Isolation of a Pericentromeric Satellite DNA Family in Chnootriba argus (Henosepilachna argus) with an Unusual Short Repeat Unit (TTAAAA) for Beetles
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction of Genomic DNA and Isolation of Repetitive C0t-1 DNA Library
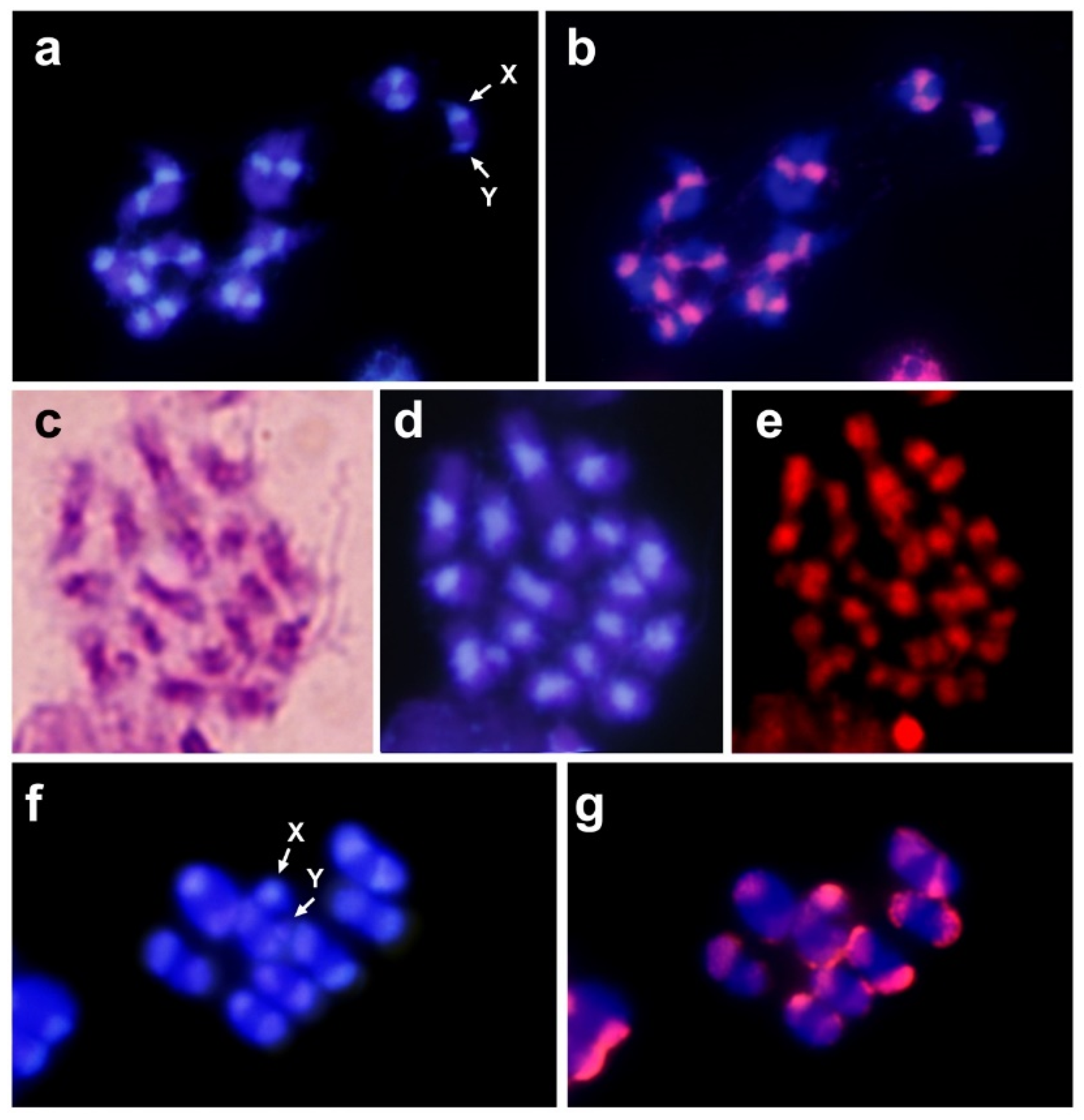
2.3. Chromosome Preparations, Probe Preparation and Fluorescence In Situ Hybridization
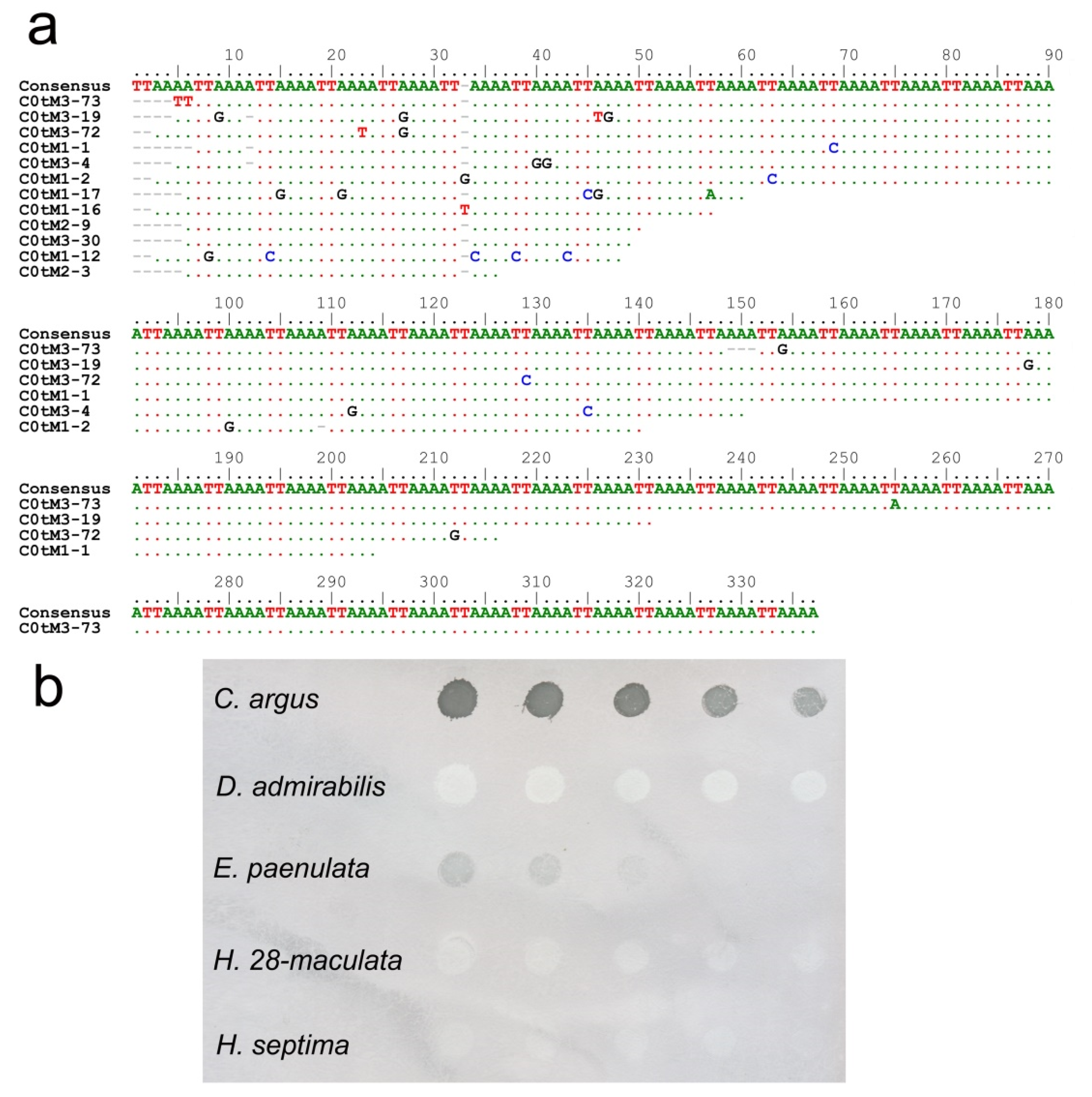
2.4. Dot-Blot Hybridization
2.5. Digestion with Restriction Endonucleases on Fixed Chromosomes
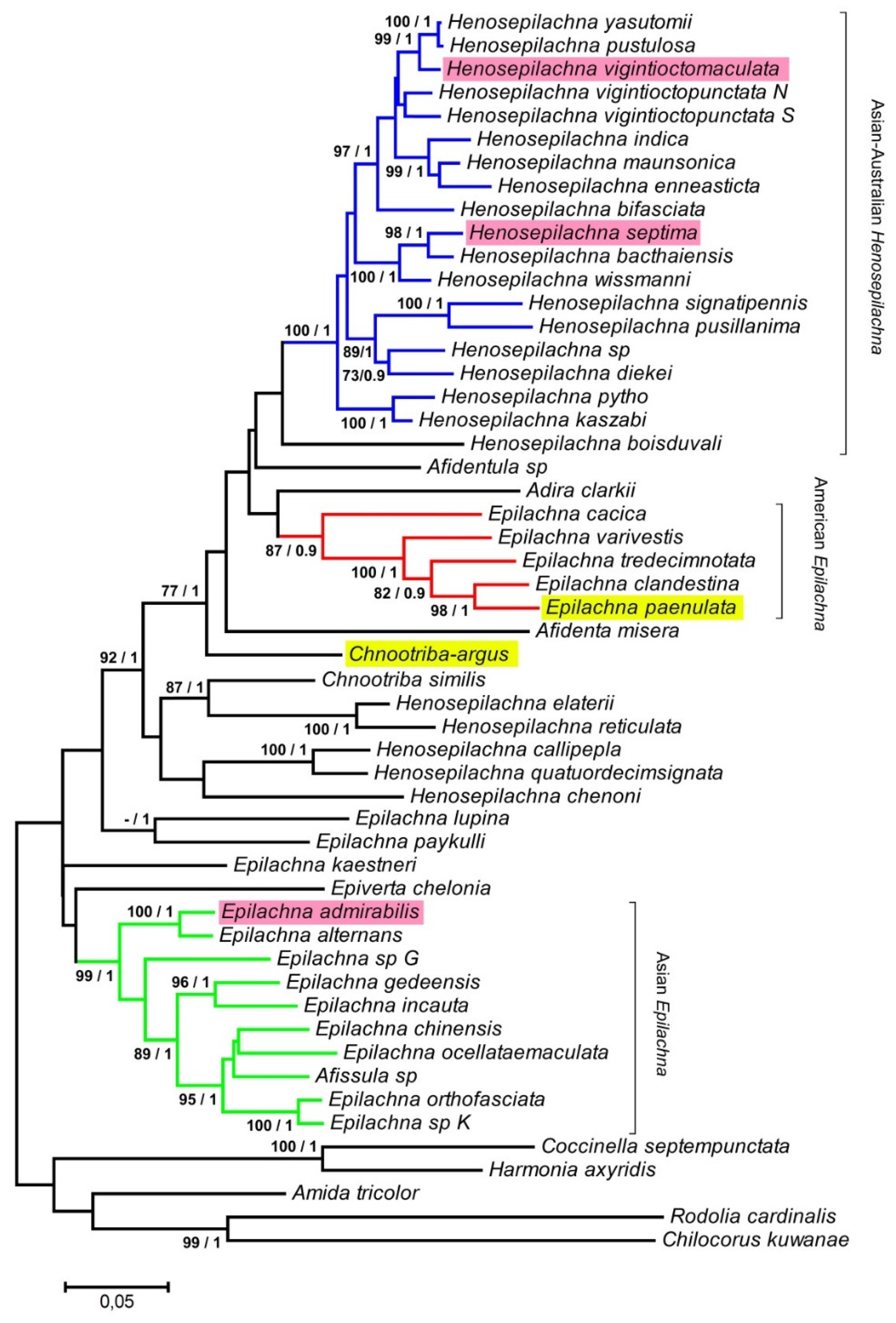
2.6. ND2 and 28S PCR Amplification and Phylogenetic Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Slipinski, A.; Tomaszewska, W. Revision of the family Cavognathidae (Coleoptera: Cucujoidea). Aust. J. Entomol. 