CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori
Abstract
1. Introduction
2. Materials and Methods
2.1. Silkworm Strain
2.2. Quantitative Real-Time PCR (qRT-PCR)
2.3. Plasmid Construction
2.4. Germline Transformation
2.5. Mutagenesis Analysis
2.6. Statistical Analysis
3. Results
3.1. BmOvo-1 Is Highly Expressed in Wing Disc and Epidermis
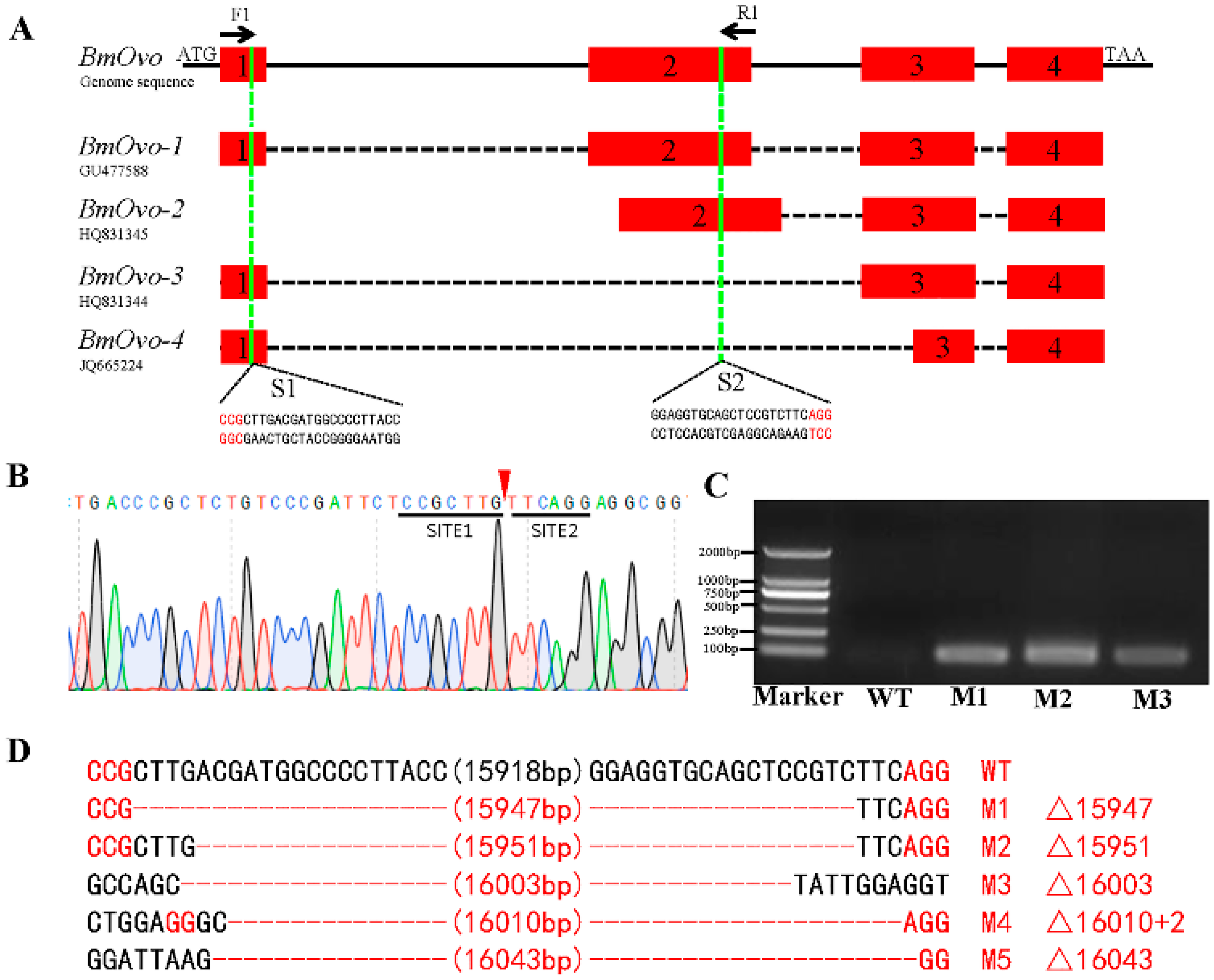
3.2. CRISPR/Cas9-Mediated Mutagenesis of BmOvo Gene
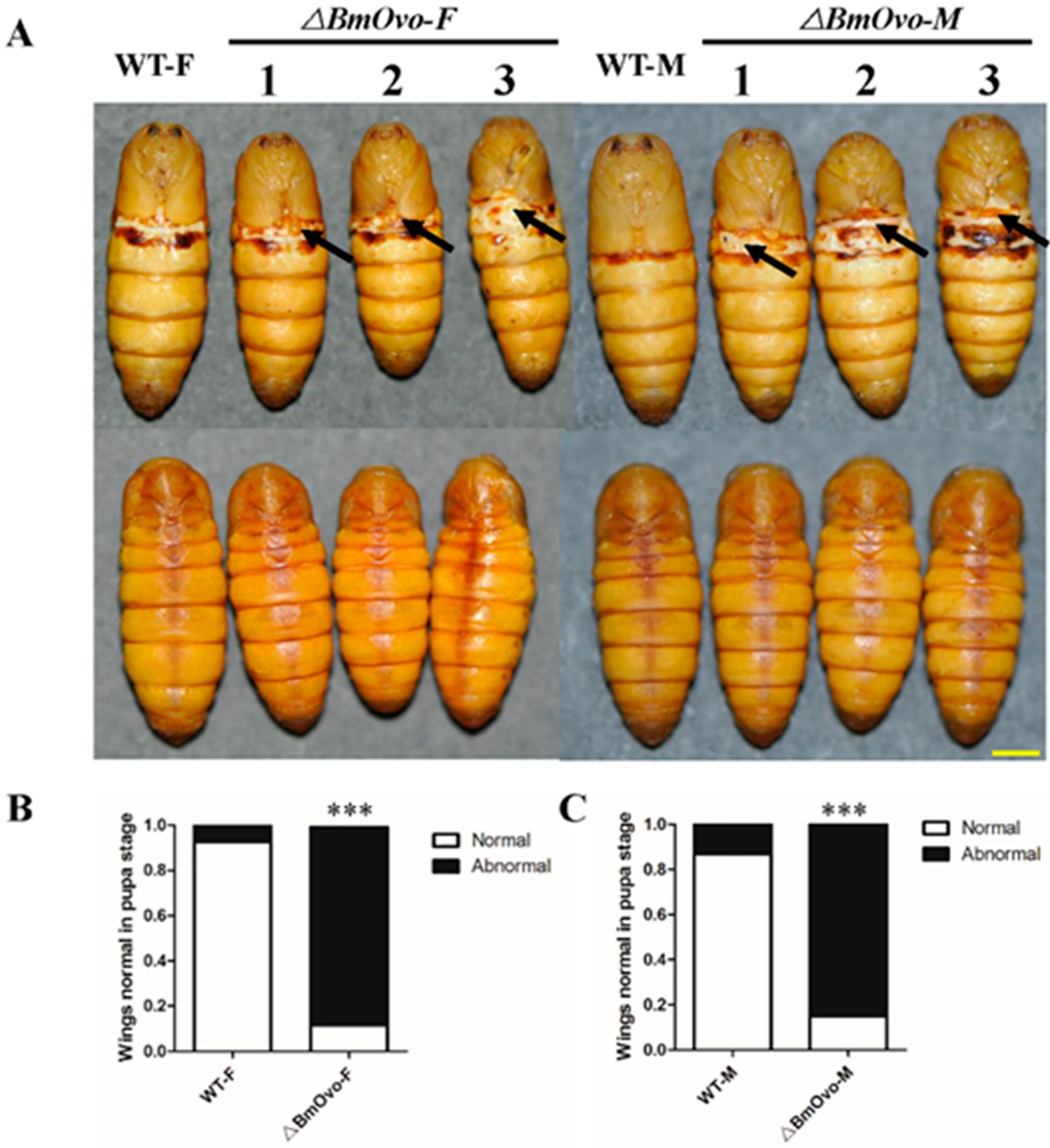
3.3. Loss of BmOvo Results in Abnormal Development of Wings and Some Other Organs
3.4. BmOvo Influences Expression of Genes Involved in Wing Development and Metamorphosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Piloto, S.; Schilling, T.F. Ovo1 links Wnt signaling with N-cadherin localization during neural crest migration. Development 2010, 137, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Lapan, S.W.; Reddien, P.W. Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. Cell Rep. 2012, 2, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Schonbaum, C.; Degenstein, L.; Bai, W.; Mahowald, A.; Fuchs, E. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 1998, 12, 3452–3463. [Google Scholar] [CrossRef] [PubMed]
- Payre, F.; Vincent, A.; Carreno, S. Ovo/Svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 1999, 400, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Delon, I.; Chanutdelalande, H.; Payre, F. The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech. Dev. 2003, 120, 747–758. [Google Scholar] [CrossRef]
- Mevelninio, M.; Terracol, R.; Kafatos, F.C. The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 1991, 10, 2259–2266. [Google Scholar] [CrossRef]
- Zhu, M.; Hu, X.; Liang, Z.; Jiang, M.; Xue, R.; Gong, Y. Functional characterization of BmOVOs in silkworm, Bombyx mori. BMC Genom. 2019, 20, 342. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, M.; Wang, J.; Liang, Y.; Mahowald, A. Multiple products from the shavenbaby-OVO gene region of Drosophila melanogaster: Relationship to genetic complexity. Mol. Cell Biol. 1994, 14, 6809–6818. [Google Scholar] [CrossRef] [PubMed]
- Bielinska, B.; Lu, J.; Sturgill, D.; Oliver, B. Core promoter sequences contribute to OVO-B regulation in the Drosophila melanogaster germline. Genetics 2005, 169, 161–172. [Google Scholar] [CrossRef]
- Casper, A.; van Doren, M. The control of sexual identity in the Drosophila germline. Development 2006, 133, 2783–2791. [Google Scholar] [CrossRef]
- Oliver, B.; Singer, J.; Laget, V.; Pennetta, G.; Pauli, D. Function of Drosophila ovo+ in germ-line sex determination depends on X-chromosome number. Development 1994, 120, 3185–3195. [Google Scholar] [PubMed]
