0414 | 0.9117 | |
| Nicotinate and nicotinamide metabolism | nicotinic acid | 1.4744 | 0.0064 | 1.3137 | 1.7465 | <0.0001 | 0.6829 |
| pyruvic acid | 1.3513 | 0.0167 | 0.5493 | ||||
| fumaric acid | 2.0018 | <0.0001 | 1.2941 | 1.018 | 0.0414 | 0.9117 | |
| nicotinamide | 1.2384 | 0.0028 | 0.5651 | 1.0728 | 0.0489 | 1.0777 | |
| Oxidative phosphorylation | phosphate | 1.3474 | 0.0149 | 2.0608 | 1.4143 | 0.0063 | 1.4409 |
| succinic acid | 1.4135 | 0.0076 | 0.6709 | ||||
| fumaric acid | 2.0018 | <0.0001 | 1.2941 | 1.018 | 0.0414 | 0.9117 | |
| pyrophosphate | 1.5484 | 0.0054 | 1.6668 | ||||
| Tryptophan metabolism | indoleacetonitrile | 1.7692 | <0.0001 | 0.4340 | |||
| indole-3-acetamide | 1.7442 | 0.0075 | 2.5556 | 1.7658 | 0.0001 | 0.5501 | |
| aminophenol | 1.2944 | 0.0150 | 0.0892 | 1.0252 | 0.0025 | 0.4543 | |
| 5-methoxytryptamine | 1.3455 | 0.0130 | 0.7748 | ||||
| hydroxyanthranilic acid | 1.3539 | 0.0141 | <0.0001 | ||||
| melatonin | 1.1751 | 0.0416 | 1.1508 | 1.3098 | 0.0166 | 1.3538 | |
| indolelactate | 1.7985 | 0.0013 | 0.7070 | ||||
| N-acetylisatin | 1.2651 | 0.0152 | 1.4572 | ||||
| Glutathione metabolism | L-glutamic acid | 1.8040 | <0.0001 | 0.7483 | |||
| putrescine | 1.5495 | 0.0007 | 0.8533 | ||||
| L-cysteine | 1.4476 | 0.0172 | 0.8256 | ||||
| glycine | 1.3452 | 0.0253 | 0.8108 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Zhang, X.; Qian, H.; Liu, M.; Zhao, G.; Xu, A. Gas Chromatography-Mass Spectrometry Based Midgut Metabolomics Reveals the Metabolic Perturbations under NaF Stress in Bombyx mori. Insects 2020, 11, 17. https://doi.org/10.3390/insects11010017
Li G, Zhang X, Qian H, Liu M, Zhao G, Xu A. Gas Chromatography-Mass Spectrometry Based Midgut Metabolomics Reveals the Metabolic Perturbations under NaF Stress in Bombyx mori. Insects. 2020; 11(1):17. https://doi.org/10.3390/insects11010017
Chicago/Turabian StyleLi, Gang, Xiao Zhang, Heying Qian, Mingzhu Liu, Guodong Zhao, and Anying Xu. 2020. "Gas Chromatography-Mass Spectrometry Based Midgut Metabolomics Reveals the Metabolic Perturbations under NaF Stress in Bombyx mori" Insects 11, no. 1: 17. https://doi.org/10.3390/insects11010017
APA StyleLi, G., Zhang, X., Qian, H., Liu, M., Zhao, G., & Xu, A. (2020). Gas Chromatography-Mass Spectrometry Based Midgut Metabolomics Reveals the Metabolic Perturbations under NaF Stress in Bombyx mori. Insects, 11(1), 17. https://doi.org/10.3390/insects11010017