2010, 49, 256–267. [Google Scholar] [CrossRef]
- Howard, N.F. Feeding of the Mexican bean beetle larva. Ann. Entomol. Soc. Am. 1941, 34, 766–769. [Google Scholar] [CrossRef]
- Katakura, H.; Nakano, S.; Kahono, S.; Abbas, I.; Noerdjito, W.A.; Nakamura, K. Epilachnine ladybird beetles (Coleoptera: Coccineliidae) of West Sumatra and West Java. In Evolutionary Biology and Population Dynamics of Herbivorous Lady Beetles in Indonesia; Nakamura, K., Katakura, H., Eds.; Published by editors: Sapporo, Japan, 1992; pp. 2–21. [Google Scholar]
- Das, R.; Mandal, S.K.; Maity, T.K. Insect pollinators of pointed gourd and effect of different artificial methods of pollination on fruit setting and subsequent development of fruits. Veg. Sci. 2009, 36, 353–355. [Google Scholar]
- Steiner, F.; Henikoff, S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 2014, 3, e02025. [Google Scholar] [CrossRef] [PubMed]
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Matsubayashi, K.W.; Ohshima, I. Genome size increase in the phytophagous ladybird beetle Henosepilachna vigintioctomaculata species complex (Coleoptera: Coccinellidae). Entomol. Sci. 2014, 18, 134–137. [Google Scholar] [CrossRef]
- Gregory, T.R. Animal Genome Size Database. Available online: http://www.genomesize.com (accessed on 8 August 2019).