- Matova, N.; Cooley, L. Comparative Aspects of Animal Oogenesis. Dev. Biol. 2001, 231, 291–320. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.; Garcia-Estefania, D.; Delon, I.; Lü, J.; Mével-Ninio, M.; Spierer, A.; Payre, F.; Pauli, D.; Oliver, B. OVO transcription factors function antagonistically in the Drosophila female germline. Development 2000, 127, 881–892. [Google Scholar] [PubMed]
- Andrews, J.; Oliver, B. Sex determination signals control OVO-B transcription in Drosophila melanogaster germ cells. Genetics 2002, 160, 537–545. [Google Scholar] [PubMed]
- Nagoshi, R.; Patton, S.; Bae, E.; Geyer, P. The somatic sex determines the requirement for ovarian tumor gene activity in the proliferation of the Drosophila germline. Development 1995, 121, 579–587. [Google Scholar] [PubMed]
- Salles, C.; Mevelninio, M.; Vincent, A.; Payre, F. A Germline-Specific Splicing Generates an Extended Ovo Protein Isoform Required for Drosophila Oogenesis. Dev. Biol. 2002, 246, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Thibert, C.; Royer, C.; Kanda, T.; Abraham, E.; Kamba, M.; Komoto, N.; Thomas, J.L.; Mauchamp, B.; Chavancy, G.; et al. Germline transformation of the silkworm Bombyx mori L. using a piggyback transposon-derived vector. Nat. Biotechnol. 2000, 18, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Tanaka, H.; Tamura, T.; Shiotsuki, T. Precocious metamorphosis in transgenic silkworms overexpressing juvenile hormone esterase. Proc. Natl. Acad. Sci. USA 2005, 102, 11751–11756. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Xu, J.; Zeng, B.; Ling, L.; You, L.; Chen, Y.; Huang, Y.; Tan, A. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013, 23, 1414–1416. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.; Li, Z.; Ling, L.; Zeng, B.; James, A.A.; Tan, A.; Huang, Y. Transcription activator-like effector nuclease (TALEN)-mediated female-specific sterility in the silkworm, Bombyx mori. Insect Mol. Boil. 2014, 23, 800–807. [Google Scholar] [CrossRef]
- Ma, S.; Chang, J.; Wang, X.; Liu, Y.; Zhang, J.; Lu, W.; Gao, J.; Shi, R.; Zhao, P.; Xia, Q. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci. Rep. 2014, 4, 4489. [Google Scholar] [CrossRef] [PubMed]