- Blackmon, H.; Demuth, J.P. Coleoptera karyotype database. Coleopt. Bull. 2015, 69, 174–175. [Google Scholar] [CrossRef]
- Mora, P.; Vela, J.; Sanllorente, O.; Palomeque, T.; Lorite, P. Molecular cytogenetic studies in the ladybird beetle Henosepilachna argus Geoffroy, 1762 (Coleoptera, Coccinellidae, Epilachninae). Comp. Cytogenet. 2015, 9, 423–434. [Google Scholar] [CrossRef]
- Mora, P.; Vela, J.; Ruiz-Mena, A.; Palomeque, T.; Lorite, P. Characterization and transcriptional analysis of a subtelomeric satellite DNA family in the ladybird beetle Henosepilachna argus (Coleoptera: Coccinellidae). Eur. J. Entomol. 2017, 114, 481–487. [Google Scholar] [CrossRef][Green Version]
- Waring, M.; Britten, R. Nucleotide sequence repetition: A rapidly reassociating fraction of mouse DNA. Science 1966, 154, 791–794. [Google Scholar] [CrossRef] [PubMed]
- Zwick, M.S.; Hanson, R.E.; Islam-Faridi, M.N.; Stelly, D.M.; Wing, R.A.; Price, H.J.; McKnight, T.D. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; de Moura, R.C.; de Souza Melo, A.; Martins, C. Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 2011, 139, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Carvalho, C.R.; Ferrari Soares, F.A.; Cabral-de-Mello, D.C. Contrasting the chromosomal organization of repetitive DNAs in two Gryllidae crickets with highly divergent karyotypes. PLoS ONE 2015, 10, e0143540. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Koji, S.; Ishida, T.A.; Matsubahashi, K.W.; Kahono, S.; Kobayashi, N.; Furukawa, K.; Viet, B.T.; Vasconcellos-Neto, J.; Lange, C.N.; et al. Phylogeny of Epilachna, Henosepilachna, and some minor genera of phytophagous ladybird beetles (Coleoptera: Coccinellidae: Coccinellinae: Epilachnini), with an analysis of ancestral biogeography and host-plant utilization. Zool. Sci. 2014, 31, 820:830. [Google Scholar] [CrossRef] [PubMed]
- Szawaryn, K.; Bocak, L.; Ślipiński, A.; Escalona, H.E.; Tomaszewska, W. Phylogeny and evolution of phytophagous ladybird beetles (Coleoptera: Coccinellidae: Epilachnini), with recognition of new genera. Syst. Entomol. 2015, 40, 547–569. [Google Scholar] [CrossRef]
- Ando, T.A. Nuclease specific for heat-denatured DNA is isolated from a product of Aspergillus oryzae. Biochim. Biophys. Acta 1966, 114, 158–168. [Google Scholar] [CrossRef]
- Vogt, V. Purification and properties of S1 nuclease from Aspergillus. Methods Enzymol. 1980, 65, 248–255. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning a Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Lorite, P.; Chica, E.; Palomeque, T. G-banding and chromosome condensation in the ant Tapinoma nigerrimum. Chromosome Res. 1996, 4, 77–79. [Google Scholar] [CrossRef]
- Lorite, P.; Carrillo, J.A.; Palomeque, T. Conservation of (TTAGG)n telomeric sequences among ants (Hymenoptera, Formicidae). J. Hered. 2002, 93, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Palomeque, T.; Muñoz-Lopez, M.; Carrillo, J.A.; Lorite, P. Characterization and evolutionary dynamics of a complex family of satellite DNA in the leaf beetle Chrysolina carnifex (Coleoptera, Chrysomelidae). Chromosome Res. 2005, 13, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Lorite, P.; Palomeque, T.; Garnería, I.; Petitpierre, E. Characterization and chromosome location of satellite DNA in the leaf beetle Chrysolina americana (Coleoptera, Chrysomelidae). Genetica 2001, 110, 143–150. [Google Scholar] [CrossRef]