- Daimon, T.; Uchibori, M.; Nakao, H.; Sezutsu, H.; Shinoda, T. Knockout silkworms reveal a dispensable role for juvenile hormones in holometabolous life cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 4226–4235. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Dong, F.; Yu, X.; Huang, L.; Jiang, Y.; Hu, Z.; Chen, P.; Lu, C.; Pan, M. Excision of Nucleopolyhedrovirus Form Transgenic Silkworm Using the CRISPR/Cas9 System. Front. Microbiol. 2018, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Sumitani, M.; Chikami, Y.; Yahata, K.; Uchino, K.; Kiuchi, T.; Suzuki, M.G. Transgenic Expression of the piRNA-Resistant Masculinizer Gene Induces Female-Specific Lethality and Partial Female-to-Male Sex Reversal in the Silkworm, Bombyx mori. PLoS Genet. 2016, 12, e1006203. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Ge, X.; Li, Z.; Zeng, B.; Xu, J.; Chen, X.; Tan, A. MiR-2 family targets awd and fng to regulate wing morphogenesis in Bombyx mori. RNA Biol. 2015, 12, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Aslam, A.F.; Liu, X.; Li, M.; Huang, Y.; Tan, A. Functional analysis of bombyx wnt1 during embryogenesis using the CRISPR/Cas9 system. J. Insect Physiol. 2015, 79, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, W.; Zeng, B.; Wang, G.; Hao, D.; Huang, Y. Deletion of the Bombyx mori odorant receptor co-receptor (BmOrco) impairs olfactory sensitivity in silkworms. Insect Biochem. Mol. Biol. 2017, 86, 58. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, S.; Zeng, B.; James, A.; Tan, A.; Huang, Y. Bombyx mori p-element somatic inhibitor (BmPSI) is a key auxiliary factor for silkworm male sex determination. PLoS Genet. 2017, 13, e1006576. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Huang, Y.; Xu, J.; Shiotsuki, T.; Bai, H.; Palli, S.R.; Huang, Y.; Tan, A. The FOXO transcription factor controls insect growth and development by regulating juvenile hormone degradation in the silkworm, Bombyx mori. J. Boil. Chem. 2017, 292, 11659–11669. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hou, C.; Bi, H.; Wang, Y.; Xu, J.; Li, M.; James, A.A.; Huang, Y.; Tan, A. Transgenic Clustered Regularly Interspaced Short Palindromic Repeat/Cas9-Mediated Viral Gene Targeting for Antiviral Therapy of Bombyx mori Nucleopolyhedrovirus. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Xue, R.; Hu, X.; Cao, G.; Huang, M.; Xue, G.; Qian, Y.; Gong, C. BmOvo-1 regulates ovary size in the silkworm, Bombyx mori. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.; Joung, J. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Hino, K.; Bono, H.; UiTei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, R.; Chen, S.; Chen, K.; Tang, L.; Yang, D.; Yang, X.; Zhang, Y.; Song, H.; Huang, Y. Identification of a germline-expression promoter for genome editing in Bombyx mori. Insect Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Fujita, K.; Sagisaka, A.; Tomimoto, K.; Imanishi, S.; Yamakawa, M. shRNA Expression Plasmids Generated by a Novel Method Efficiently Induce Gene-Specific Knockdown in a Silkworm Cell Line. Mol. Biotechnol. 2009, 41, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Xu, J.; Tan, A.; Huang, Y. CRISPR/Cas9-mediated targeted gene mutagenesis in Spodoptera litura. Insect Sci. 2016, 23, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; You, L.; Yan, D.; James, A.; Huang, Y.; Tan, A. Bombyx mori histone methyltransferase BmAsh2 is essential for silkworm piRNA-mediated sex determination. PLoS Genet. 2018, 14, e1007245. [Google Scholar] [CrossRef] [PubMed]
- Trautwein, M.; Wiegmann, B.M.; Beutel, R.G.; Kjer, K.M.; Yeates, D.K. Advances in Insect Phylogeny at the Dawn of the Post genomic Era. Annu. Rev. Entomol. 2012, 57, 449–468. [Google Scholar] [CrossRef]
- Shimmi, O.; Matsuda, S.; Hatakeyama, M. Insights into the molecular mechanisms underlying diversified wing venation among insects. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140264. [Google Scholar] [CrossRef]
- Ewing, A.W.; Bennetclark, H.C. The Courtship Songs of Drosophila. Behaviour 1968, 31, 288–301. [Google Scholar] [CrossRef]
- Ding, Y.; Berrocal, A.; Morita, T.; Longden, K.D.; Stern, D.L. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 2016, 536, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Brakefield, P.M.; French, V. Butterfly wings: The evolution of development of colour patterns. BioEssays 1999, 21, 391–401. [Google Scholar] [CrossRef]
- Nijhout, H.F. The Development and Evolution of Butterfly Wing Patterns; Smithsonian Institution Scholarly Press: Washington, DC, USA, 1991. [Google Scholar]
- Li, X.; Fan, D.; Zhang, W.; Liu, G.S.; Zhang, L.; Zhao, L.; Wang, W. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 2015, 6, 8212. [Google Scholar] [CrossRef] [PubMed]
- Beldade, P.; Brakefield, P.M. The genetics and evo-devo of butterfly wing patterns. Nat. Rev. Genet. 2002, 3, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Mcmillan, W.O.; Monteiro, A.; Kapan, D.D. Development and evolution on the wing. Trends Ecol. Evol. 2002, 17, 125–133. [Google Scholar] [CrossRef]
- Gäde, G.; Hoffmann, K.H.; Spring, J.H. Hormonal regulation in insects: Facts, gaps, and future directions. Physiol. Rev. 1997, 77, 963–1032. [Google Scholar] [CrossRef]
- Jindra, M.; Palli, S.R.; Riddiford, L.M. The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 2013, 58, 181–204. [Google Scholar] [CrossRef] [PubMed]