- Lorite, P.; Garcia, M.F.; Carrillo, J.A.; Palomeque, T. Restriction endonuclease chromosome banding in Tapinoma nigerrimum (Hymenoptera, Formicidae). Hereditas 1999, 131, 197–201. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.; Frati, F.; Stewart, J.; Beckenbach, A. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Hasegawa, E.; Kasuya, E. Phylogenetic analysis of the insect order Odonata using 28S and 16S rDNA sequences: A comparison between data sets with different evolutionary rates. Entomol. Sci. 2006, 9, 55–66. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 1, 1572–1574. [Google Scholar] [CrossRef]
- Tomaszewska, W.; Szawaryn, K. Epilachnini (Coleoptera: Coccinellidae)—A revision of the World Genera. J. Insect Sci. 2016, 16, 1–91. [Google Scholar] [CrossRef]
- Smith, S.G. Chromosome numbers of Coleoptera. Heredity 1953, 7, 31–48. [Google Scholar] [CrossRef]
- Camacho, J.P.; Ruiz-Ruano, F.J.; Martín-Blázquez, R.; López-León, M.D.; Cabrero, J.; Lorite, P.; Cabral-de-Mello, D.C.; Bakkali, M. A step to the gigantic genome of the desert locust: Chromosome sizes and repeated DNAs. Chromosoma 2015, 124, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Vicari, M.R.; Nogaroto, V.; Noleto, R.B.; Cestari, M.M.; Cioffi, M.B.; Almeida, M.C.; Moreira-Filho, O.; Bertollo, L.A.; Artoni, R.F. Satellite DNA and chromosomes in Neotropical fishes: Methods, applications and perspectives. J. Fish Biol. 2010, 76, 1094–1116. [Google Scholar] [CrossRef] [PubMed]
- Ugarković, D.; Petitpierre, E.; Juan, C.; Plohl, M. Satellite DNAs in Tenebrionid species: Structure, organization and evolution. Croat. Chem. Acta 1995, 68, 627–638. [Google Scholar]
- Palomeque, T.; Lorite, P. Satellite DNA in insects: A review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Petitpierre, E.; Juan, C. Characterization of the heterochromatin of the darkling beetle Misolampus goudoti: Cloning of two satellite DNA families and digestion of chromosomes with restriction enzymes. Hereditas 1993, 119, 179–185. [Google Scholar] [CrossRef]
- Mravinac, B.; Ugarkovic, E.; Franjevic, D.; Plohl, M. Long inversely oriented subunits form a complex monomer of Tribolium brevicornis satellite DNA. J. Mol. Evol. 2005, 60, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Lorite, P.; Torres, M.I.; Palomeque, T. Characterization of two unrelated satellite DNA families in the Colorado potato beetle Leptinotarsa decemlineata (Coleoptera, Chrysomelidae). Bull. Entomol. Res. 2013, 103, 538–546. [Google Scholar] [CrossRef]
- Gall, J.G.; Atherton, D.D. Satellite DNA sequences in Drosophila virilis. J. Mol. Biol. 1974, 85, 633–664. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Transcribing centromeres: Noncoding RNAs and kinetochore assembly. Trends Genet. 2018, 34, 587–599. [Google Scholar] [CrossRef]
- Burgtorf, C.; Bünemann, H.A. telomere-like satellite (GGGTCAT)n comprises 4% of genomic DNA of Drosophila hydei and is located mainly in centromeric heterochromatin of all large acrocentric autosomes. Gene 1993, 137, 287–291. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Mora, P.; Vela, J.; Cuadrado, A.; Sánchez, A.; Palomeque, T.; Lorite, P. Comparative repeatome analysis on Triatoma infestans Andean and Non-Andean lineages, main vector of Chagas disease. PLoS ONE 2017, 12, e0181635. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Mora, P.; Vela, J.; Palomeque, T.; Sánchez, A.; Panzera, F.; Lorite, P. Comparative analysis of repetitive DNA between the main vectors of Chagas disease: Triatoma infestans and Rhodnius prolixus. Int. J. Mol. Sci. 2018, 24, 1277. [Google Scholar] [CrossRef] [PubMed]