- Zera, A.J. Juvenile Hormone and the endocrine regulation of wing polymorphism in insects: New insights from circadian and functional genomic studies in Gryllus crickets. Physiol. Entomol. 2016, 41, 313–326. [Google Scholar] [CrossRef]
- Ou, J.; Deng, H.; Zheng, S.; Huang, L.; Feng, Q.; Liu, L. Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC Genom. 2014, 15, 820. [Google Scholar] [CrossRef]
- Klein, T. Wing disc development in the fly: The early stages. Curr. Opin. Genet. Dev. 2001, 11, 470–475. [Google Scholar] [CrossRef]
- Truman, J.; Riddiford, L. Endocrine Insights Into The Evolution Of Metamorphosis In Insects. Annu. Rev. Entomol. 2002, 47, 467–500. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Zhang, J.; Li, Y.; Zheng, S.C.; Liu, L.; Huang, L.; Feng, Q. Homeodomain POU and Abd-A proteins regulate the transcription of pupal genes during metamorphosis of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2012, 109, 12598–12603. [Google Scholar] [CrossRef] [PubMed]
- Rebers, J.; Riddiford, L. Structure and expression of a manduca sexta larval cuticle gene homologous to drosophila cuticle genes. J. Mol. Biol. 1988, 203, 411–423. [Google Scholar] [CrossRef]
- Iconomidou, V.; Willis, J.; Hamodrakas, S. Unique features of the structural model of ‘hard’ cuticle proteins: Implications for chitin-protein interactions and cross-linking in cuticle. Insect Biochem. Mol. Biol. 2005, 35, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Mita, K.; Quan, G.-X.; Shimada, T.; Okano, K.; Kanke, E.; Kawasaki, H. Mass isolation of cuticle protein cDNAs from wing discs of Bombyx mori and their characterizations. Insect Biochem. Mol. Biol. 2001, 31, 1019–1028. [Google Scholar] [CrossRef]
- Noji, T.; Ote, M.; Takeda, M.; Mita, K.; Shimada, T.; Kawasaki, H. Isolation and comparison of different ecdysone-responsive cuticle protein genes in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 2003, 33, 671–679. [Google Scholar] [CrossRef]
- Deng, H.; Zheng, S.; Yang, X.; Liu, L.; Feng, Q. Transcription factors BmPOUM2 and BmβFTZ-F1 are involved in regulation of the expression of the wing cuticle protein gene BmWCP4 in the silkworm, Bombyx mori. Insect Mol. Biol. 2011, 20, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Li, Y.; Zhang, J.; Liu, L.; Feng, Q. Analysis of expression and chitin-binding activity of the wing disc cuticle protein BmWCP4 in the silkworm, Bombyx mori. Insect Sci. 2016, 23, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Mevelninio, M.; Terracol, R.; Salles, C.; Vincent, A.; Payre, F. OVO, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech. Dev. 1995, 49, 83–95. [Google Scholar] [CrossRef]
- Nakao, H. A Bombyx homolog of ovo is a segmentation gene that acts downstream of bm-wnt1 (bombyx wnt1 homolog). Gene Expr. Patterns 2018, 27, 1–7. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Shiotsuki, T.; Wang, Z.; Xu, X.; Huang, Y.; Li, M.; Li, K.; Tan, A. Depletion of juvenile hormone esterase extends larval growth in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 72–79. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence (5′ to 3′) | Primer Purpose |
|---|---|---|
| Ovo-sgRNA1-F | TATCGTGCTCTACAAGTGGTAAGGGGCCATCGTCAAG GTTTTAGAGCTAGAAATAG | Preparation of sgRNA template |
| Ovo-sgRNA2-F | TATCGTGCTCTACAAGTGGAGGTGCAGCTCCGTCTTC GTTTTAGAGCTAGAAATAG | Preparation of sgRNA template |
| sgRNA-R | TAGATATCAAGCTGCTAGAAAAAAAAGCACCGACTC GGTGCC | Preparation of sgRNA template |
| F1 | ATGCCGAAAATCTTCTGGATTAAG | Detection of mutations |
| R1 | GTTTTTGGTTGATGGACCGAGTGT | Detection of mutations |
| Ovo-1-qF | GCCCCTTACCGCTCCTTTCG | qRT-PCR |
| Ovo-1-qR | ATCGCCTCCAAGAATCGATG | qRT-PCR |
| RP49-qF | TCAATCGGATCGCTATGACA | qRT-PCR |
| RP49-qR | ATGACGGGTCTTCTTGTTGG | qRT-PCR |
| WCP10-qF | TGGAGCACGCCTTCATATCA | qRT-PCR |
| WCP10-qR | GGACGGTGTAAACTTTGCCA | qRT-PCR |
| WCP5-qF | GCAGCCCCTTTGATTCAACA | qRT-PCR |
| WCP5-qR | CGTGTTGGGACTTGTGATCG | qRT-PCR |
| WCP4-qF | AGTCCACGAGGCTTCTTC | qRT-PCR |
| WCP4-qR | CCTTGCGGAATGAACCA | qRT-PCR |
| Wnt1-qF | CAGGGAATTCGTTGATACCG | qRT-PCR |
| Wnt1-qR | TCATCCAGCAAGTCTTCACG | qRT-PCR |
| FTZ-F1-qF | ATGCGTCGCCGAAAGAGCCT | qRT-PCR |
| FTZ-F1-qR | ATTGCGACCACCGCGCATAC | qRT-PCR |
| USP-qF | ACACTTCGGGCAGCTAGAA | qRT-PCR |
| USP-qR | TCCGCGAGTCTACGTTCTCT | qRT-PCR |
| E74-qF | GCACAAGAACAAGCCAGACA | qRT-PCR |
| E74-qR | GTCGATCTCGACGATGTCCT | qRT-PCR |
| BRC-qF | AAAGGCCTCCCTGAAGAGAC | qRT-PCR |
| BRC-qR | CGCGACTTGTGGTAGGTGTA | qRT-PCR |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bi, H.; Xu, X.; Li, X.; Zhang, Y.; Huang, Y.; Li, K.; Xu, J. CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori. Insects 2019, 10, 254. https://doi.org/10.3390/insects10080254
Bi H, Xu X, Li X, Zhang Y, Huang Y, Li K, Xu J. CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori. Insects. 2019; 10(8):254. https://doi.org/10.3390/insects10080254
Chicago/Turabian StyleBi, Honglun, Xia Xu, Xiaowei Li, Yong Zhang, Yongping Huang, Kai Li, and Jun Xu. 2019. "CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori" Insects 10, no. 8: 254. https://doi.org/10.3390/insects10080254
APA StyleBi, H., Xu, X., Li, X., Zhang, Y., Huang, Y., Li, K., & Xu, J. (2019). CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori. Insects, 10(8), 254. https://doi.org/10.3390/insects10080254