- Heckmann, S.; Macas, J.; Kumke, K.; Fuchs, J.; Schubert, V.; Ma, L.; Novak, P.; Neumann, P.; Taudien, S.; Platzer, M.; et al. The holocentric species Luzula elegans shows an interplay between centromere and large-scale genome organization. Plant J. 2013, 73, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Pavlek, M.; Gelfand, Y.; Plohl, M.; Meštrović, N. Genome wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res. 2015, 22, 387–401. [Google Scholar] [CrossRef]
- Palacios-Gimenez, O.M.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.E.S.; Ramos, E.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; Castillo-Martínez, J.; Cabrero, J.; Gómez, R.; Camacho, J.P.M.; López-León, M.D. High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 2018, 127, 323–340. [Google Scholar] [CrossRef]
- Leitão, A.; Raquel, C.; Santos, S.; Guedes-Pinto, H.; Boudry, P. Interspecific hybridization in oysters: Restriction enzyme digestion chromosome banding confirms Crassostrea angulata X Crassostrea gigas F1 hybrids. J. Exp. Mar. Biol. Ecol. 2007, 343, 253–260. [Google Scholar] [CrossRef][Green Version]
- Bitencourt, J.A.; Affonso, P.R.; Giuliano-Caetano, L.; Dias, A.L. Heterochromatin heterogeneity in Hypostomus prope unae (Steindachner, 1878) (Siluriformes, Loricariidae) from Northeastern Brazil. Comp. Cytogen. 2011, 5, 329–344. [Google Scholar] [CrossRef]
- Fernandes, A.; Werneck, H.A.; Pompolo, S.G.; Lopes, D.M. Evidence of separate karyotype evolutionary pathway in Euglossa orchid bees by cytogenetic analyses. An. Acad. Bras. Ciênc. 2013, 85, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Nieddu, M.; Pichiri, G.; Melis, V.; Mezzanotte, R. The impact of StuI digestion in situ on FISH to human chromosomes with satellite DNA probes. Heredity 2003, 90, 298–301. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Juan, C.; Gosálvez, J.; Mezzanotte, R.; Petitpierre, E. Cytological and biochemical characterization of the in situ endonuclease digestion of fixed Tenebrio molitor chromosomes. Chromosoma 1991, 100, 432–438. [Google Scholar] [CrossRef]
- Ugarković, D.; Plohl, M.; Petitpierre, E.; Lucijanić-Justić, V.; Juan, C. Tenebrio obscurus satellite DNA is resistant to cleavage by restriction endonucleases in situ. Chromosome Res. 1994, 2, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Drets, M.E.; Corbella, E.; Panzera, F.; Folle, G.A. C-banding and non-homologous association II. The “parachute” Xyp sex bivalent and the behaviour of heterochromatic segments in Epilachna paenulata. Chromosoma 1983, 88, 249–255. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora, P.; Vela, J.; Ruiz-Mena, A.; Palomeque, T.; Lorite, P. Isolation of a Pericentromeric Satellite DNA Family in Chnootriba argus (Henosepilachna argus) with an Unusual Short Repeat Unit (TTAAAA) for Beetles. Insects 2019, 10, 306. https://doi.org/10.3390/insects10090306
Mora P, Vela J, Ruiz-Mena A, Palomeque T, Lorite P. Isolation of a Pericentromeric Satellite DNA Family in Chnootriba argus (Henosepilachna argus) with an Unusual Short Repeat Unit (TTAAAA) for Beetles. Insects. 2019; 10(9):306. https://doi.org/10.3390/insects10090306
Chicago/Turabian StyleMora, Pablo, Jesús Vela, Areli Ruiz-Mena, Teresa Palomeque, and Pedro Lorite. 2019. "Isolation of a Pericentromeric Satellite DNA Family in Chnootriba argus (Henosepilachna argus) with an Unusual Short Repeat Unit (TTAAAA) for Beetles" Insects 10, no. 9: 306. https://doi.org/10.3390/insects10090306
APA StyleMora, P., Vela, J., Ruiz-Mena, A., Palomeque, T., & Lorite, P. (2019). Isolation of a Pericentromeric Satellite DNA Family in Chnootriba argus (Henosepilachna argus) with an Unusual Short Repeat Unit (TTAAAA) for Beetles. Insects, 10(9), 306. https://doi.org/10.3390/insects10090306





